Proteomic-Based Analysis of Hypoxia- and Physioxia-Responsive Proteins and Pathways in Diffuse Large B-Cell Lymphoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culturing under Physioxia and Hypoxia
2.2. Sample Preparation for Mass Spectrometry
2.3. LC–MS/MS
2.4. MS/MS Data Analysis and Statistical Analysis
2.5. Bioinformatic Analysis
3. Results
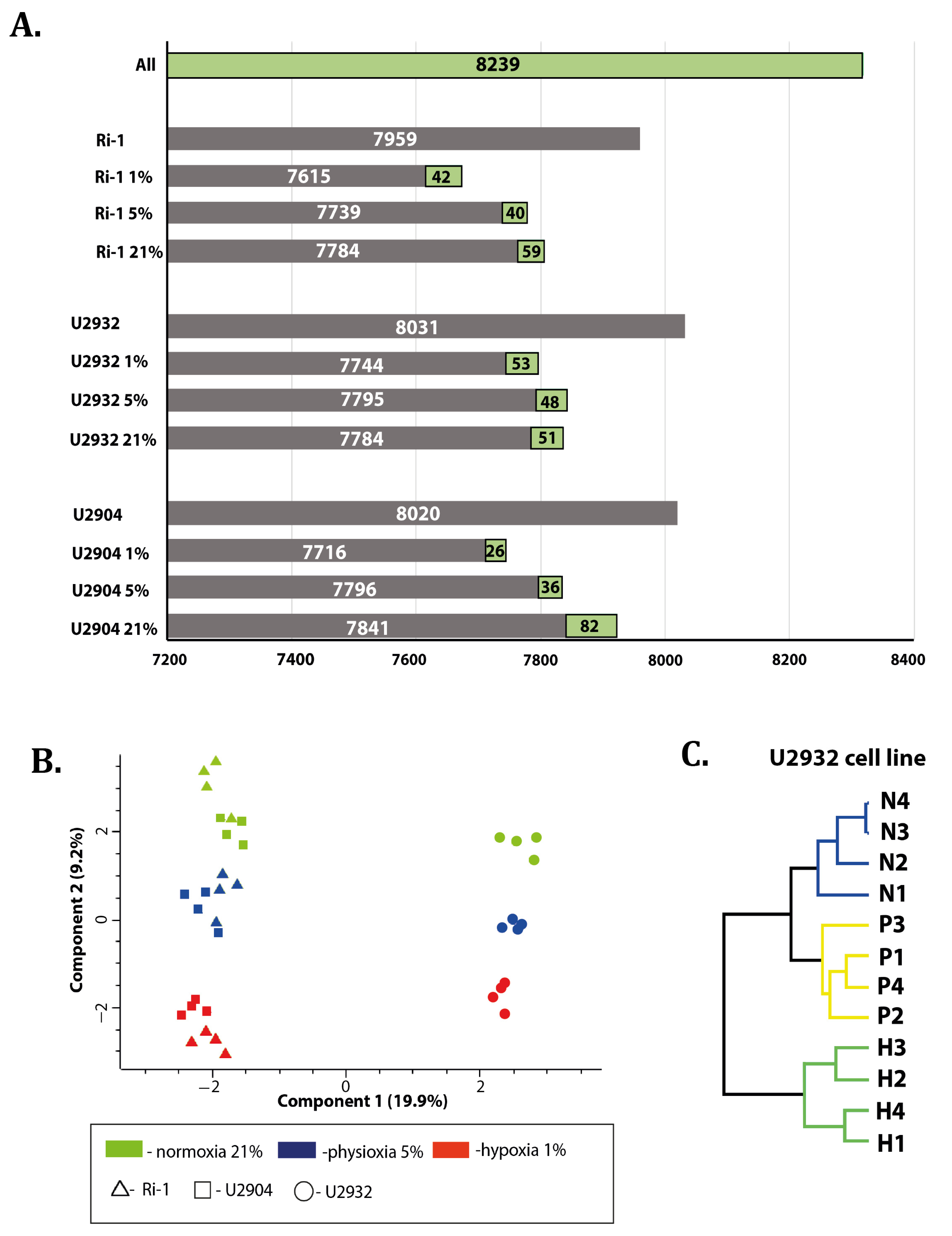
3.1. Overview of Data
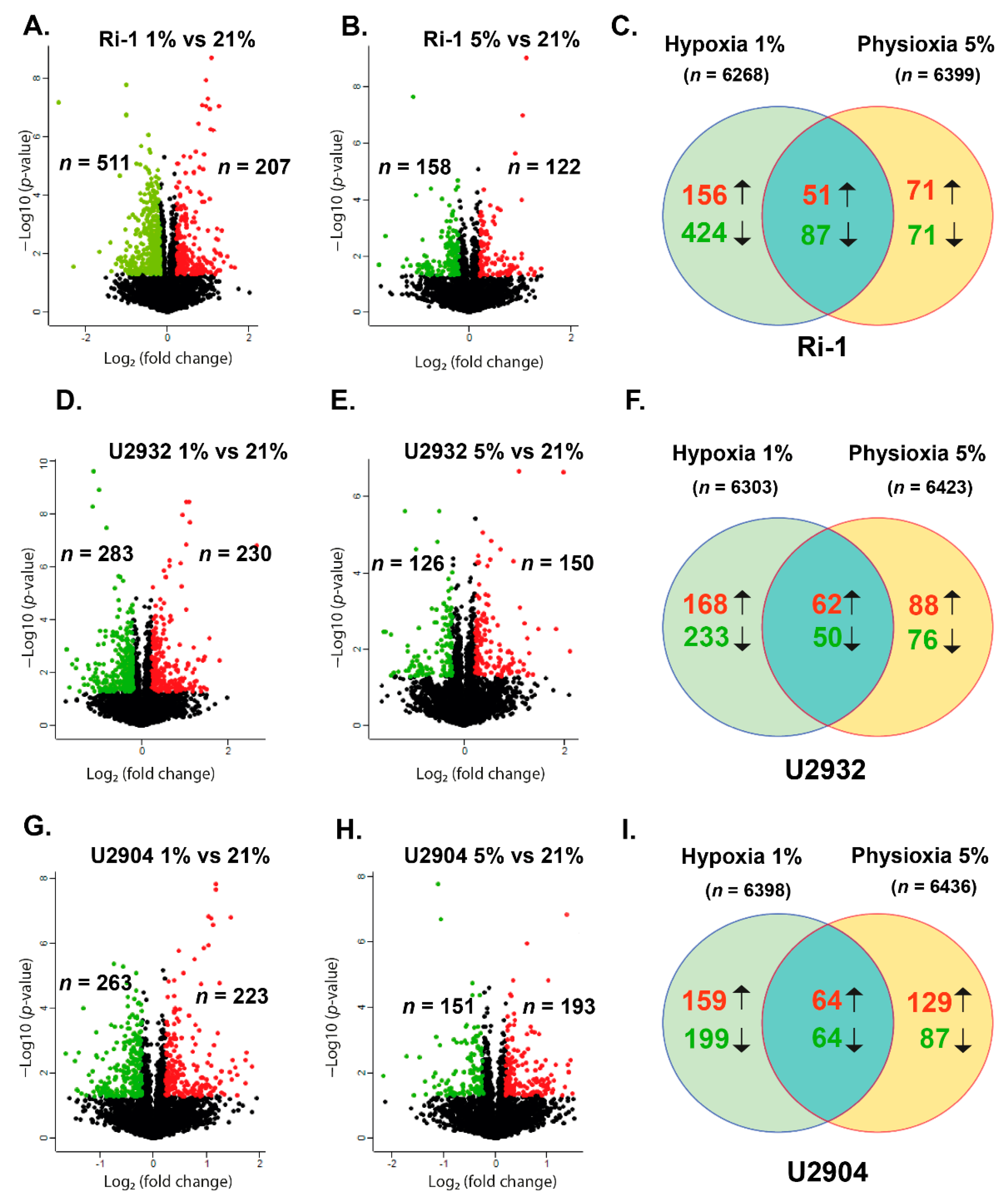
3.2. DLBCL Shows Extensive Changes in Proteomic Profile under Hypoxia and Physioxia When Compared to Normoxia
Response to Both: Hypoxia and Physioxia Is Cell Line Specific
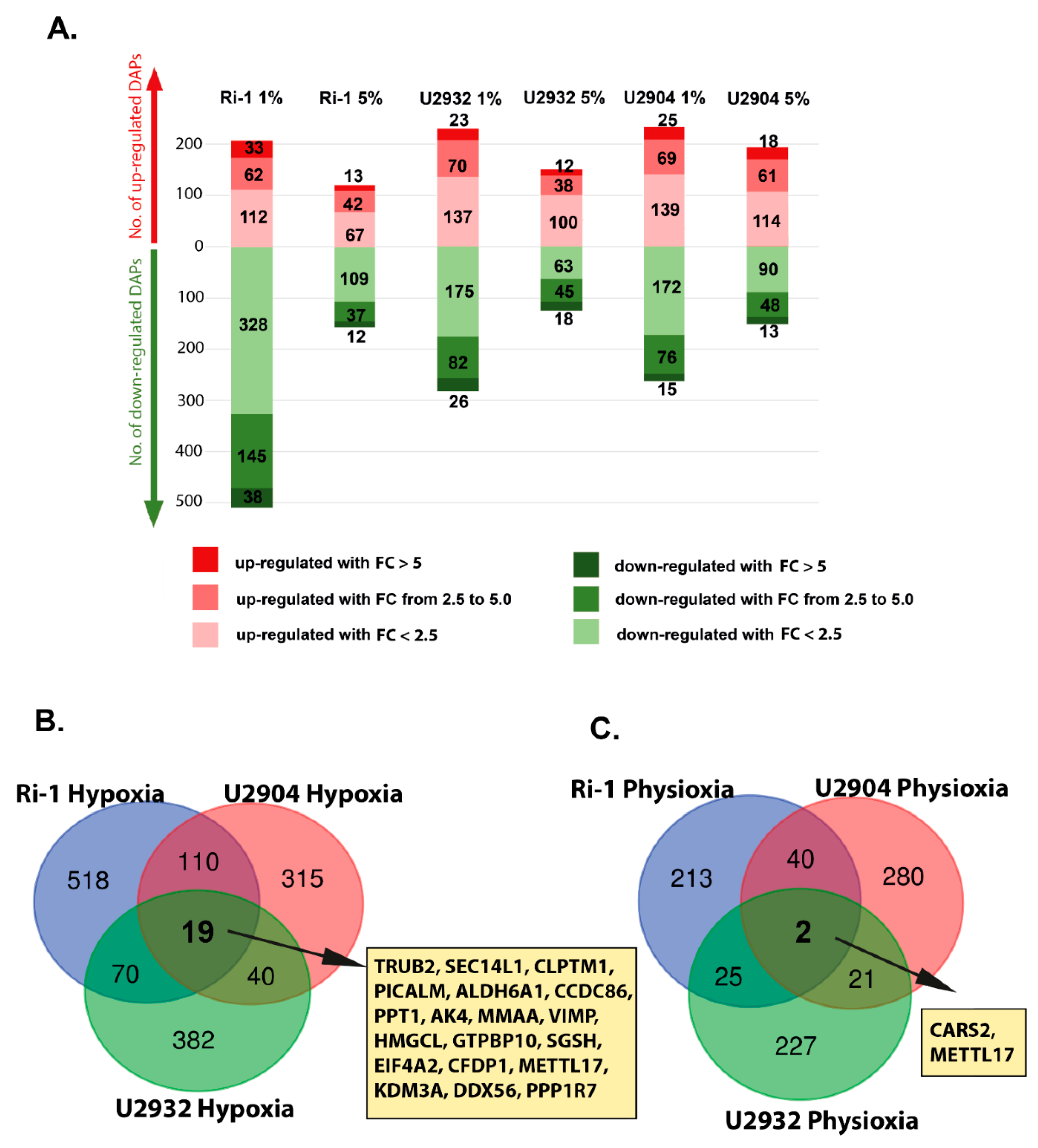
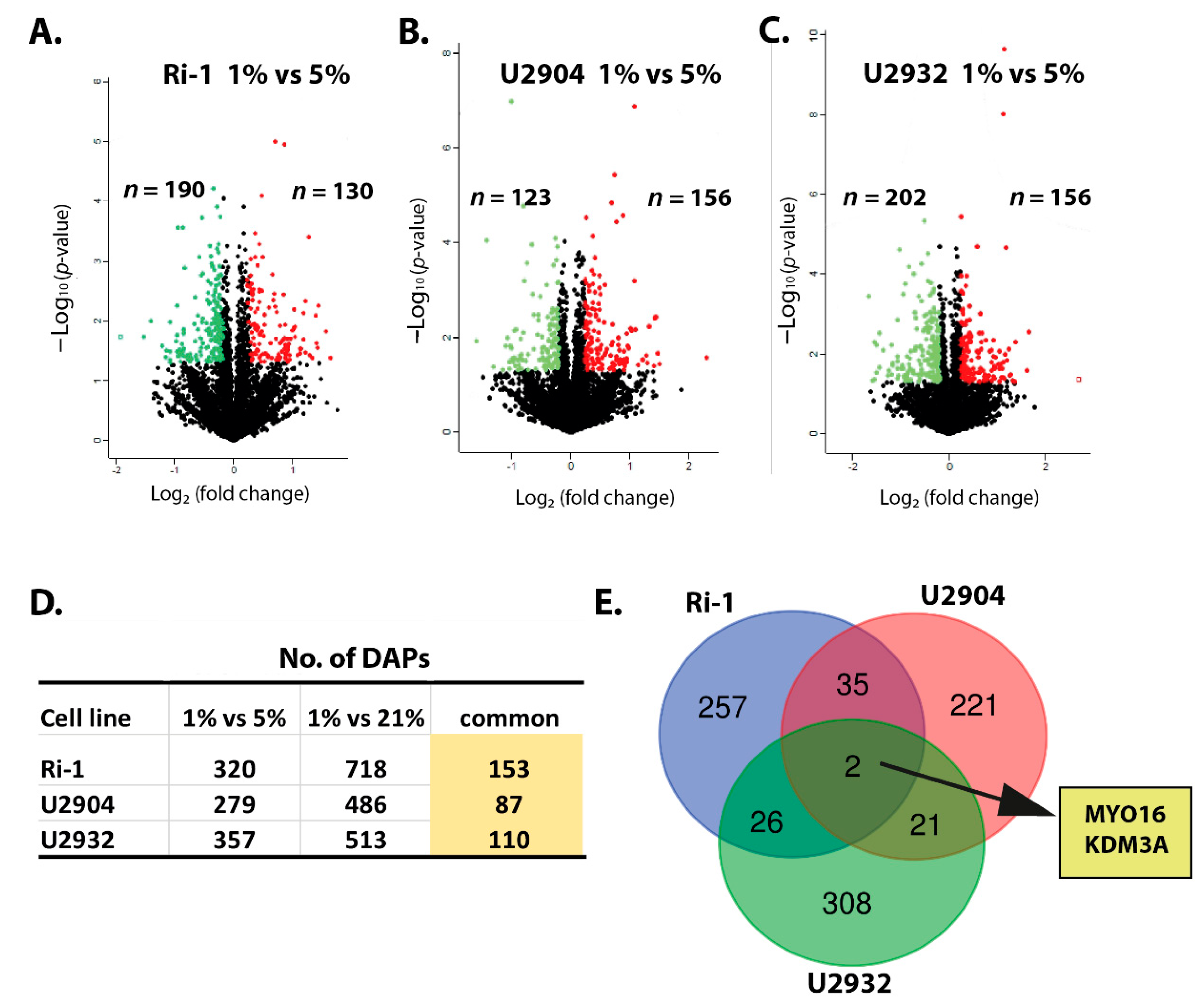
3.3. Hypoxic Treatment Exhibit Lower Magnitude of Changes When Compared with Physioxia
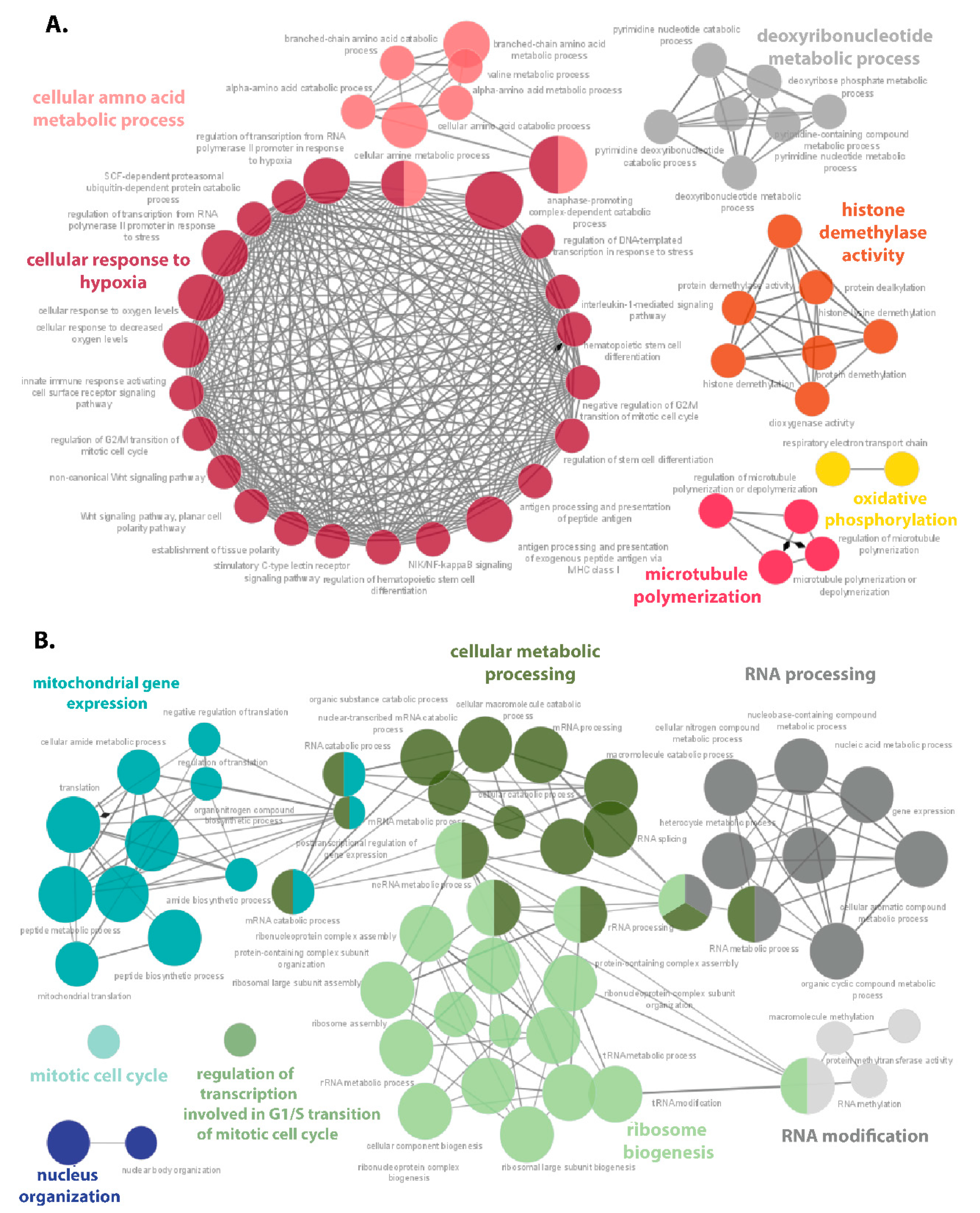
3.4. Functional Enrichment Analysis
3.4.1. Upregulated Proteins Reveals Changes in Metabolism and HIF-1 Signaling Pathway under Hypoxia
3.4.2. Ribosome Biogenesis, Translation, and Mitochondrial Gene Expression Decrease in in DLBCL under Hypoxia and Physioxia
3.5. Hypoxia-Responsive Proteins Make a Complex Protein–Protein Interaction Network in DLBCL
3.6. Hypoxia Induces Prominent Changes in Translation in DLBCL
3.7. Hub Proteins Associated with Hypoxia and Physioxia Are Frequently Related to Ribosome Biogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mattei, G.; Giusti, S.; Ahluwalia, A. Design Criteria for Generating Physiologically Relevant In Vitro Models in Bioreactors. Processes 2014, 2, 548–569. [Google Scholar] [CrossRef] [Green Version]
- Matolay, O.; Méhes, G. Sustain, Adapt, and Overcome-Hypoxia Associated Changes in the Progression of Lymphatic Neoplasia. Front. Oncol. 2019, 9, 1277. [Google Scholar] [CrossRef] [Green Version]
- Spencer, J.A.; Ferraro, F.; Roussakis, E.; Klein, A.; Wu, J.; Runnels, J.M.; Zaher, W.; Mortensen, L.J.; Alt, C.; Turcotte, R.; et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 2014, 508, 269–273. [Google Scholar] [CrossRef] [Green Version]
- Caldwell, C.C.; Kojima, H.; Lukashev, D.; Armstrong, J.; Farber, M.; Apasov, S.G.; Sitkovsky, M.V. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J. Immunol. 2001, 167, 6140–6149. [Google Scholar] [CrossRef]
- Bhalla, K.; Jaber, S.; Nahid, M.N.; Underwood, K.; Beheshti, A.; Landon, A.; Bhandary, B.; Bastian, P.; Evens, A.M.; Haley, J.; et al. Role of hypoxia in Diffuse Large B-cell Lymphoma: Metabolic repression and selective translation of HK2 facilitates development of DLBCL. Sci. Rep. 2018, 8, 744. [Google Scholar] [CrossRef] [Green Version]
