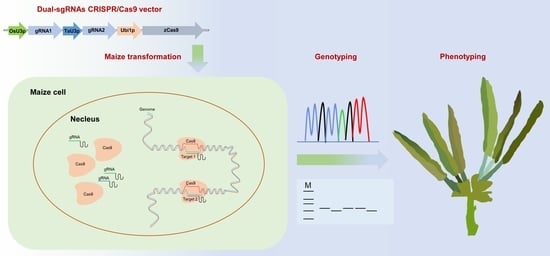
Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Characterization of Mutant Phenotypes
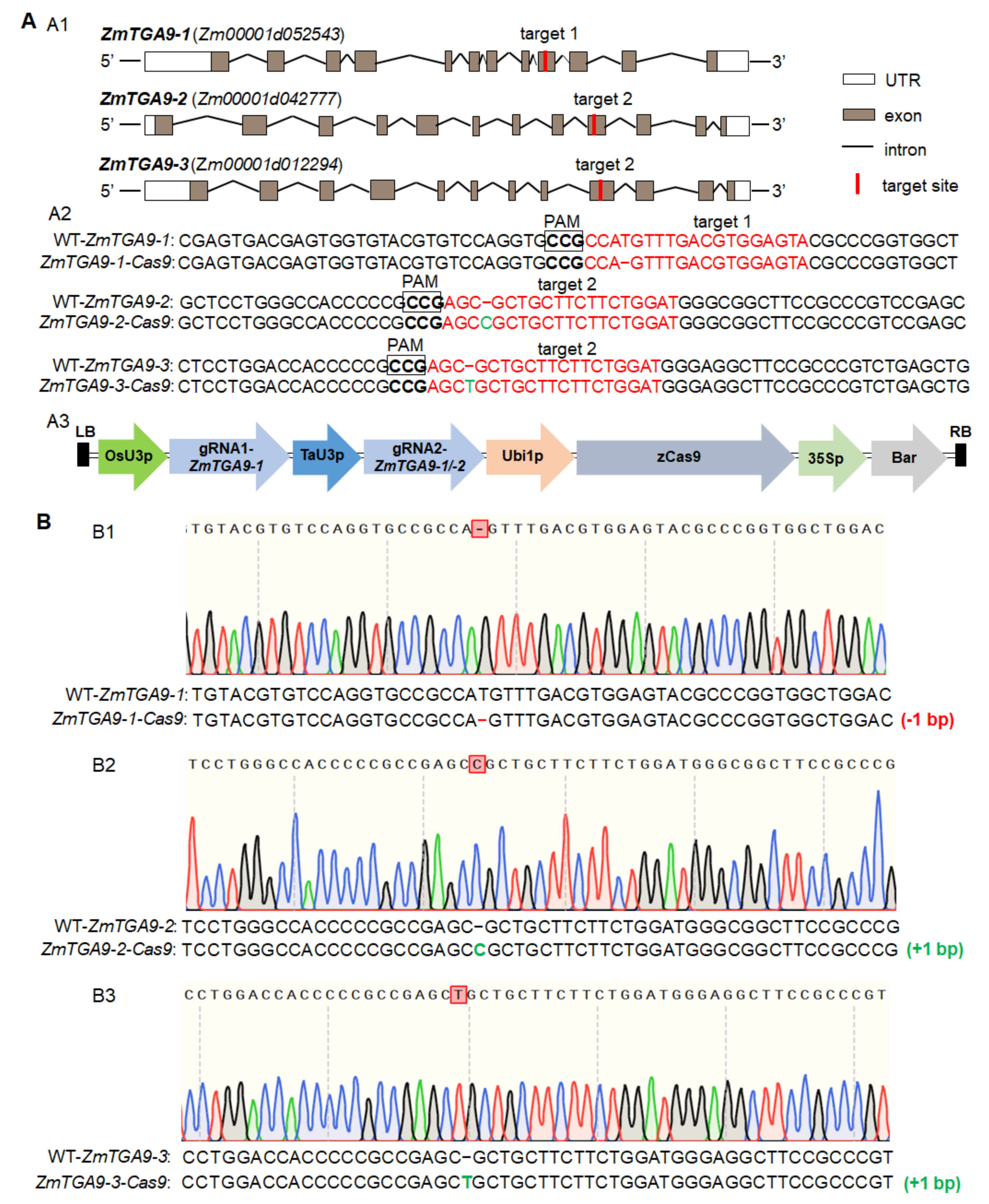
2.3. Plasmid Construction
2.4. Maize Genetic Transformation
2.5. Genotyping Maize Mutant Plants
2.6. Statistical Analysis
3. Results
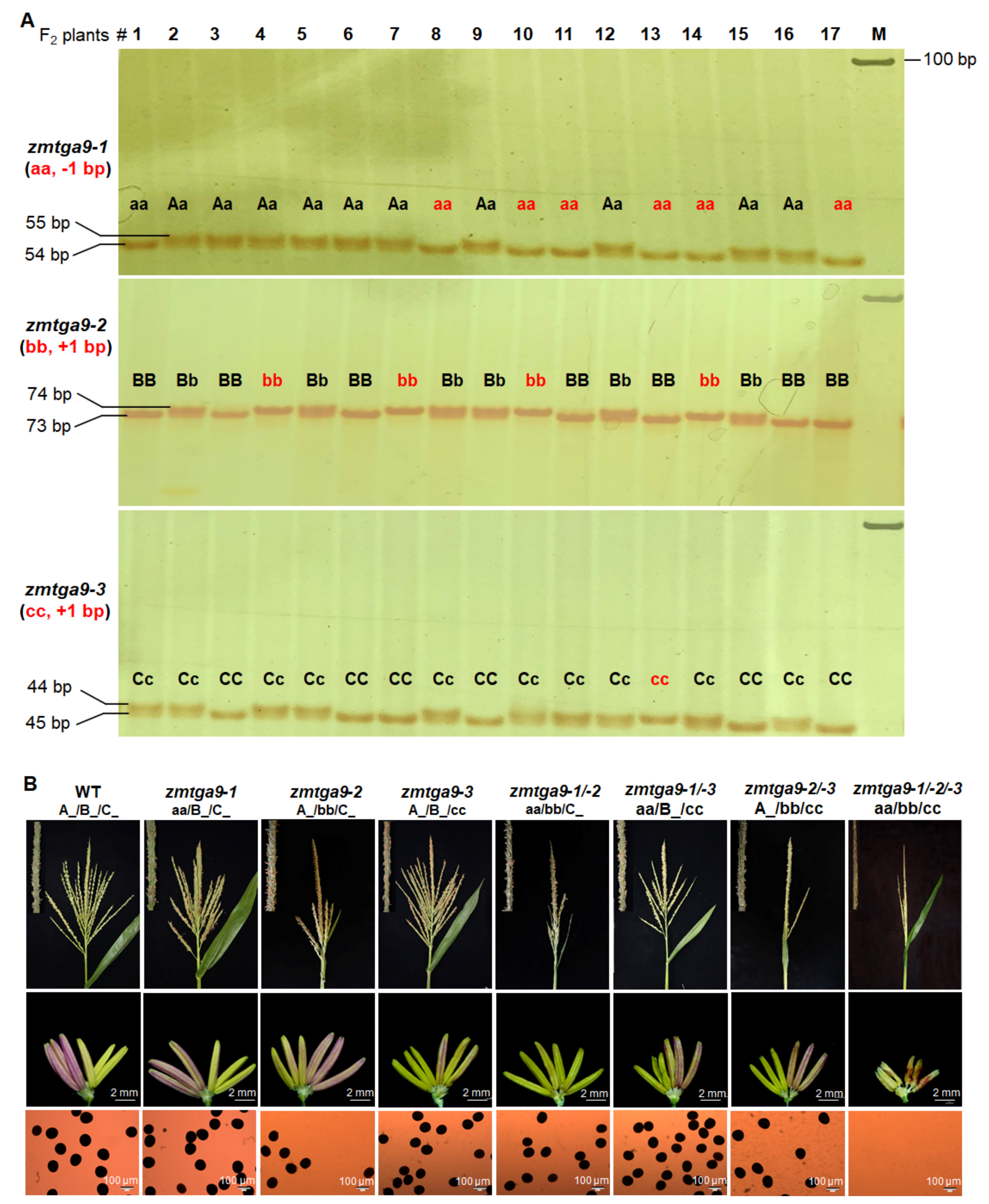
3.1. ZmTGA9-1, ZmTGA9-2 and ZmTGA9-3 Display Completely Functional Redundancy to Control Anther and Pollen Development in Maize
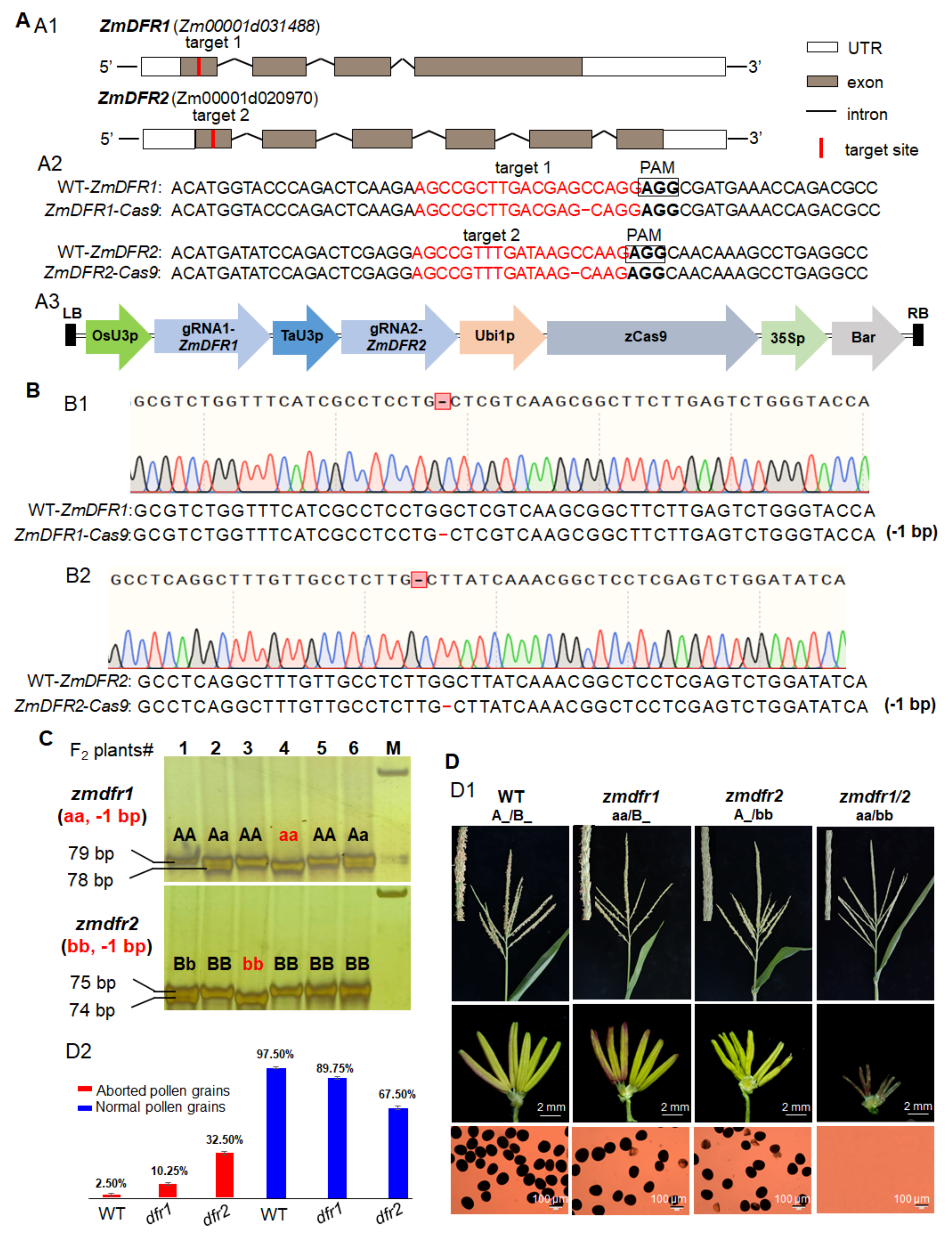
3.2. ZmDFR1 and ZmDFR2 Have Partially Redundant Functions in Controlling Maize Male Fertility
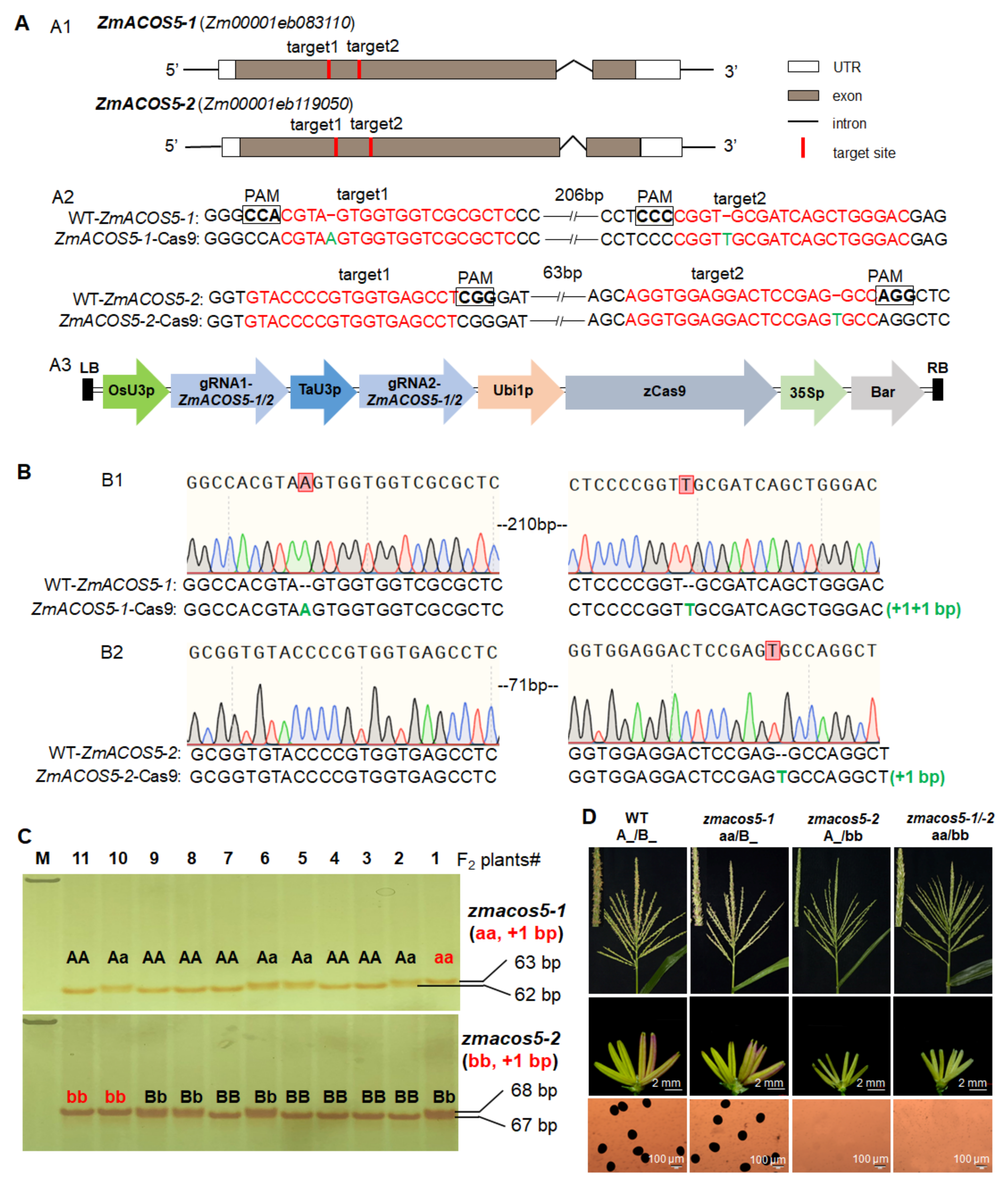
3.3. ZmACOS5-1 and ZmACOS5-2 Display No Functional Redundancy in Maize Male Fertility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Cas | CRISPR-associated protein |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| GMS | genic male sterility |
| PAGE | polyacrylamide gel electrophoresis |
| TFs | transcription factors |
| WT | wild type |
References
- Schnable, J.C. Genome evolution in maize: From genomes back to genes. Annu. Rev. Plant Biol. 2015, 66, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Wu, S.; Li, Z.; Dong, Z.; An, X.; Ma, B.; Tian, Y.; Li, J. Maize genic male-sterility genes and their applications in hybrid breeding: Progress and perspectives. Mol. Plant 2019, 12, 321–342. [Google Scholar] [CrossRef] [PubMed]
- Perez-Prat, E.; van Lookeren Campagne, M.M. Hybrid seed production and the challenge of propagating male-sterile plants. Trends Plant Sci. 2002, 7, 199–203. [Google Scholar] [CrossRef]
- Virmani, S.; Ilyas-Ahmed, M. Environment-sensitive genic male sterility (EGMS) in crops. Adv. Agron. 2001, 72, 139–195. [Google Scholar] [CrossRef]
