MNase Digestion Protection Patterns of the Linker DNA in Chromatosomes
Abstract
:1. Introduction
2. Materials and Methods
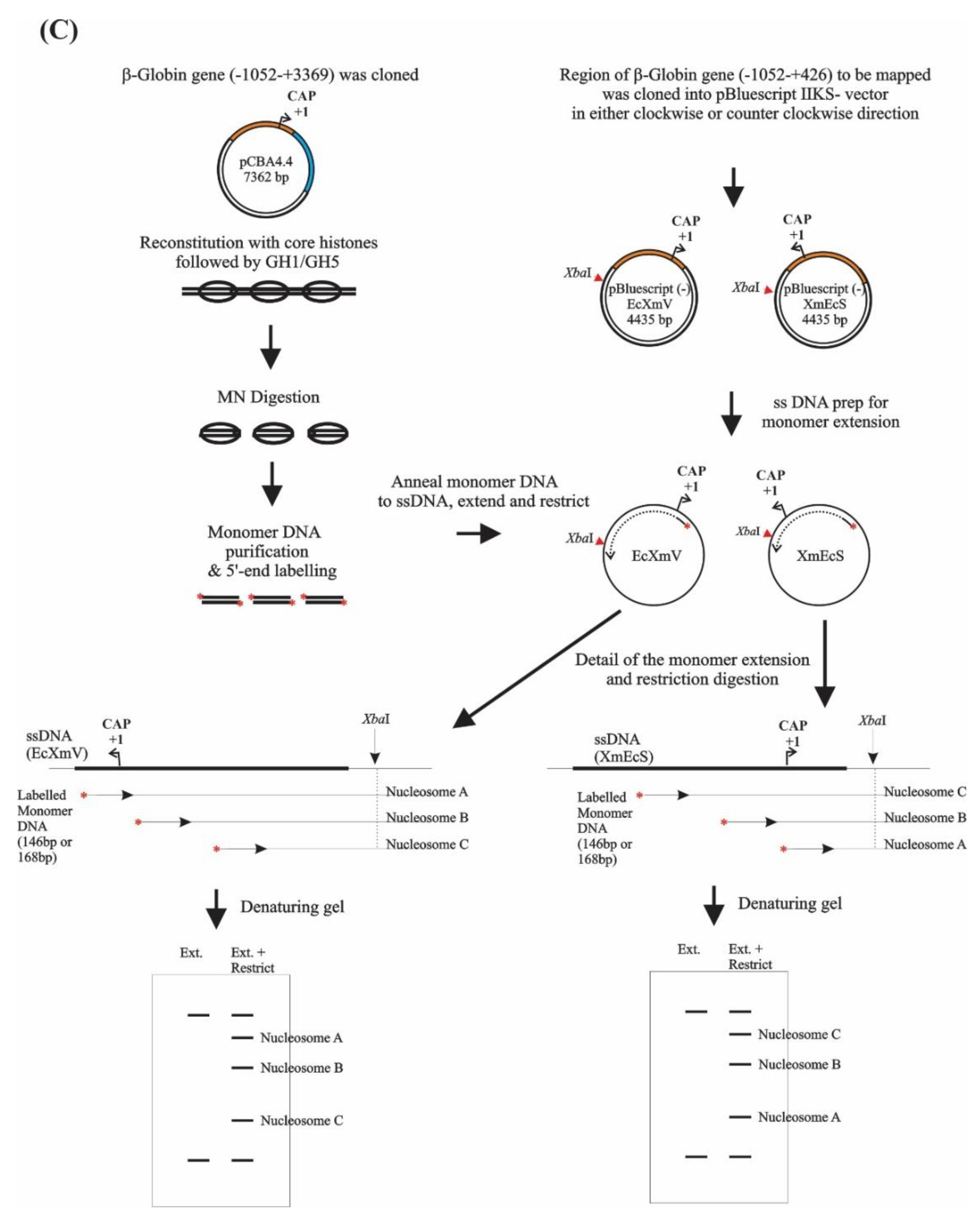
2.1. Phagemid Construction and Preparation of Single Strand DNA
2.2. In Vitro Nucleosome Reconstitution and Titration of GH1/GH5 to Reconstituted Chromatin
2.3. In Vitro Nucleosome Reconstitution at 37 °C
2.4. Monomer Extension
2.5. Analysis of the Nucleosome Mapping
3. Results
3.1. DNA Sequence-Directed Nucleosome Positioning Is Determined by the Core Histone Octamer and Is Not Altered by the Addition of Linker Histone Globular Domains
3.2. Effect of Reconstitution Conditions on Nucleosome Positioning
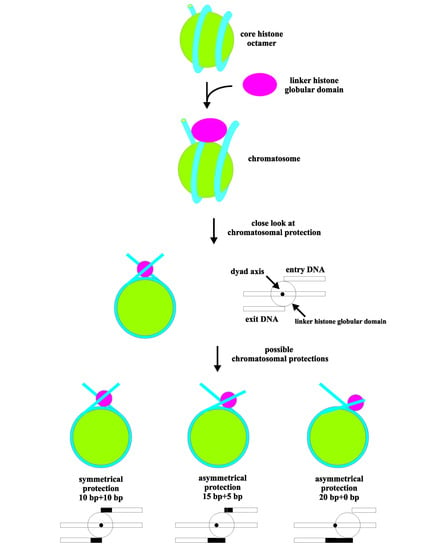
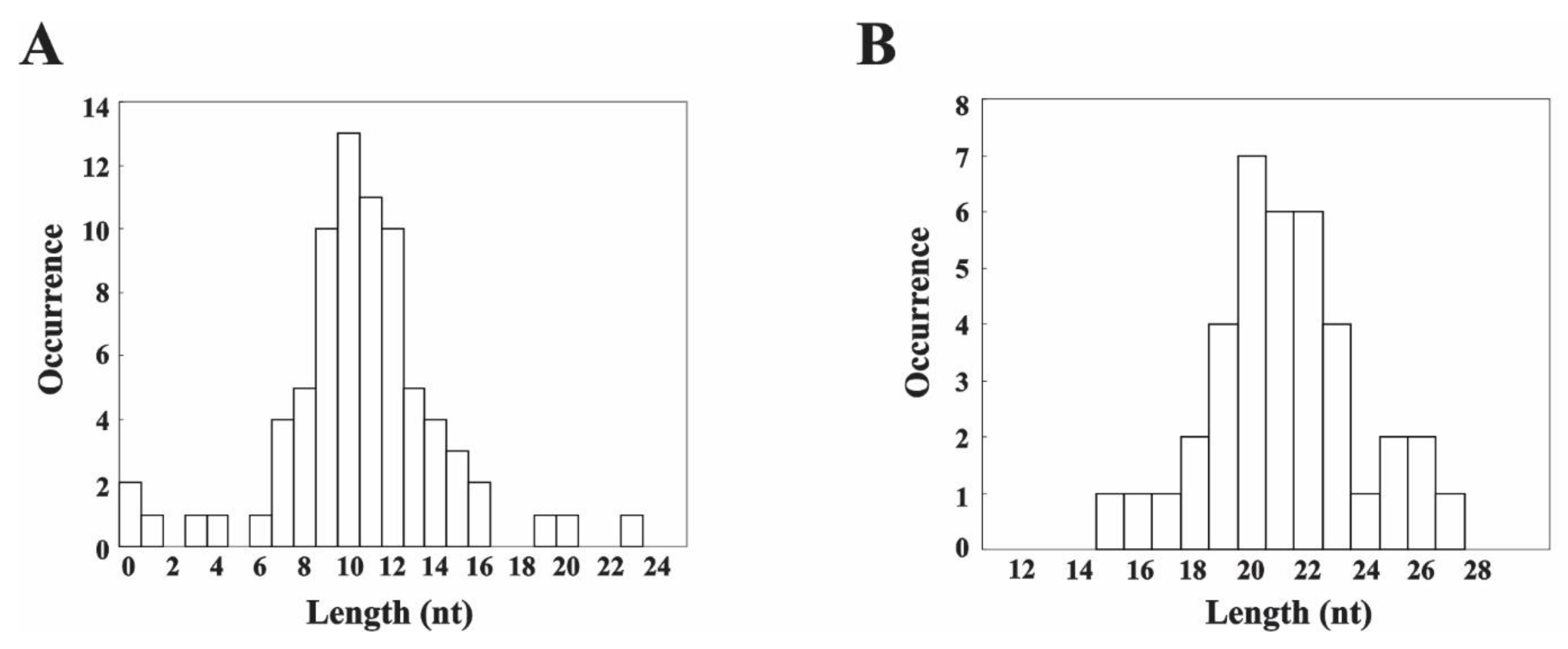
3.3. MNase Digestion Patterns of the Linker DNA in Chromatosomes
3.4. Effects of the Linker Histones Tails on the Nucleosome Positioning
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kornberg, R.D. Chromatin structure: A repeating unit of histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Kornberg, R.D.; Lorch, Y. Primary Role of the Nucleosome. Mol. Cell 2020, 79, 371–375. [Google Scholar] [CrossRef]
- Prajapati, H.K.; Ocampo, J.; Clark, D.J. Interplay among ATP-Dependent Chromatin Remodelers Determines Chromatin Organisation in Yeast. Biology 2020, 9, 190. [Google Scholar] [CrossRef]
- Sundaramoorthy, R.; Owen-Hughes, T. Chromatin remodelling comes into focus. F1000Research 2020, 9, 1011. [Google Scholar] [CrossRef]
- Gamarra, N.; Narlikar, G.J. Collaboration through chromatin: Mechanisms of molecular motors at the interface of transcription and chromatin structure. J. Mol. Biol. 2021, 166876. [Google Scholar] [CrossRef]
- Noll, M.; Kornberg, R.D. Action of micrococcal nuclease on chromatin and the location of histone H1. J. Mol. Biol. 1977, 109, 393–404. [Google Scholar] [CrossRef]
- Simpson, R.T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 1978, 17, 5524–5531. [Google Scholar] [CrossRef] [PubMed]
- Allan, J.; Hartman, P.G.; Crane-Robinson, C.; Aviles, F.X. The structure of histone H1 and its location in chromatin. Nature 1980, 288, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, C.L.; Ghosh, R.P. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010, 2, a000596. [Google Scholar] [CrossRef] [PubMed]
- Satchwell, S.C.; Drew, H.R.; Travers, A.A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986, 191, 659–675. [Google Scholar] [CrossRef]
