PAX7 Balances the Cell Cycle Progression via Regulating Expression of Dnmt3b and Apobec2 in Differentiating PSCs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pluripotent Stem Cell Lines
2.2. Preparation of Feeder Cells
2.3. Genotyping
2.4. Karyotyping
2.5. In Vitro Differentiation of PSCs
2.6. In Vivo Differentiation of PSCs–Teratomas Formation
2.7. Histological Analysis
2.8. Muscle Isolation
2.9. RNA Isolation, RT-PCR and qPCR
2.10. Immunolocalization
2.11. Global DNA Methylation Measurement
2.12. Data Analysis
3. Results
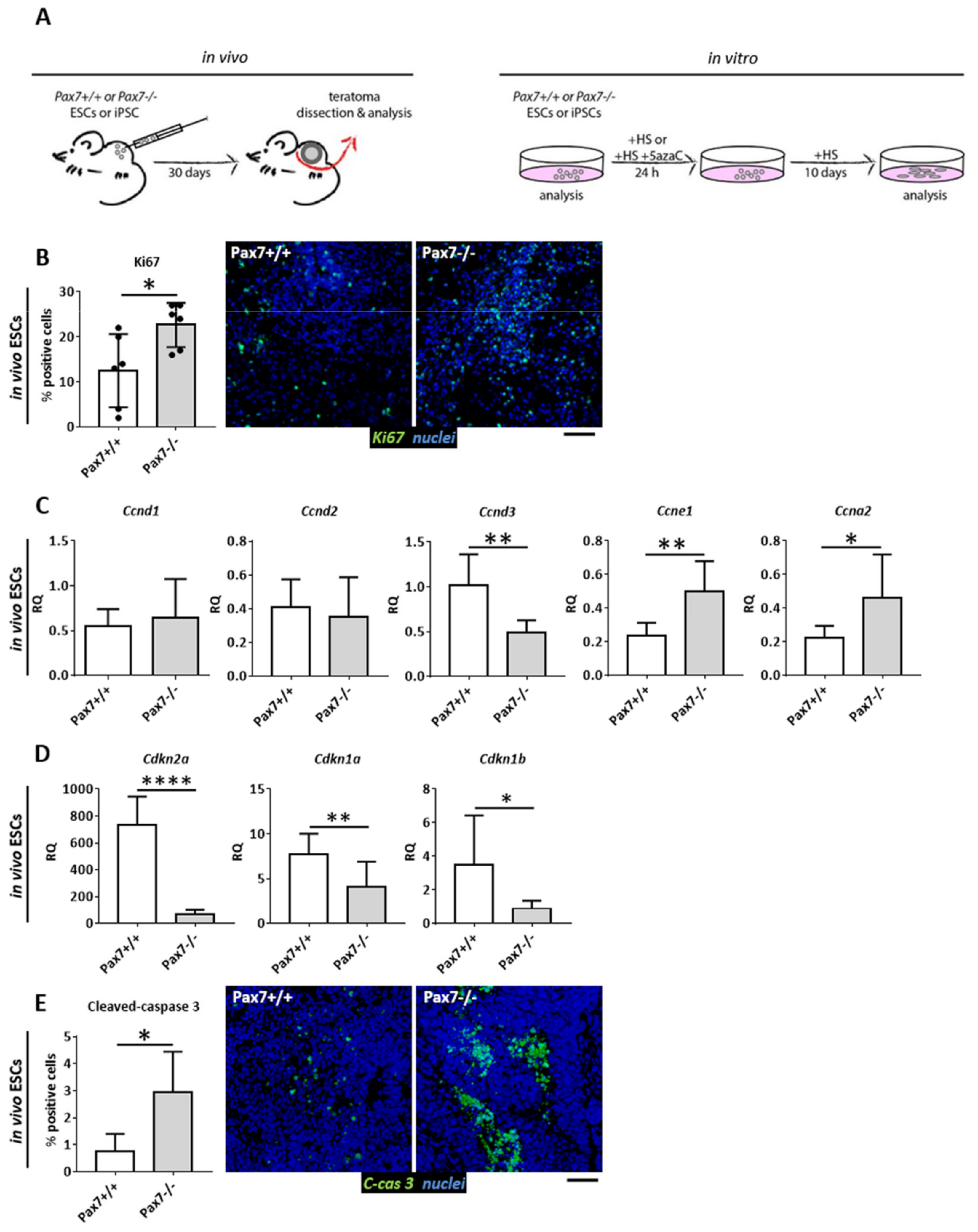
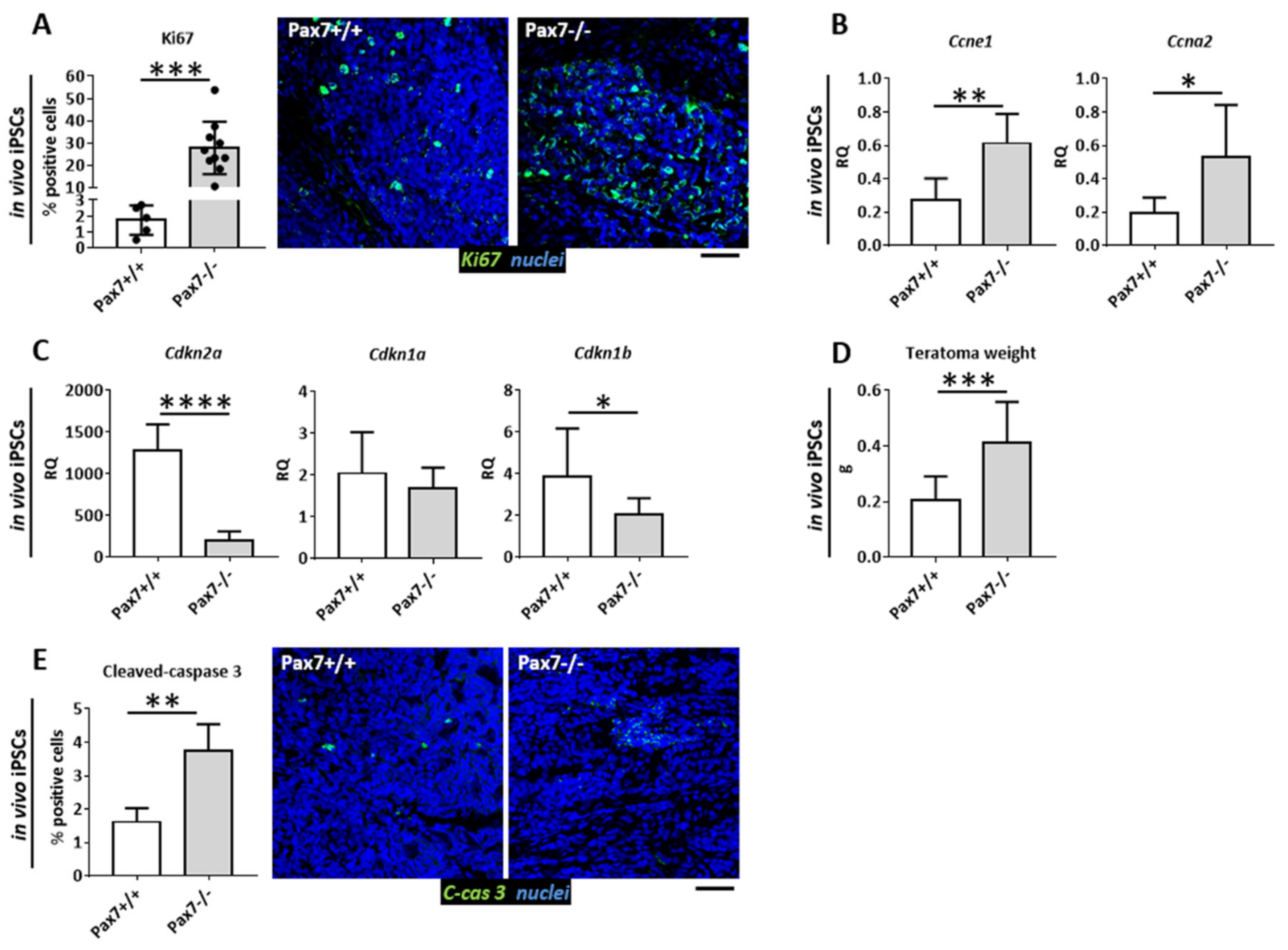
3.1. Proliferative Status of Cells Building Pax7+/+ and Pax7−/− Teratomas
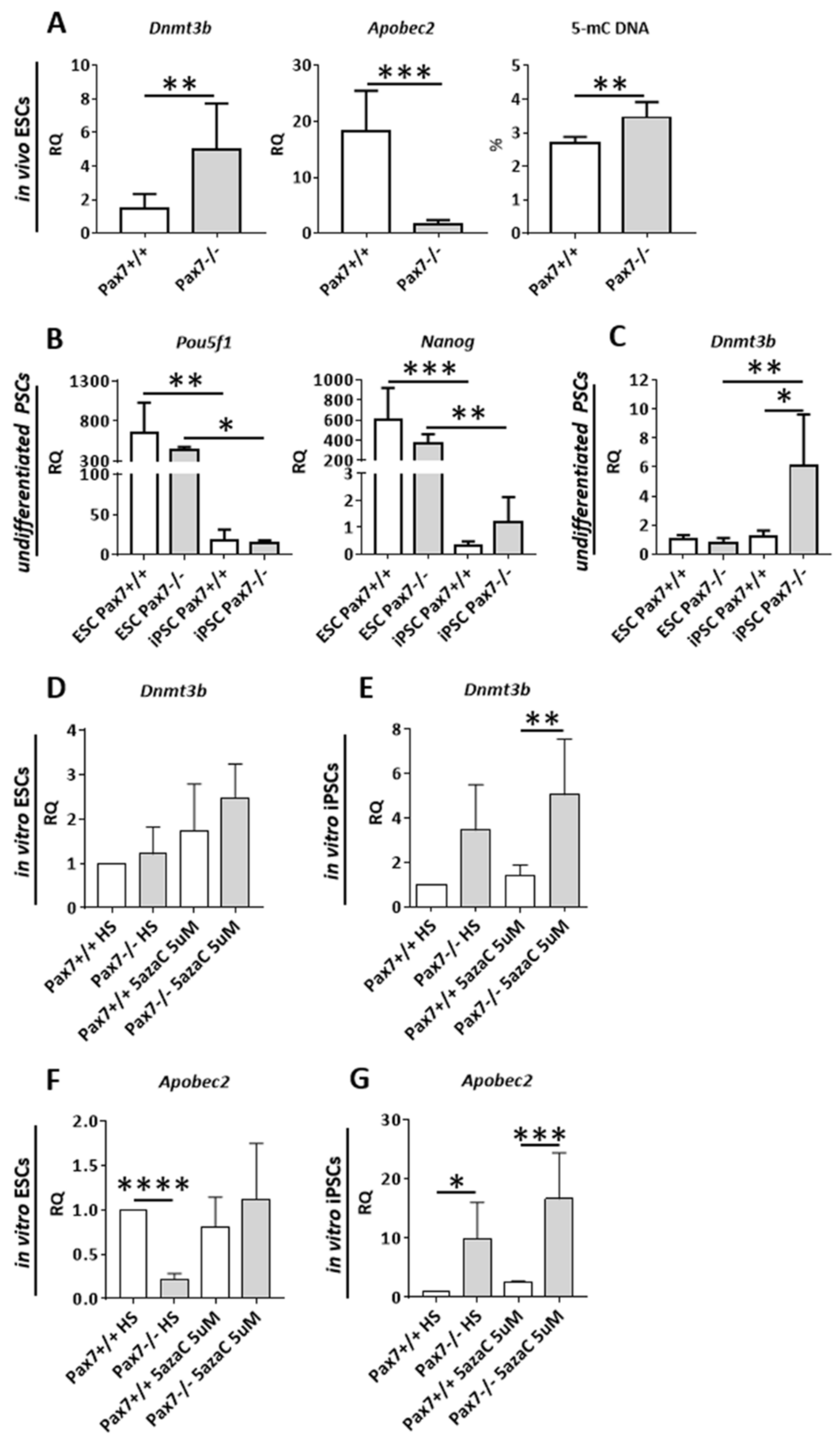
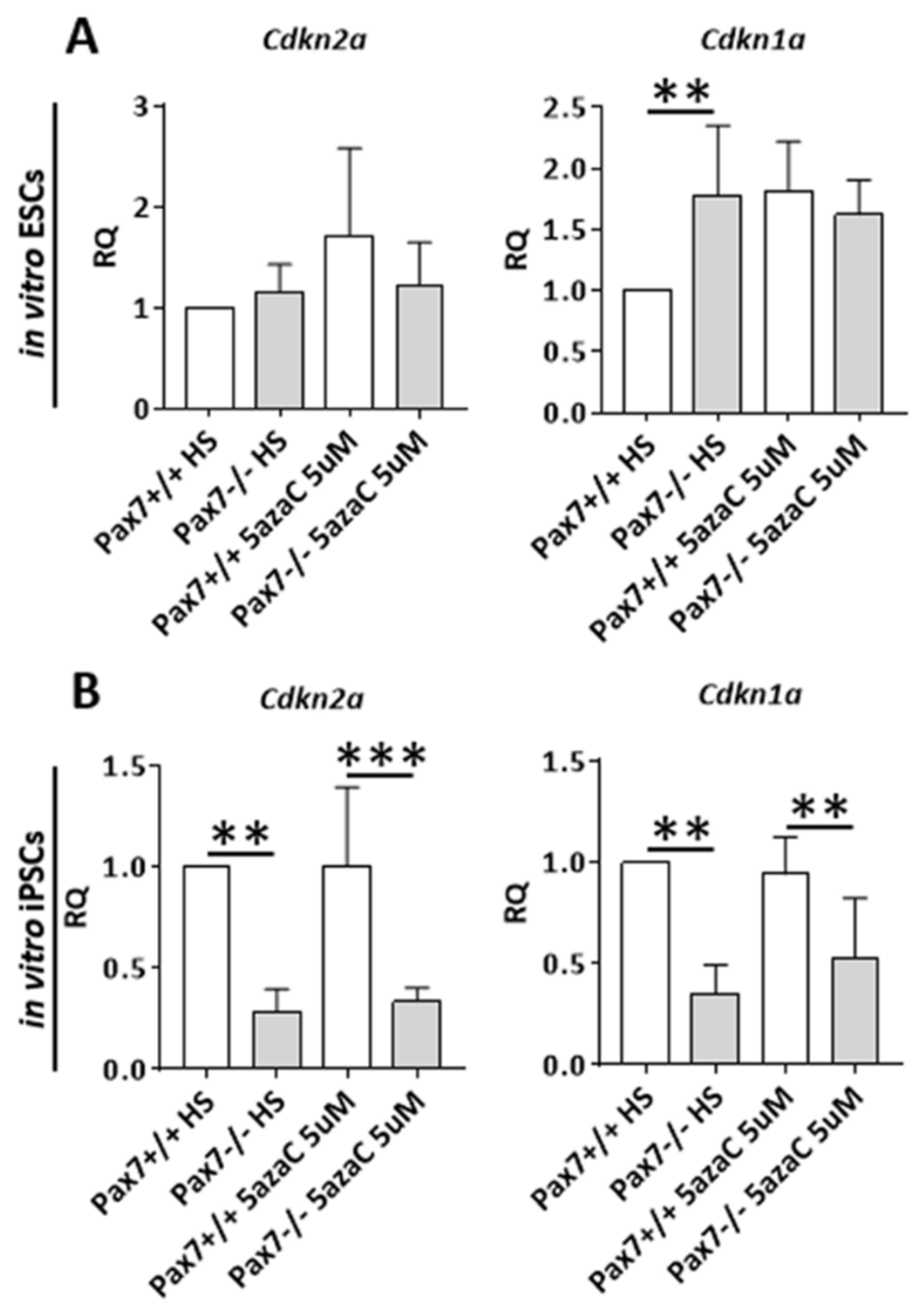
3.2. Relation between DNA Methylation, PAX7, and Cell Cycle Regulation in In Vitro and In Vivo Differentiating Pax7+/+ and Pax7−/− PSCs
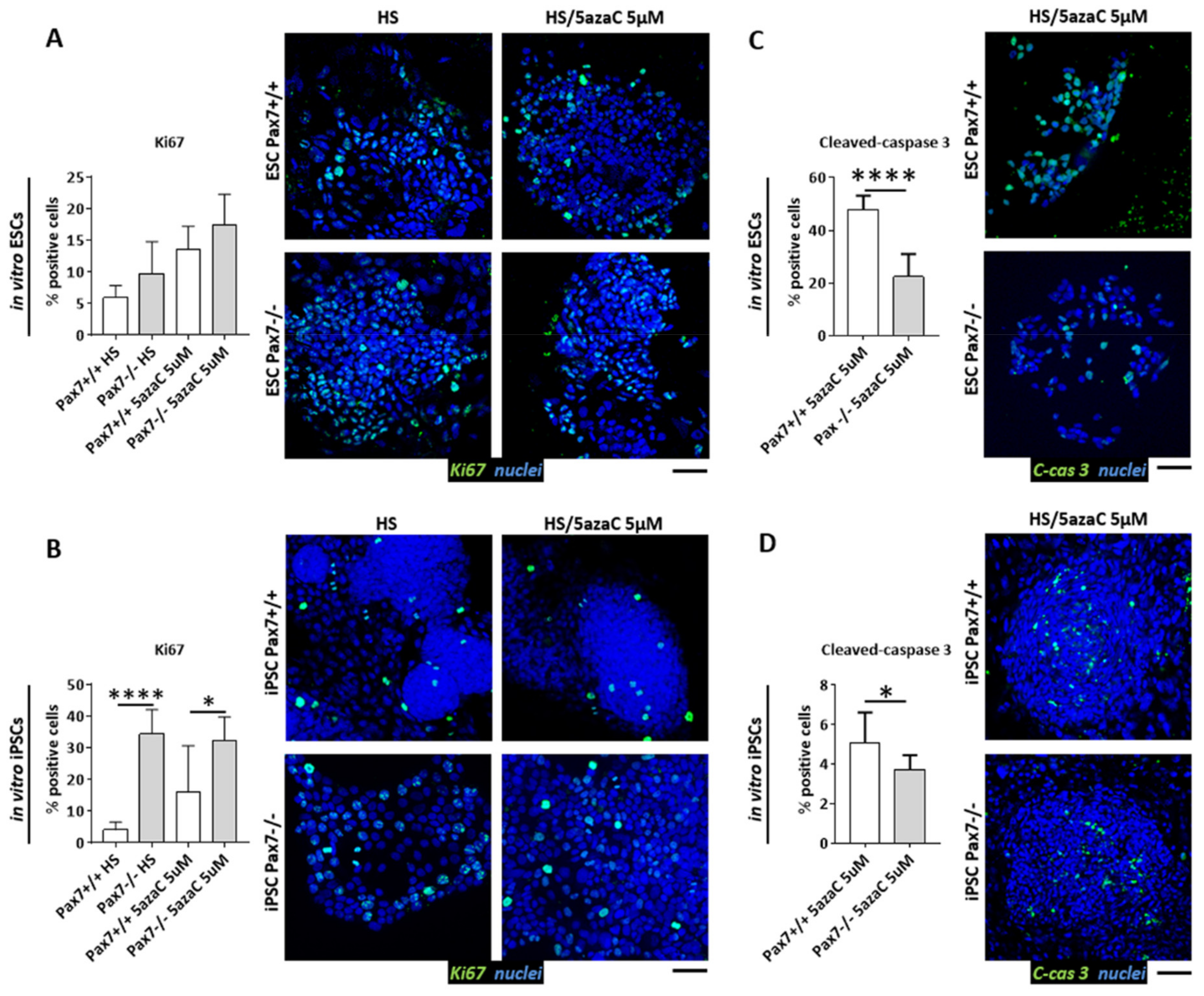
3.3. Proliferation of Pax7+/+ and Pax7−/− Differentiating PSC
3.4. Dnmt3a, Apobec2, and CDKIs in Pax7+/+ and Pax7−/− Skeletal Muscles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magli, A.; Incitti, T.; Kiley, J.; Swanson, S.A.; Darabi, R.; Rinaldi, F.; Selvaraj, S.; Yamamoto, A.; Tolar, J.; Yuan, C.; et al. PAX7 Targets, CD54, Integrin α9β1, and SDC2, Allow Isolation of Human ESC/iPSC-Derived Myogenic Progenitors. Cell Rep. 2017, 19, 2867–2877. [Google Scholar] [CrossRef] [Green Version]
- Darabi, R.; Arpke, R.W.; Irion, S.; Dimos, J.T.; Grskovic, M.; Kyba, M.; Perlingeiro, R.C.R. Human ES- and iPS-Derived Myogenic Progenitors Restore DYSTROPHIN and Improve Contractility upon Transplantation in Dystrophic Mice. Cell Stem Cell 2012, 10, 610–619. [Google Scholar] [CrossRef] [Green Version]
- Darabi, R.; Pan, W.; Bosnakovski, D.; Baik, J.; Kyba, M.; Perlingeiro, R.C. Functional myogenic engraftment from mouse iPS cells. Stem Cell Rev. 2011, 7, 948–957. [Google Scholar] [CrossRef] [Green Version]
- Helinska, A.; Krupa, M.; Archacka, K.; Czerwinska, A.M.; Streminska, W.; Janczyk-Ilach, K.; Ciemerych, M.A.; Grabowska, I. Myogenic potential of mouse embryonic stem cells lacking functional Pax7 tested in vitro by 5-azacitidine treatment and in vivo in regenerating skeletal muscle. Eur. J. Cell Biol. 2017, 96, 47–60. [Google Scholar] [CrossRef]
- Watanabe, S.; Hirai, H.; Asakura, Y.; Tastad, C.; Verma, M.; Keller, C.; Dutton, J.R.; Asakura, A. MyoD gene suppression by Oct4 is required for reprogramming in myoblasts to produce induced pluripotent stem cells. Stem Cells 2011, 29, 505–516. [Google Scholar] [CrossRef] [Green Version]