- Wu, Y.; Fox, T.W.; Trimnell, M.R.; Wang, L.; Xu, R.j.; Cigan, A.M.; Huffman, G.A.; Garnaat, C.W.; Hershey, H.; Albertsen, M.C. Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 2016, 14, 1046–1054. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, S.; An, X.; Xie, K.; Dong, Z.; Zhou, Y.; Xu, L.; Fang, W.; Liu, S.; Liu, S.; et al. Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 2018, 16, 459–471. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, X. Breeding with dominant genic male-sterility genes to boost crop grain yield in the post-heterosis utilization era. Mol. Plant 2021, 14, 531–534. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid metabolism: Critical roles in male fertility and other aspects of reproductive development in plants. Mol. Plant 2020, 13, 955–983. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Z.; Liu, X.; Zhu, T.; Xie, K.; Hou, Q.; Yan, T.; Niu, C.; Zhang, S.; Yang, M.; et al. ZmFAR1 and ZmABCG26 Regulated by microRNA Are Essential for Lipid Metabolism in Maize Anther. Int. J. Mol. Sci. 2021, 22, 7916. [Google Scholar] [CrossRef]
- Jiang, Y.; An, X.; Li, Z.; Yan, T.; Zhu, T.; Xie, K.; Liu, S.; Hou, Q.; Zhao, L.; Wu, S.; et al. CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotechnol. J. 2021, 19, 1769–1784. [Google Scholar] [CrossRef]
- Hyams, G.; Abadi, S.; Lahav, S.; Avni, A.; Halperin, E.; Shani, E.; Mayrose, I. CRISPys: Optimal sgRNA design for editing multiple members of a gene family using the CRISPR system. J. Mol. Biol. 2018, 430, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Sun, K.; An, X. CRISPR/Cas System: Applications and Prospects for Maize Improvement. ACS Agric. Sci. Technol. 2022. [Google Scholar] [CrossRef]