- Estrada, J.C.; Albo, C.; Benguría, A.; Dopazo, A.; López-Romero, P.; Carrera-Quintanar, L.; Roche, E.; Clemente, E.P.; Enríquez, J.A.; Bernad, A.; et al. Culture of human mesenchymal stem cells at low oxygen tension improves growth and genetic stability by activating glycolysis. Cell Death Differ. 2012, 19, 743–755. [Google Scholar] [CrossRef] [Green Version]
- Antebi, B.; Rodriguez, L.A., 2nd; Walker, K.P., 3rd; Asher, A.M.; Kamucheka, R.M.; Alvarado, L.; Mohammadipoor, A.; Cancio, L.C. Short-term physiological hypoxia potentiates the therapeutic function of mesenchymal stem cells. Stem Cell Res. Ther. 2018, 9, 265. [Google Scholar] [CrossRef] [Green Version]
- Tomecka, E.; Lech, W.; Zychowicz, M.; Sarnowska, A.; Murzyn, M.; Oldak, T.; Domanska-Janik, K.; Buzanska, L.; Rozwadowska, N. Assessment of the Neuroprotective and Stemness Properties of Human Wharton’s Jelly-Derived Mesenchymal Stem Cells under Variable (5% vs. 21%) Aerobic Conditions. Cells 2021, 10, 717. [Google Scholar] [CrossRef]
- Pattappa, G.; Johnstone, B.; Zellner, J.; Docheva, D.; Angele, P. The Importance of Physioxia in Mesenchymal Stem Cell Chondrogenesis and the Mechanisms Controlling Its Response. Int. J. Mol. Sci. 2019, 20, 484. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Tang, Q.; Zhang, Y.; Yu, M.; Jing, W.; Tian, W. Physioxia: A more effective approach for culturing human adipose-derived stem cells for cell transplantation. Stem Cell Res. Ther. 2018, 9, 148. [Google Scholar] [CrossRef]
- Gomes, A.; Guillaume, L.; Grimes, D.R.; Fehrenbach, J.; Lobjois, V.; Ducommun, B. Oxygen Partial Pressure Is a Rate-Limiting Parameter for Cell Proliferation in 3D Spheroids Grown in Physioxic Culture Condition. PLoS ONE 2016, 11, e0161239. [Google Scholar]
- Hasan, N.M.; Adams, G.E.; Joiner, M.C.; Marshall, J.F.; Hart, I.R. Hypoxia facilitates tumour cell detachment by reducing expression of surface adhesion molecules and adhesion to extracellular matrices without loss of cell viability. Br. J. Cancer 1998, 77, 1799–1805. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wen, Y.; Bi, P.; Lai, X.; Liu, X.S.; Liu, X.; Kuang, S. Hypoxia promotes satellite cell self-renewal and enhances the efficiency of myoblast transplantation. Development 2012, 139, 2857–2865. [Google Scholar] [CrossRef] [Green Version]
- Dennis, J.E.; Whitney, G.A.; Rai, J.; Fernandes, R.J.; Kean, T.J. Physioxia Stimulates Extracellular Matrix Deposition and Increases Mechanical Properties of Human Chondrocyte-Derived Tissue-Engineered Cartilage. Front. Bioeng. Biotechnol. 2020, 8, 590743. [Google Scholar] [CrossRef]
- Timpano, S.; Guild, B.D.; Specker, E.J.; Melanson, G.; Medeiros, P.J.; Sproul, S.L.J.; Uniacke, J. Physioxic human cell culture improves viability, metabolism, and mitochondrial morphology while reducing DNA damage. FASEB J. 2019, 33, 5716–5728. [Google Scholar] [CrossRef]