- Archacka, K.; Denkis, A.; Brzoska, E.; Swierczek, B.; Tarczyluk, M.; Janczyk-Ilach, K.; Ciemerych, M.A.; Moraczewski, J. Competence of In Vitro Cultured Mouse Embryonic Stem Cells for Myogenic Differentiation and Fusion with Myoblasts. Stem Cells Dev. 2014, 23, 2455–2468. [Google Scholar] [CrossRef] [Green Version]
- Chal, J.; Al Tanoury, Z.; Oginuma, M.; Moncuquet, P.; Gobert, B.; Miyanari, A.; Tassy, O.; Guevara, G.; Hubaud, A.; Bera, A.; et al. Recapitulating early development of mouse musculoskeletal precursors of the paraxial mesoderm in vitro. Development 2018, 145, dev.157339. [Google Scholar] [CrossRef] [Green Version]
- Chal, J.; Pourquie, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef] [Green Version]
- Archacka, K.; Brzoska, E.; Ciemerych, M.A.; Czerwinska, A.M.; Grabowska, I.; Kowalski, K.K.; Zimowska, M. Pluripotent and Mesenchymal Stem Cells—Challenging Sources for Derivation of Myoblast. In Cardiac Cell Culture Technologies; Brzozka, Z., Jastrzebska, E., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 109–154. [Google Scholar]
- Desbaillets, I.; Ziegler, U.; Groscurth, P.; Gassmann, M. Embryoid bodies: An in vitro model of mouse embryogenesis. Exp. Physiol. 2000, 85, 645–651. [Google Scholar] [CrossRef]
- Itskovitz-Eldor, J.; Schuldiner, M.; Karsenti, D.; Eden, A.; Yanuka, O.; Amit, M.; Soreq, H.; Benvenisty, N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000, 6, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Grabowska, I.; Archacka, K.; Czerwinska, A.M.; Krupa, M.; Ciemerych, M.A. Mouse and human pluripotent stem cells and the means of their myogenic differentiation. In Mouse Development; Kubiak, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; Results and Problems in Cell Differentiation; Volume 55, pp. 321–356. [Google Scholar]
- Bem, J.; Grabowska, I.; Daniszewski, M.; Zawada, D.; Czerwinska, A.M.; Bugajski, L.; Piwocka, K.; Fogtman, A.; Ciemerych, M.A. Transient MicroRNA Expression Enhances Myogenic Potential of Mouse Embryonic Stem Cells. Stem Cells 2018, 36, 655–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czerwinska, A.M.; Nowacka, J.; Aszer, M.; Gawrzak, S.; Archacka, K.; Fogtman, A.; Iwanicka-Nowicka, R.; Janczyk-Ilach, K.; Koblowska, M.; Ciemerych, M.A.; et al. Cell cycle regulation of embryonic stem cells and mouse embryonic fibroblasts lacking functional Pax7. Cell Cycle 2016, 15, 2931–2942. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Jones, W.K.; Gulick, J.; Doetschman, T.; Robbins, J. Myosin heavy chain gene expression in mouse embryoid bodies. An in vitro developmental study. J. Biol. Chem. 1991, 266, 22419–22426. [Google Scholar] [CrossRef]