- Wei, X.; Pu, A.; Liu, Q.; Leng, Y.; Fu, Z.; Wu, F.; An, X.; Long, Y. Commercialization and Supervision Policies of Gene Edited Crops in China and Other Main Countries. ACS Agric. Sci. Technol. 2021. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Minkenberg, B.; Wheatley, M.; Yang, Y. CRISPR/Cas9-enabled multiplex genome editing and its application. Prog. Mol. Biol. Transl. Sci. 2017, 149, 111–132. [Google Scholar] [CrossRef]
- Jouanin, A.; Gilissen, L.J.; Schaart, J.G.; Leigh, F.J.; Cockram, J.; Wallington, E.J.; Boyd, L.A.; Van Den Broeck, H.C.; Van der Meer, I.M.; America, A.; et al. CRISPR/Cas9 gene editing of gluten in wheat to reduce gluten content and exposure—Reviewing methods to screen for coeliac safety. Front. Nutr. 2020, 7, 51. [Google Scholar] [CrossRef]
- Tang, X.; Liu, G.; Zhou, J.; Ren, Q.; You, Q.; Tian, L.; Xin, X.; Zhong, Z.; Liu, B.; Zheng, X.; et al. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 2018, 19, 84. [Google Scholar] [CrossRef]
- Duan, K.; Cheng, Y.; Ji, J.; Wang, C.; Wei, Y.; Wang, Y. Large chromosomal segment deletions by CRISPR/LbCpf1-mediated multiplex gene editing in soybean. J. Integr. Plant Biol. 2021, 63, 1620–1631. [Google Scholar] [CrossRef]
- Lee, K.; Zhang, Y.; Kleinstiver, B.P.; Guo, J.A.; Aryee, M.J.; Miller, J.; Malzahn, A.; Zarecor, S.; Lawrence-Dill, C.J.; Joung, J.K.; et al. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 2019, 17, 362–372. [Google Scholar] [CrossRef]