- Jiang, M.; Bennani, N.N.; Feldman, A.L. Lymphoma classification update: B-cell non-Hodgkin lymphomas. Expert. Rev. Hematol. 2017, 10, 405–415. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Adams, H.J.A.; de Klerk, J.M.H.; Fijnheer, R.; Dubois, S.V.; Nievelstein, R.A.J.; Kwee, T.C. Prognostic value of tumor necrosisat CT in diffuselarge B-cell lymphoma. Eur. J. Radiol. 2015, 84, 372–377. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, V.; Li, C.H.; Bristow, R.G.; Boutros, P.C.; PCAWG Consortium. Divergent mutational processes distinguish hypoxic and normoxic tumours. Nat. Commun. 2020, 11, 737. [Google Scholar] [CrossRef] [Green Version]
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Holcik, M.; Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.O.; Juul, N.O.; Rønning, O.W. Regulation of protein metabolism of human cells during and after acute hypoxia. Cancer Res. 1986, 46, 4346–4351. [Google Scholar]
- Ricci, J.E.; Chiche, J. Metabolic Reprogramming of Non-Hodgkin’s B-Cell Lymphomas and Potential Therapeutic Strategies. Front. Oncol. 2018, 8, 556. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Wang, J.; Alqassim, E.; Portwood, S.; Cortes Gomez, E.; Maguire, O.; Basse, P.H.; Wang, E.S.; Segal, B.H.; Baysal, B.E. Mitochondrial hypoxic stress induces wide spread RNA editing by APOBEC3G in natural killer cells. Genome Biol. 2019, 20, 37. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yao, Y.; Zhang, S.; Liu, Y.; Guo, H.; Ahmed, M.; Bell, T.; Zhang, H.; Han, G.; Lorence, E. Metabolic reprogramming toward oxidative phosphorylation identifies a therapeutic target for mantle cell lymphoma. Sci. Transl. Med. 2019, 11, eaau1167. [Google Scholar] [CrossRef] [PubMed]
- Clapper, E.; Di Trapani, G.; Tonissen, K.F. The regulation of bcr-abl in hypoxia is through the mTOR pathway. Leuk. Lymphoma. 2021, 62, 967–978. [Google Scholar] [CrossRef]
- Burrows, N.; Bashford-Rogers, R.J.M.; Bhute, V.J.; Peñalver, A.; Ferdinand, J.R.; Stewart, B.J.; Smith, J.E.G.; Deobagkar-Lele, M.; Giudice, G.; Connor, T.M.; et al. Dynamic regulation of hypoxia-inducible factor-1α activity is essential for normal B cell development. Nat. Immunol. 2020, 21, 1408–1420. [Google Scholar] [CrossRef]
- Duś-Szachniewicz, K.; Rymkiewicz, G.; Agrawal, A.K.; Kołodziej, P.; Wiśniewski, J.R. Large-Scale Proteomic Analysis of Follicular Lymphoma Reveals Extensive Remodeling of Cell Adhesion Pathway and Identifies Hub Proteins Related to the Lymphomagenesis. Cancers 2021, 13, 630. [Google Scholar] [CrossRef]