- Satchwell, S.C.; Travers, A.A. Asymmetry and polarity of nucleosomes in chicken erythrocyte chromatin. EMBO J. 1989, 8, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Nikitina, T.; Zhao, J.; Fleury, T.J.; Bhattacharyya, R.; Bouhassira, E.E.; Stein, A.; Woodcock, C.L.; Skoultchi, A.I. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 2005, 123, 1199–1212. [Google Scholar] [CrossRef] [Green Version]
- Routh, A.; Sandin, S.; Rhodes, D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA 2008, 105, 8872–8877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoma, F.; Losa, R.; Koller, T. Involvement of the domains of histones H1 and H5 in the structural organization of soluble chromatin. J. Mol. Biol. 1983, 167, 619–640. [Google Scholar] [CrossRef]
- Allan, J.; Mitchell, T.; Harborne, N.; Bohm, L.; Crane-Robinson, C. Roles of H1 domains in determining higher-order chromatin structure and H1 location. J. Mol. Biol. 1986, 187, 591–601. [Google Scholar] [CrossRef]
- Izzo, A.; Kamieniarz-Gdula, K.; Schneider, R. The histone H1 family: Specific members, specific functions? Biol. Chem. 2008, 389, 333–343. [Google Scholar] [CrossRef]
- Hergeth, S.P.; Schneider, R. The H1 linker histones: Multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015, 16, 1439–1453. [Google Scholar] [CrossRef] [Green Version]
- Bustin, M.; Catez, F.; Lim, J.-H. The dynamics of histone H1 function in chromatin. Mol. Cell 2005, 17, 617–620. [Google Scholar] [CrossRef]
- Bednar, J.; Hamiche, A.; Dimitrov, S. H1–nucleosome interactions and their functional implications. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 436–443. [Google Scholar] [CrossRef]
- Fyodorov, D.V.; Zhou, B.-R.; Skoultchi, A.I.; Bai, Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 192–206. [Google Scholar] [CrossRef]
- Barra, J.L.; Rhounim, L.; Rossignol, J.L.; Faugeron, G. Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol. Cell. Biol. 2000, 20, 61–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgieva, M.; Roguev, A.; Balashev, K.; Zlatanova, J.; Miloshev, G. Hho1p, the linker histone of Saccharomyces cerevisiae, is important for the proper chromatin organization in vivo. Biochim. Biophys. Acta. 2012, 1819, 366–374. [Google Scholar] [CrossRef]
- Uzunova, K.; Georgieva, M.; Miloshev, G. Saccharomyces cerevisiae linker histone-Hho1p maintains chromatin loop organization during ageing. Oxidative Med. Cell. Longev. 2013, 2013, 437146. [Google Scholar] [CrossRef]