- Yin, X.; Biswal, A.K.; Dionora, J.; Perdigon, K.M.; Balahadia, C.P.; Mazumdar, S.; Chater, C.; Lin, H.-C.; Coe, R.A.; Kretzschmar, T.; et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017, 36, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, M.; Albertsen, M.C.; Young, J.K.; Cigan, A.M. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol. Biol. 2018, 97, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Z.; He, G.; Ma, L.; Deng, X.W. CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J. Genet. Genomics 2020, 47, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhu, T.; Liu, S.; Jiang, Y.; Liu, H.; Zhang, Y.; Xie, K.; Li, J.; An, X.; Wan, X. Genome-wide analyses on transcription factors and their potential microRNA regulators involved in maize male fertility. Crop J. 2021, 9, 1248–1262. [Google Scholar] [CrossRef]
- An, X.; Ma, B.; Duan, M.; Dong, Z.; Liu, R.; Yuan, D.; Hou, Q.; Wu, S.; Zhang, D.; Liu, D.; et al. Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. USA 2020, 117, 23499–23509. [Google Scholar] [CrossRef]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Frame, B.R.; Shou, H.; Chikwamba, R.K.; Zhang, Z.; Xiang, C.; Fonger, T.M.; Pegg, S.E.K.; Li, B.; Nettleton, D.S.; Pei, D.; et al. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 2002, 129, 13–22. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, D.; Tian, Y.; Wu, S.; An, X.; Dong, Z.; Zhang, S.; Bao, J.; Li, Z.; Li, J.; et al. Map-Based Cloning, Phylogenetic, and Microsynteny Analyses of ZmMs20 Gene Regulating Male Fertility in Maize. Int. J. Mol. Sci. 2019, 20, 1411. [Google Scholar] [CrossRef]
- Lawan, Z.; Yeye, M.; Ishiyaku, M.; Bugaje, S.; Ahmed, H.; Shaibu, A. Genetic analysis of male sterility genes in different A and B sorghum lines. Afr. Crop Sci. J. 2018, 26, 107–115. [Google Scholar] [CrossRef]