- Reinders, J.; Altenbuchinger, M.; Limm, K.; Schwarzfischer, P.; Scheidt, T.; Strasser, L.; Richter, J.; Szczepanowski, M.; Huber, C.G.; Klapper, W.; et al. Platform independent protein-based cell-of-origin subtyping of diffuse large B-cell lymphoma in formalin-fixed paraffin-embedded tissue. Sci. Rep. 2020, 10, 7876. [Google Scholar] [CrossRef]
- Deeb, S.J.; D’Souza, R.C.; Cox, J.; Schmidt-Supprian, M.; Mann, M. Super-SILAC allows classification of diffuselarge B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell Proteom. 2012, 11, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, V.; Hoey, C.; Liu, L.Y.; Lalonde, E.; Ray, J.; Livingstone, J.; Lesurf, R.; Shiah, Y.J.; Vujcic, T.; Huang, X.; et al. Molecular landmarks of tumor hypoxia acrosscancertypes. Nat. Genet. 2019, 51, 308–318. [Google Scholar] [CrossRef]
- Shin, D.Y.; Huang, X.; Gil, C.H.; Aljoufi, A.; Ropa, J.; Broxmeyer, H.E. Physioxia enhances T-cell development ex vivo from human hematopoietic stem and progenitor cells. Stem Cells 2020, 38, 1454–1466. [Google Scholar]
- Wiśniewski, J.R.; Gaugaz, F.Z. Fast and sensitive total protein and Peptide assays for proteomic analysis. Anal. Chem. 2015, 87, 4110–4116. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Mann, M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal. Chem. 2012, 84, 2631–2637. [Google Scholar] [CrossRef]
- Wiśniewski, J.R. Quantitative Evaluation of Filter Aided Sample Preparation (FASP) and Multienzyme Digestion FASP Protocols. Anal. Chem. 2016, 88, 5438–5443. [Google Scholar] [CrossRef] [Green Version]
- Ishihama, Y.; Rappsilber, J.; Andersen, J.S.; Mann, M. Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A. 2002, 979, 233–239. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Rakus, D. Multi-enzyme digestion FASP and the ‘Total Protein Approach’-based absolute quantification of the Escherichia coli proteome. J. Proteom. 2014, 109, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Cox, J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol. Biol. 2018, 1711, 133–148. [Google Scholar]
- Mlecnik, B.; Galon, J.; Bindea, G. Comprehensive functional analysis of large lists of genes and proteins. J. Proteom. 2018, 171, 2–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Bioinformatics and Evolutionary Genomics Group. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 25 March 2021).
- Bolland, H. eIF4A2 is a Regulator of Hypoxic Translation and Colorectal Tumour Cell Survival. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2020. [Google Scholar]