- Mazaleyrat, K.; Badja, C.; Broucqsault, N.; Chevalier, R.; Laberthonniere, C.; Dion, C.; Baldasseroni, L.; El-Yazidi, C.; Thomas, M.; Bachelier, R.; et al. Multilineage Differentiation for Formation of Innervated Skeletal Muscle Fibers from Healthy and Diseased Human Pluripotent Stem Cells. Cells 2020, 9, 1531. [Google Scholar] [CrossRef]
- Muntener, M.; Kagi, U.; Stevens, L.C.; Walt, H. Innervation and maturation of muscular tissue in testicular teratomas in strain 129/Sv-ter mice. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1990, 59, 223–229. [Google Scholar] [CrossRef]
- Quattrocelli, M.; Palazzolo, G.; Floris, G.; Schoffski, P.; Anastasia, L.; Orlacchio, A.; Vandendriessche, T.; Chuah, M.K.; Cossu, G.; Verfaillie, C.; et al. Intrinsic cell memory reinforces myogenic commitment of pericyte-derived iPSCs. J. Pathol. 2011, 223, 593–603. [Google Scholar] [CrossRef]
- Kazuki, Y.; Hiratsuka, M.; Takiguchi, M.; Osaki, M.; Kajitani, N.; Hoshiya, H.; Hiramatsu, K.; Yoshino, T.; Kazuki, K.; Ishihara, C.; et al. Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol. Ther. 2010, 18, 386–393. [Google Scholar] [CrossRef]
- Lezmi, E.; Weissbein, U.; Golan-Lev, T.; Nissim-Rafinia, M.; Meshorer, E.; Benvenisty, N. The Chromatin Regulator ZMYM2 Restricts Human Pluripotent Stem Cell Growth and Is Essential for Teratoma Formation. Stem Cell Rep. 2020, 15, 1275–1286. [Google Scholar] [CrossRef]
- Blum, B.; Bar-Nur, O.; Golan-Lev, T.; Benvenisty, N. The anti-apoptotic gene survivin contributes to teratoma formation by human embryonic stem cells. Nat. Biotechnol. 2009, 27, 281–287. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czerwinska, A.M.; Grabowska, I.; Archacka, K.; Bem, J.; Swierczek, B.; Helinska, A.; Streminska, W.; Fogtman, A.; Iwanicka-Nowicka, R.; Koblowska, M.; et al. Myogenic Differentiation of Mouse Embryonic Stem Cells That Lack a Functional Pax7 Gene. Stem Cells Dev. 2016, 25, 285–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florkowska, A.; Meszka, I.; Zawada, M.; Legutko, D.; Proszynski, T.J.; Janczyk-Ilach, K.; Streminska, W.; Ciemerych, M.A.; Grabowska, I. Pax7 as molecular switch regulating early and advanced stages of myogenic mouse ESC differentiation in teratomas. Stem Cell Res. Ther. 2020, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, A.; Stoykova, A.; Torres, M.; Gruss, P. Dysgenesis of cephalic neural crest derivatives in Pax7-/- mutant mice. Development 1996, 122, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Hutcheson, D.A.; Zhao, J.; Merrell, A.; Haldar, M.; Kardon, G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for β-catenin. Genes Dev. 2009, 23, 997–1013. [Google Scholar] [CrossRef] [Green Version]