- Caron, F.; Thomas, J.O. Exchange of histone H1 between segments of chromatin. J. Mol. Biol. 1981, 146, 513–537. [Google Scholar] [CrossRef]
- Thomas, J.O.; Rees, C. Exchange of histones H1 and histone H5 between chromatin fragments—A preference of H5 for higher-order structures. Eur. J. Biochem. 1983, 134, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Misteli, T.; Gunjan, A.; Hock, R.; Bustin, M.; Brown, D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature 2000, 408, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.H.; Goutte-Gattat, D.; Becker, N.; Meyer, S.; Shukla, M.S.; Hayes, J.J.; Everaers, R.; Angelov, D.; Bednar, J.; Dimitrov, S. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. USA 2010, 107, 9620–9625. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.; Roberts, V.A. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc. Natl. Acad. Sci. USA 2006, 103, 8384–8389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, S.; Becker, N.B.; Syed, S.H.; Goutte-Gattat, D.; Shukla, M.S.; Hayes, J.J.; Angelov, D.; Bednar, J.; Dimitrov, S.; Everaers, R. From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: Nanoscale modeling of the nucleosomal stem. Nucleic Acids Res. 2011, 39, 9139–9154. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.B.; Gerchman, S.E.; Ramakrishnan, V.; Travers, A.; Muyldermans, S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature 1998, 395, 402–405. [Google Scholar] [CrossRef]
- Bharath, M.M.; Chandra, N.R.; Rao, M.R. Molecular modeling of the chromatosome particle. Nucleic Acids Res. 2003, 31, 4264–4274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.T.; Izard, T.; Misteli, T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat. Struct. Mol. Biol. 2006, 13, 250–255. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.; Li, Q.; Levi, B.; Shi, Y.; Wolffe, A.P. Structural and functional features of a specific nucleosome containing a recognition element for the thyroid hormone receptor. EMBO J. 1997, 16, 7130–7145. [Google Scholar] [CrossRef] [Green Version]
- An, W.; Leuba, S.; van Holde, K.; Zlatanova, J. Linker histone protects linker DNA on only one side of the core particle and in a sequence-dependent manner. Proc. Natl. Acad. Sci. USA 1998, 95, 3396–3401. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.-R.; Feng, H.; Kato, H.; Dai, L.; Yang, Y.; Zhou, Y.; Bai, Y. Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. USA 2013, 110, 19390–19395. [Google Scholar] [CrossRef] [Green Version]