- Li, K.; Guo, W.; Li, Z.; Wang, Y.; Sun, B.; Xu, D.; Ling, J.; Song, H.; Liao, Y.; Wang, T.; et al. ALDH2 Repression Promotes Lung Tumor Progression via Accumulated Acetaldehyde and DNA Damage. Neoplasia 2019, 21, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Agell, L.; Hernández, S.; Nonell, L.; Lorenzo, M.; Puigdecanet, E.; de Muga, S.; Juanpere, N.; Bermudo, R.; Fernández, P.L.; Lorente, J.A.; et al. 12-gene expression signature is associated with aggressive histological in prostate cancer: SEC14L1 and TCEB1 genes are potential markers of progression. Am. J. Pathol. 2012, 181, 1585–1594. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Yuan, B.; Cao, J.; Zhao, J.; Ding, L.; Chen, L.; Ying, S.; Jiang, L.; Lin, J.; Xu, X.; et al. Methyltransferase-like 17 physically and functionally interacts with estrogen receptors. IUBMB Life 2015, 67, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Jeon, Y.H.; Cho, H.Y.; Lee, S.W.; Kim, G.W.; Lee, D.H.; Kwon, S.H. Advances in Histone Demethylase KDM3A as a Cancer Therapeutic Target. Cancers 2020, 12, 1098. [Google Scholar] [CrossRef]
- Rebecca, V.W.; Nicastri, M.C.; Fennelly, C.; Chude, C.I.; Barber-Rotenberg, J.S.; Ronghe, A.; McAfee, Q.; McLaughlin, N.P.; Zhang, G.; Goldman, A.R.; et al. PPT1 Promotes Tumor Growth and Is the Molecular Target of Chloroquine Derivatives in Cancer. Cancer Discov. 2019, 9, 220–229. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Kim, D.; Kim, H.L.; Choi, M.; Koh, Y.; Yoon, S.S. Oncogenic effects of germline variants in lysosomal storage disease genes. Genet. Med. 2019, 21, 2695–2705. [Google Scholar] [CrossRef]
- Jan, Y.H.; Lai, T.C.; Yang, C.J.; Lin, Y.F.; Huang, M.S.; Hsiao, M. Adenylate kinase 4 modulates oxidative stress and stabilizes HIF-1α to drive lung adenocarcinoma metastasis. J. Hematol. Oncol. 2019, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- López-Nieva, P.; Fernández-Navarro, P.; Graña-Castro, O.; Andrés-León, E.; Santos, J.; Villa Morales, M.; Cobos-Fernández, M.Á.; González-Sánchez, L.; Malumbres, M.; Salazar Roa, M.; et al. Detection of novel fusion-transcripts by RNA-Seq in T-cell lymphoblastic lymphoma. Sci. Rep. 2019, 9, 5179. [Google Scholar] [CrossRef]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nature reviews. Mol. Cell. Biol. 2020, 21, 268–283. [Google Scholar]
- Lai, M.C.; Chang, C.M.; Sun, H.S. Hypoxia Induces Autophagy through Translational Up-Regulation of Lysosomal Proteins in Human Colon Cancer Cells. PLoS ONE 2016, 11, e0153627. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, S.; Kitadate, A.; Abe, F.; Takahashi, N.; Tagawa, H. Hypoxia-inducible KDM3A addiction in multiple myeloma. Blood Adv. 2018, 2, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Spriggs, K.A.; Bushell, M.; Willis, A.E. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010, 40, 228–237. [Google Scholar] [CrossRef]
- Chee, N.T.; Lohse, I.; Brothers, S.P. mRNA-to-protein translation in hypoxia. Mol. Cancer 2019, 18, 49. [Google Scholar] [CrossRef]