- Kuang, S.; Charge, S.B.; Seale, P.; Huh, M.; Rudnicki, M.A. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 2006, 172, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 is required for the specification of myogenic satellite cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef] [Green Version]
- Oustanina, S.; Hause, G.; Braun, T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004, 23, 3430–3439. [Google Scholar] [CrossRef] [Green Version]
- Worl, J.; Breuer, C.; Neuhuber, W.L. Deletion of Pax7 changes the tunica muscularis of the mouse esophagus from an entirely striated into a mixed phenotype. Dev. Dyn. 2009, 238, 864–874. [Google Scholar] [CrossRef]
- Wang, J.; Conboy, I. Embryonic vs. adult myogenesis: Challenging the ‘regeneration recapitulates development’ paradigm. J. Mol. Cell Biol. 2009, 2, 1–4. [Google Scholar] [CrossRef]
- Relaix, F.; Zammit, P.S. Satellite cells are essential for skeletal muscle regeneration: The cell on the edge returns centre stage. Development 2012, 139, 2845–2856. [Google Scholar] [CrossRef] [Green Version]
- Relaix, F.; Montarras, D.; Zaffran, S.; Gayraud-Morel, B.; Rocancourt, D.; Tajbakhsh, S.; Mansouri, A.; Cumano, A.; Buckingham, M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 2006, 172, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Bober, E.; Franz, T.; Arnold, H.H.; Gruss, P.; Tremblay, P. Pax-3 is required for the development of limb muscles: A possible role for the migration of dermomyotomal muscle progenitor cells. Development 1994, 120, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.A.; Ordahl, C.P. Pax-3 expression in segmental mesoderm marks early stages in myogenic cell specification. Development 1994, 120, 785–796. [Google Scholar] [CrossRef]
- Braun, T.; Bober, E.; Rudnicki, M.A.; Jaenisch, R.; Arnold, H.H. MyoD expression marks the onset of skeletal myogenesis in Myf-5 mutant mice. Development 1994, 120, 3083–3092. [Google Scholar] [CrossRef]
- Tajbakhsh, S.; Rocancourt, D.; Cossu, G.; Buckingham, M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 1997, 89, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005, 435, 948–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004, 18, 1088–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKinnell, I.W.; Ishibashi, J.; Le Grand, F.; Punch, V.G.; Addicks, G.C.; Greenblatt, J.F.; Dilworth, F.J.; Rudnicki, M.A. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 2008, 10, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, C.A.; Gnocchi, V.F.; White, R.B.; Boldrin, L.; Perez-Ruiz, A.; Relaix, F.; Morgan, J.E.; Zammit, P.S. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS ONE 2009, 4, e4475. [Google Scholar] [CrossRef] [Green Version]