- Song, F.; Chen, P.; Sun, D.; Wang, M.; Dong, L.; Liang, D.; Xu, R.-M.; Zhu, P.; Li, G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 2014, 344, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.R.; Jiang, J.; Feng, H.; Ghirlando, R.; Xiao, T.S.; Bai, Y. Structural Mechanisms of Nucleosome Recognition by Linker Histones. Mol. Cell. 2015, 59, 628–638. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.-R.; Feng, H.; Ghirlando, R.; Li, S.; Schwieters, C.D.; Bai, Y. A small number of residues can determine if linker histones are bound on or off dyad in the chromatosome. J. Mol. Biol. 2016, 428, 3948–3959. [Google Scholar] [CrossRef]
- Bednar, J.; Garcia-Saez, I.; Boopathi, R.; Cutter, A.R.; Papai, G.; Reymer, A.; Syed, S.H.; Lone, I.N.; Tonchev, O.; Crucifix, C.; et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell. 2017, 66, 384–397. [Google Scholar] [CrossRef] [Green Version]
- Yenidunya, A.; Davey, C.; Clark, D.; Felsenfeld, G.; Allan, J. Nucleosome positioning on chicken and human globin gene promoters in vitro novel mapping techniques. J. Mol. Biol. 1994, 237, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Davey, C.; Pennings, S.; Meersseman, G.; Wess, T.J.; Allan, J. Periodicity of strong nucleosome positioning sites around the chicken adult β-globin gene may encode regularly spaced chromatin. Proc. Natl. Acad. Sci. USA 1995, 92, 11210–11214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davey, C.; Pennings, S.; Allan, J. CpG methylation remodels chromatin structure in vitro. J. Mol. Biol. 1997, 267, 276–288. [Google Scholar] [CrossRef]
- Shen, C.-H.; Leblanc, B.P.; Alfieri, J.A.; Clark, D.J. Remodeling of yeast CUP1 chromatin involves activator-dependent repositioning of nucleosomes over the entire gene and flanking sequences. Mol. Cell. Biol. 2001, 21, 534–547. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.-H.; Clark, D.J. DNA sequence plays a major role in determining nucleosome positions in yeast CUP1 chromatin. J. Biol. Chem. 2001, 276, 35209–35216. [Google Scholar] [CrossRef] [Green Version]