- Hershey, J.W. The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta 2015, 1849, 792–800. [Google Scholar] [CrossRef]
- Ali, M.U.; Ur Rahman, M.S.; Jia, Z.; Jiang, C. Eukaryotic translation initiation factors and cancer. Tumor Biol. 2017, 39, 1010428317709805. [Google Scholar] [CrossRef] [Green Version]
- Horvilleur, E.; Sbarrato, T.; Hill, K.; Spriggs, R.V.; Screen, M.; Goodrem, P.J.; Sawicka, K.; Chaplin, L.C.; Touriol, C.; Packham, G.; et al. A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia 2014, 28, 1092–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Maiti, P.; Barrientos, A. Mitochondrial ribosomes in cancer. Semin. Cancer Biol. 2017, 47, 67–81. [Google Scholar] [CrossRef]
- Obre, E.; Rossignol, R. Emerging concepts in bioenergetics and cancer research: Metabolic flexibility, coupling, symbiosis, switch, oxidative tumors, metabolic remodeling, signaling and bioenergetic therapy. Int. J. Biochem. Cell Biol. 2015, 59, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Huang, G.; Li, H.; Zhang, H. Abnormal Expression of Mitochondrial Ribosomal Proteins and Their Encoding Genes with Cell Apoptosis and Diseases. Int. J. Mol. Sci. 2020, 21, 8879. [Google Scholar] [CrossRef]
- Aasebø, E.; Berven, F.S.; Hovland, R.; Døskeland, S.O.; Bruserud, Ø.; Selheim, F.; Hernandez-Valladares, M. The Progression of Acute Myeloid Leukemia from First Diagnosis to Chemoresistant Relapse: A Comparison of Proteomic and Phosphoproteomic Profiles. Cancers 2020, 12, 1466. [Google Scholar] [CrossRef]
- Wu, D.; Zhao, J.; Ma, H.; Wang, M.C. Integrating transcriptome-wide association study and copy number variation study identifies candidate genes and pathways for diffuse non-Hodgkin’s lymphoma. Cancer Genet. 2020, 243, 7–10. [Google Scholar] [CrossRef]
- Pelletier, J.; Thomas, G.; Volarević, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef]
- Hagner, P.R.; Mazan-Mamczarz, K.; Dai, B.; Balzer, E.M.; Corl, S.; Martin, S.S.; Zhao, X.F.; Gartenhaus, R.B. Ribosomal protein S6 is highly expressed in non-Hodgkin lymphoma and associates with mRNA containing a 5’ terminal oligopyrimidine tract. Oncogene 2011, 30, 1531–1541. [Google Scholar] [CrossRef] [Green Version]
- Derenzini, M.; Montanaro, L.; Trerè, D. Ribosome biogenesis and cancer. Acta Histochem. 2017, 119, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Sulima, S.O.; Hofman, I.J.F.; De Keersmaecker, K.; Dinman, J.D. How ribosomes translate cancer. Cancer Discov. 2017, 7, 1069–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derenzini, E.; Agostinelli, C.; Rossi, A.; Rossi, M.; Scellato, F.; Melle, F.; Pileri, S. Genomic alterations of ribosomal protein genes in diffuse large B cell lymphoma. Br. J. Haematol. 2018, 185, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, E.; Rossi, A.; Treré, D. Treating hematological malignancies with drugs inhibiting ribosome biogenesis: When and why. J. Hematol. Oncol. 2018, 11, 75. [Google Scholar] [CrossRef]