- Murmu, J.; Bush, M.J.; DeLong, C.; Li, S.; Xu, M.; Khan, M.; Malcolmson, C.; Fobert, P.R.; Zachgo, S.; Hepworth, S.R. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 2010, 154, 1492–1504. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Zhang, H.; Arikit, S.; Huang, K.; Nan, G.-L.; Walbot, V.; Meyers, B.C. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 2015, 112, 3146–3151. [Google Scholar] [CrossRef] [PubMed]
- Grienenberger, E.; Kim, S.S.; Lallemand, B.; Geoffroy, P.; Heintz, D.; Souza, C.d.A.; Heitz, T.; Douglas, C.J.; Legrand, M. Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 2010, 22, 4067–4083. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Qu, S.; Tucker, M.R.; Zhang, D.; Liang, W.; Shi, J. Ostkpr1 functions in anther cuticle development and pollen wall formation in rice. BMC Plant Biol. 2019, 19, 104. [Google Scholar] [CrossRef]
- De Azevedo Souza, C.; Kim, S.S.; Koch, S.; Kienow, L.; Schneider, K.; McKim, S.M.; Haughn, G.W.; Kombrink, E.; Douglas, C.J. A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 2009, 21, 507–525. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Guo, Z.; Shi, Q.; Xiong, S.; Zhang, C.; Zhu, J.; Yang, Z. OsACOS12, an orthologue of Arabidopsis acyl-CoA synthetase5, plays an important role in pollen exine formation and anther development in rice. BMC Plant Biol. 2016, 16, 256. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.-G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef]
- An, X.; Dong, Z.; Tian, Y.; Xie, K.; Wu, S.; Zhu, T.; Zhang, D.; Zhou, Y.; Niu, C.; Ma, B.; et al. ZmMs30 encoding a novel GDSL lipase is essential for male fertility and valuable for hybrid breeding in maize. Mol. Plant 2019, 12, 343–359. [Google Scholar] [CrossRef]