- Walmsley, M.E.; Buckle, R.S.; Allan, J.; Patient, R.K. A chicken red cell inhibitor of transcription associated with the terminally differentiated state. J. Cell Biol. 1991, 114, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.J.; Smith, M.; Jessee, J.; Bloom, F. DH11S: An Escherichia coli strain for preparation of single-stranded DNA from phagemid vectors. BioTechniques 1992, 12, 718–721. [Google Scholar]
- Bell, O.; Tiwari, V.K.; Thomä, N.H.; Schübeler, D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011, 12, 554–564. [Google Scholar] [CrossRef]
- Fraser, R.M.; Keszenman-Pereyra, D.; Simmen, M.W.; Allan, J. High-resolution mapping of sequence-directed nucleosome positioning on genomic DNA. J. Mol. Biol. 2009, 390, 292–305. [Google Scholar] [CrossRef]
- Guertin, M.J.; Lis, J.T. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Curr. Opin. Genet. Dev. 2013, 23, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Pugh, B.F. Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.L.; Rando, O.J. Mechanisms underlying nucleosome positioning in vivo. Annu. Rev. Biophys. 2014, 43, 41–63. [Google Scholar] [CrossRef]
- Brouwer, T.; Pham, C.; Kaczmarczyk, A.; de Voogd, W.-J.; Botto, M.; Vizjak, P.; Mueller-Planitz, F.; van Noort, J. A critical role for linker DNA in higher-order folding of chromatin fibers. Nucleic Acids Res. 2021, 49, 2537–2551. [Google Scholar] [CrossRef]
- Pennings, S.; Meersseman, G.; Bradbury, E. Mobility of positioned nucleosomes on 5S rDNA. J. Mol. Biol. 1991, 220, 101–110. [Google Scholar] [CrossRef]
- Pennings, S.; Meersseman, G.; Bradbury, E.M. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl. Acad. Sci. USA 1994, 91, 10275–10279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meersseman, G.; Pennings, S.; Bradbury, E. Mobile nucleosomes—A general behavior. EMBO J. 1992, 11, 2951–2959. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.R.; Feng, H.; Kale, S.; Fox, T.; Khant, H.; de Val, N.; Ghirlando, R.; Panchenko, A.R.; Bai, Y. Distinct structures and dynamics of chromatosomes with different human linker histone isoforms. Mol. Cell. 2021, 81, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Martin, S.; Thomas, J. A stable alpha-helical element in the carboxy-terminal domain of free and chromatin-bound histone H1 from sea urchin sperm. EMBO J. 1989, 8, 2591–2599. [Google Scholar] [CrossRef]
- Ring, D.; Cole, R.D. Close contacts between H1 histone molecules in nuclei. J. Biol. Chem. 1983, 258, 5361–5364. [Google Scholar] [CrossRef]