- Olguin, H.C.; Olwin, B.B. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: A potential mechanism for self-renewal. Dev. Biol. 2004, 275, 375–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernasconi, M.; Remppis, A.; Fredericks, W.J.; Rauscher, F.J.; Schafer, B.W. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 13164–13169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [Green Version]
- So, A.Y.; Jung, J.W.; Lee, S.; Kim, H.S.; Kang, K.S. DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS ONE 2011, 6, e19503. [Google Scholar] [CrossRef] [Green Version]
- Megiorni, F.; Camero, S.; Ceccarelli, S.; McDowell, H.P.; Mannarino, O.; Marampon, F.; Pizer, B.; Shukla, R.; Pizzuti, A.; Marchese, C.; et al. DNMT3B in vitro knocking-down is able to reverse embryonal rhabdomyosarcoma cell phenotype through inhibition of proliferation and induction of myogenic differentiation. Oncotarget 2016, 7, 79342–79356. [Google Scholar] [CrossRef] [Green Version]
- Blanc, R.S.; Vogel, G.; Chen, T.; Crist, C.; Richard, S. PRMT7 Preserves Satellite Cell Regenerative Capacity. Cell Rep. 2016, 14, 1528–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharyya, S.; Sharma, S.M.; Cheng, A.S.; Ladner, K.J.; He, W.; Kline, W.; Wang, H.; Ostrowski, M.C.; Huang, T.H.; Guttridge, D.C. TNF inhibits Notch-1 in skeletal muscle cells by Ezh2 and DNA methylation mediated repression: Implications in duchenne muscular dystrophy. PLoS ONE 2010, 5, e12479. [Google Scholar] [CrossRef] [PubMed]
- Carrio, E.; Magli, A.; Munoz, M.; Peinado, M.A.; Perlingeiro, R.; Suelves, M. Muscle cell identity requires Pax7-mediated lineage-specific DNA demethylation. BMC Biol. 2016, 14, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, W.; Hong, S.H.; Chan, B.H.; Rudolph, F.B.; Clark, S.C.; Chan, L. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem. Biophys. Res. Commun. 