- Zhu, T.; Li, Z.; An, X.; Long, Y.; Xue, X.; Xie, K.; Ma, B.; Zhang, D.; Guan, Y.; Niu, C.; et al. Normal structure and function of endothecium chloroplasts maintained by ZmMs33-mediated lipid biosynthesis in tapetal cells are critical for anther development in maize. Mol. Plant 2020, 13, 1624–1643. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, S.; Niu, C.; Liu, D.; Yan, T.; Tian, Y.; Liu, S.; Xie, K.; Li, Z.; Wang, Y.; et al. ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 2021, 72, 4298–4318. [Google Scholar] [CrossRef]
- Zhang, M.; Wei, H.; Hao, P.; Wu, A.; Ma, Q.; Zhang, J.; Wang, H.; Fu, X.; Ma, L.; Lu, J.; et al. GhGPAT12/25 Are Essential for the Formation of Anther Cuticle and Pollen Exine in Cotton (Gossypium hirsutum L.). Front. Plant Sci. 2021, 12, 667739. [Google Scholar] [CrossRef] [PubMed]
- Shou, H.; Frame, B.R.; Whitham, S.A.; Wang, K. Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol. Breed. 2004, 13, 201–208. [Google Scholar] [CrossRef]
- Fellenberg, C.; Vogt, T. Evolutionarily conserved phenylpropanoid pattern on angiosperm pollen. Trends Plant Sci. 2015, 20, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J.; Talle, B.; Wilson, Z.A. Anther and pollen development: A conserved developmental pathway. J. Integr. Plant Biol. 2015, 57, 876–891. [Google Scholar] [CrossRef] [PubMed]
| F2 Lines | Mutation Types | F2 Plants | Expected Ratio | Observed Ratio | Χ2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZmTGA9-1/-2/-3-Cas9 | tga9-1 (aa): −1 bp tga9-2 (bb): +1 bp tga9-3 (cc): +1 bp | Genotypes | A_/B_/C_ | aa/B_/C_ | A_/bb/C_ | A_/B_/cc | aa/bb/C_ | aa/B_/cc | A_/bb/cc | aa/bb/cc | 27:9:9:9:3:3:3:1 | 36:15:15:8:2:3:5:1 | 4.43 |