- Bousquet, P.A.; Sandvik, J.A.; Arntzen, M.Ø.; JeppesenEdin, N.F.; Christoffersen, S.; Krengel, U.; Pettersen, E.O.; Thiede, B. Hypoxia Strongly Affects Mitochondrial Ribosomal Proteins and Translocases, as Shown by Quantitative Proteomics of HeLa Cells. Int. J. Proteomics. 2015, 2015, 678527. [Google Scholar] [CrossRef] [Green Version]
- Koshiji, M.; Kageyama, Y.; Pete, E.A.; Horikawa, I.; Barrett, J.C.; Huang, L.E. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004, 23, 1949–1956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Druker, J.; Wilson, J.W.; Child, F.; Shakir, D.; Fasanya, T.; Rocha, S. Role of Hypoxia in the Control of the Cell Cycle. Int. J. Mol. Sci. 2021, 22, 4874. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.; Gong, Y.; Kim, H.; Chen, Y. Proteome dynamics analysis identifies functional roles of SDE2 and hypoxia in DNA damage response in prostate cancer cells. NAR Cancer 2020, 2, zcaa010. [Google Scholar] [CrossRef] [PubMed]
- Duś-Szachniewicz, K.; Drobczyński, S.; Ziółkowski, P.; Kołodziej, P.; Walaszek, K.M.; Korzeniewska, A.K.; Agrawal, A.; Kupczyk, P.; Woźniak, M. Physiological Hypoxia (Physioxia) Impairs the Early Adhesion of Single Lymphoma Cell to Marrow Stromal Cell and Extracellular Matrix. Optical Tweezers Study. Int. J. Mol. Sci. 2018, 19, 1880. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Goerne, T.; Zelmer, A.; Guzman, R.; Kapfhammer, J.P.; Wellmann, S.; Zhu, X. The RNA Binding Protein RBM3 Promotes Neural Stem Cell (NSC) Proliferation Under Hypoxia. Front. Cell Dev. Biol. 2019, 7, 288. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Tang, W.G.; Fan, J.; Xu, Y.; Sun, H.X. Differentially expressed miRNAs in hepatocellular carcinoma cells under hypoxic conditions are associated with transcription and phosphorylation. Oncol. Lett. 2018, 15, 467–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semenza, G.L. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci. 2012, 33, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Stewart, M.; Talks, K.; Leek, R.; Turley, H.; Pezzella, F.; Harris, A.; Gatter, K. Expression of angiogenic factors and hypoxia inducible factors HIF 1, HIF 2 and CA IX in non-Hodgkin’s lymphoma. Histopathology 2002, 40, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Miar, A.; Arnaiz, E.; Bridges, E.; Beedie, S.; Cribbs, A.P.; Downes, D.J.; Beagrie, R.; Rehwinkel, J.; Harris, A.L. Hypoxia induces transcriptional and translational downregulation of the type I interferon (IFN) pathway in multiple cancer cell types. Cancer Res. 2020, 80, 5245–5256. [Google Scholar] [CrossRef]