- Lennard, A.C.; Thomas, J.O. The arrangement of H5 molecules in extended and condensed chicken erythrocyte chromatin. EMBO J. 1985, 4, 3455–3462. [Google Scholar] [CrossRef]
- Stützer, A.; Liokatis, S.; Kiesel, A.; Schwarzer, D.; Sprangers, R.; Söding, J.; Selenko, P.; Fischle, W. Modulations of DNA Contacts by Linker Histones and Post-translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Mol. Cell. 2016, 61, 247–259. [Google Scholar] [CrossRef] [Green Version]
- Hayes, J.J. Site-directed cleavage of DNA by a linker histone-Fe(II) EDTA conjugate—localization of a globular domain binding site within a nucleosome. Biochemistry 1996, 35, 11931–11937. [Google Scholar] [CrossRef]
- Hayes, J.J.; Pruss, D.; Wolffe, A.P. Contacts of the globular domain of histone H5 and core histones with DNA in a chromatosome. Proc. Natl. Acad. Sci. USA 1994, 91, 7817–7821. [Google Scholar] [CrossRef] [Green Version]
- Pruss, D.; Bartholomew, B.; Persinger, J.; Hayes, J.; Arents, G.; Moudrianakis, E.N.; Wolffe, A.P. An asymmetric model for the nucleosome—A binding site for linker histones inside the DNA gyres. Science 1996, 274, 614–617. [Google Scholar] [CrossRef]
- Travers, A.A.; Muyldermans, S.V. A DNA sequence for positioning chromatosomes. J. Mol. Biol. 1996, 257, 486–491. [Google Scholar] [CrossRef]
- Woods, D.C.; Wereszczynski, J. Elucidating the influence of linker histone variants on chromatosome dynamics and energetics. Nucleic Acids Res. 2020, 48, 3591–3604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Vogirala, V.K.; Soman, A.; Berezhnoy, N.V.; Liu, Z.B.; Wong, A.S.W.; Korolev, N.; Su, C.-J.; Sandin, S.; Nordenskiöld, L. Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome. Sci. Rep. 2021, 11, 380. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, C.-H.; Allan, J. MNase Digestion Protection Patterns of the Linker DNA in Chromatosomes. Cells 2021, 10, 2239. https://doi.org/10.3390/cells10092239
Shen C-H, Allan J. MNase Digestion Protection Patterns of the Linker DNA in Chromatosomes. Cells. 2021; 10(9):2239. https://doi.org/10.3390/cells10092239
Chicago/Turabian StyleShen, Chang-Hui, and James Allan. 2021. "MNase Digestion Protection Patterns of the Linker DNA in Chromatosomes" Cells 10, no. 9: 2239. https://doi.org/10.3390/cells10092239
APA StyleShen, C.-H., & Allan, J. (2021). MNase Digestion Protection Patterns of the Linker DNA in Chromatosomes. Cells, 10(9), 2239. https://doi.org/10.3390/cells10092239