1999, 260, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, H.; Sato, Y.; Matsuyoshi, Y.; Suzuki, T.; Mizunoya, W.; Nakamura, M.; Tatsumi, R.; Ikeuchi, Y. Fluorescence microscopy data on expression of Paired Box Transcription Factor 7 in skeletal muscle of APOBEC2 knockout mice. Data Brief 2018, 17, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, H.; Sato, Y.; Suzuki, T.; Mizunoya, W.; Nakamura, M.; Tatsumi, R.; Ikeuchi, Y. APOBEC2 negatively regulates myoblast differentiation in muscle regeneration. Int. J. Biochem. Cell Biol. 2017, 85, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Robertson, E. Embryo-derived stem cell lines. In Teratocarcinomas and Embryonic Stem Cells: A Practical Approach; Robertson, E., Ed.; Oxford University Press: Oxford, UK, 1987; pp. 71–112. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gerdes, J.; Lemke, H.; Baisch, H.; Wacker, H.H.; Schwab, U.; Stein, H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 1984, 133, 1710–1715. [Google Scholar]
- Hiromura, K.; Pippin, J.W.; Fero, M.L.; Roberts, J.M.; Shankland, S.J. Modulation of apoptosis by the cyclin-dependent kinase inhibitor p27(Kip1). J. Clin. Investig. 1999, 103, 597–604. [Google Scholar] [CrossRef] [Green Version]
- Musch, T.; Oz, Y.; Lyko, F.; Breiling, A. Nucleoside drugs induce cellular differentiation by caspase-dependent degradation of stem cell factors. PLoS ONE 2010, 5, e10726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.K.; Wang, Y.; Karandikar, A.; Wang, Q.; Gai, H.; Liu, A.L.; Peng, C.; Sheng, H.Z. Skeletal myogenesis by human embryonic stem cells. Cell Res. 2006, 16, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Relaix, F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Biol. 2015, 44, 115–125. [Google Scholar] [CrossRef]
- Ciemerych, M.A.; Archacka, K.; Grabowska, I.; Przewozniak, M. Cell cycle regulation during proliferation and differentiation of mammalian muscle precursor cells. In Cell Cycle in Development; Kubiak, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Results and Problems in Cell Differentiation; Volume 53, pp. 473–527. [Google Scholar]
- Martins-Taylor, K.; Schroeder, D.I.; LaSalle, J.M.; Lalande, M.; Xu, R.H. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics 2012, 7, 71–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Walsh, G.; Liu, D.D.; Lee, J.J.; Mao, L. Expression of Delta DNMT3B variants and its association with promoter methylation of p16 and RASSF1A in primary non-small cell lung cancer. Cancer Res. 2006, 66, 8361–8366. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.S.; Lee, H.; Chen, R.A.; Ho, M.L.; Lin, C.Y.; Chen, Y.H.; Tsai, Y.Y.; Chou, M.C.; Cheng, Y.W. An association of DNMT3b protein expression with P16INK4a promoter hypermethylation in non-smoking female lung cancer with human papillomavirus infection. Cancer Lett. 2005, 226, 77–84. [Google Scholar] [CrossRef]