| Number | 36 | 15 | 15 | 8 | 2 | 3 | 5 | 1 | |||||
| Phenotypes | fertile | fertile | fertile | fertile | fertile | fertile | fertile | complete sterility | |||||
| ZmDFR1/2-Cas9 | dfr1 (aa): −1 bp dfr2 (bb): −1 bp | Genotypes | A_/B_ | aa/B_ | A_/bb | aa/bb | 9:3:3:1 | 11:5:5:1 | 3.36 | ||||
| Number | 80 | 35 | 34 | 7 | |||||||||
| Phenotypes | fertile | partial sterility | partial sterility | complete sterility | |||||||||
| ZmACOS5-1/-2-Cas9 | acos5-1 (aa): +2 bp acos5-2 (bb): +1 bp | Genotypes | A_/B_ | aa/B_ | A_/bb | aa/bb | 9:3:3:1 | 13:6:5:1 | 4.3 | ||||
| Number | 76 | 36 | 29 | 6 | |||||||||
| Phenotypes | fertile | fertile | complete sterility | complete sterility | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhang, S.; Jiang, Y.; Yan, T.; Fang, C.; Hou, Q.; Wu, S.; Xie, K.; An, X.; Wan, X. Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize. Cells 2022, 11, 439. https://doi.org/10.3390/cells11030439
Liu X, Zhang S, Jiang Y, Yan T, Fang C, Hou Q, Wu S, Xie K, An X, Wan X. Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize. Cells. 2022; 11(3):439. https://doi.org/10.3390/cells11030439
Chicago/Turabian StyleLiu, Xinze, Shaowei Zhang, Yilin Jiang, Tingwei Yan, Chaowei Fang, Quancan Hou, Suowei Wu, Ke Xie, Xueli An, and Xiangyuan Wan. 2022. "Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize" Cells 11, no. 3: 439. https://doi.org/10.3390/cells11030439
APA StyleLiu, X., Zhang, S., Jiang, Y., Yan, T., Fang, C., Hou, Q., Wu, S., Xie, K., An, X., & Wan, X. (2022). Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize. Cells, 11(3), 439. https://doi.org/10.3390/cells11030439