- Fang, J.Y.; Yang, L.; Zhu, H.Y.; Chen, Y.X.; Lu, J.; Lu, R.; Cheng, Z.H.; Xiao, S.D. 5-Aza-2′-deoxycitydine induces demethylation and up-regulates transcription of p16INK4A gene in human gastric cancer cell lines. Chin. Med. J. 2004, 117, 99–103. [Google Scholar] [PubMed]
- Kiess, M.; Gill, R.M.; Hamel, P.A. Expression of the positive regulator of cell cycle progression, cyclin D3, is induced during differentiation of myoblasts into quiescent myotubes. Oncogene 1995, 10, 159–166. [Google Scholar] [PubMed]
- De Santa, F.; Albini, S.; Mezzaroma, E.; Baron, L.; Felsani, A.; Caruso, M. pRb-dependent cyclin D3 protein stabilization is required for myogenic differentiation. Mol. Cell. Biol. 2007, 27, 7248–7265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurung, R.; Parnaik, V.K. Cyclin D3 promotes myogenic differentiation and Pax7 transcription. J. Cell. Biochem. 2012, 113, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, S.; Marusawa, H.; Matsumoto, T.; Ueda, Y.; Matsumoto, Y.; Endo, Y.; Takai, A.; Chiba, T. Excessive activity of apolipoprotein B mRNA editing enzyme catalytic polypeptide 2 (APOBEC2) contributes to liver and lung tumorigenesis. Int. J. Cancer 2012, 130, 1294–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, C.; Elsaeidi, F.; Goldman, D. Injury-dependent Muller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J. Neurosci. 2012, 32, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Probst, H.C.; Tatsumi, R.; Ikeuchi, Y.; Neuberger, M.S.; Rada, C. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J. Biol. Chem. 2010, 285, 7111–7118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Ohtsubo, H.; Nihei, N.; Kaneko, T.; Sato, Y.; Adachi, S.I.; Kondo, S.; Nakamura, M.; Mizunoya, W.; Iida, H.; et al. Apobec2 deficiency causes mitochondrial defects and mitophagy in skeletal muscle. FASEB J. 2018, 32, 1428–1439. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Cooper, J.B. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell 2010, 7, 258–262. [Google Scholar] [CrossRef] [Green Version]
- Bock, C.; Kiskinis, E.; Verstappen, G.; Gu, H.; Boulting, G.; Smith, Z.D.; Ziller, M.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011, 144, 439–452. [Google Scholar] [CrossRef] [Green Version]
- Humpherys, D.; Eggan, K.; Akutsu, H.; Hochedlinger, K.; Rideout, W.M., 3rd; Biniszkiewicz, D.; Yanagimachi, R.; Jaenisch, R. Epigenetic instability in ES cells and cloned mice. Science 2001, 293, 95–97. [Google Scholar] [CrossRef]
- Bar, S.; Benvenisty, N. Epigenetic aberrations in human pluripotent stem cells. EMBO J. 2019, 38, e101033. [Google Scholar] [CrossRef]
- Polo, J.M.; Anderssen, E.; Walsh, R.M.; Schwarz, B.A.; Nefzger, C.M.; Lim, S.M.; Borkent, M.; Apostolou, E.; Alaei, S.; Cloutier, J.; et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012, 151, 1617–1632. [Google Scholar] [CrossRef] [Green Version]
- Zammit, P.S.; Relaix, F.; Nagata, Y.; Ruiz, A.P.; Collins, C.A.; Partridge, T.A.; Beauchamp, J.R. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006, 119, 1824–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seale, P.; Ishibashi, J.; Scime, A.; Rudnicki, M.A. Pax7 is necessary and sufficient for the myogenic specification of CD45+:Sca1+ stem cells from injured muscle. PLoS Biol. 2004, 2, e130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Close, B.; Banister, K.; Baumans, V.; Bernoth, E.M.; Bromage, N.; Bunyan, J.; Erhardt, W.; Flecknell, P.; Gregory, N.; Hackbarth, H.; et al. Recommendations for euthanasia of experimental animals: Part 2. DGXT of the European Commission. Lab. Anim. 1997, 31, 1–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Close, B.; Banister, K.; Baumans, V.; Bernoth, E.M.; Bromage, N.; Bunyan, J.; Erhardt, W.; Flecknell, P.; Gregory, N.; Hackbarth, H.; et al. Recommendations for euthanasia of experimental animals: Part 1. DGXI of the European Commission. Lab. Anim. 1996, 30, 293–316. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Florkowska, A.; Meszka, I.; Nowacka, J.; Granica, M.; Jablonska, Z.; Zawada, M.; Truszkowski, L.; Ciemerych, M.A.; Grabowska, I. PAX7 Balances the Cell Cycle Progression via Regulating Expression of Dnmt3b and Apobec2 in Differentiating PSCs. Cells 2021, 10, 2205. https://doi.org/10.3390/cells10092205
Florkowska A, Meszka I, Nowacka J, Granica M, Jablonska Z, Zawada M, Truszkowski L, Ciemerych MA, Grabowska I. PAX7 Balances the Cell Cycle Progression via Regulating Expression of Dnmt3b and Apobec2 in Differentiating PSCs. Cells. 2021; 10(9):2205. https://doi.org/10.3390/cells10092205
Chicago/Turabian StyleFlorkowska, Anita, Igor Meszka, Joanna Nowacka, Monika Granica, Zuzanna Jablonska, Magdalena Zawada, Lukasz Truszkowski, Maria A. Ciemerych, and Iwona Grabowska. 2021. "PAX7 Balances the Cell Cycle Progression via Regulating Expression of Dnmt3b and Apobec2 in Differentiating PSCs" Cells 10, no. 9: 2205. https://doi.org/10.3390/cells10092205
APA StyleFlorkowska, A., Meszka, I., Nowacka, J., Granica, M., Jablonska, Z., Zawada, M., Truszkowski, L., Ciemerych, M. A., & Grabowska, I. (2021). PAX7 Balances the Cell Cycle Progression via Regulating Expression of Dnmt3b and Apobec2 in Differentiating PSCs. Cells, 10(9), 2205. https://doi.org/10.3390/cells10092205