- Minton, D.R.; Fu, L.; Mongan, N.P.; Shevchuk, M.M.; Nanus, D.M.; Gudas, L.J. Role of NADH Dehydrogenase (Ubiquinone) 1 Alpha Subcomplex 4-Like 2 in Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. 2016, 22, 2791–2801. [Google Scholar] [CrossRef] [Green Version]
- Mucaj, V.; Shay, J.E.; Simon, M.C. Effects of hypoxia and HIFs on cancer metabolism. Int. J. Hematol. 2012, 95, 464–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belisario, D.C.; Kopecka, J.; Pasino, M.; Akman, M.; De Smaele, E.; Donadelli, M.; Riganti, C. Hypoxia Dictates Metabolic Rewiring of Tumors: Implications for Chemoresistance. Cells 2020, 9, 2598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, Y.; Shi, X.; Zhou, M.; Bao, L.; Hatanpaa, K.J.; Patel, T.; DeBerardinis, R.J.; Wang, Y.; Luo, W. Regulation of branched-chain amino acid metabolism by hypoxia-inducible factor in glioblastoma. Cell. Mol. Life Sci. 2021, 78, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Filippopoulou, C.; Simos, G.; Chachami, G. The Role of Sumoylation in the Response to Hypoxia: An Overview. Cells 2020, 9, 2359. [Google Scholar] [CrossRef]
- Wang, P.; Song, M.; Zeng, Z.L.; Zhu, C.F.; Lu, W.H.; Yang, J.; Ma, M.Z.; Huang, A.M.; Hu, Y.; Huang, P. Identification of NDUFAF1 in mediating K-Ras induced mitochondrial dysfunction by a proteomic screening approach. Oncotarget 2015, 6, 3947–3962. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Chang, Y.M.; Polton, G.; Stell, A.J.; Szladovits, B.; Macfarlane, M.; Peters, L.M.; Priestnall, S.L.; Bacon, N.J.; Kow, K. Gene Expression Profiling of B Cell Lymphoma in Dogs Reveals Dichotomous Metabolic Signatures Distinguished by Oxidative Phosphorylation. Front. Oncol. 2020, 10, 307. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Liu, H.; Qing, G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target Ther. 2018, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.J.; Qiu, B.; Nakazawa, M.S.; Qing, G.; Simon, M.C. MYC degradation under low O2 tension promotes survival by evading hypoxia-induced cell death. Mol. Cell Biol. 2013, 33, 3494–3504. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Xue, M.; Chung, D.C. c-Myc is regulated by HIF-2α in chronic hypoxia and influences sensitivity to 5-FU in colon cancer. Oncotarget 2016, 7, 78910–78917. [Google Scholar] [CrossRef] [PubMed]



| Cell Line | Origin | Subtype | Features |
|---|---|---|---|
| Ri-1 | transformed SLL (Richter Syndrome) | ABC | MYC R |
| U2904 | transformed FL (Kiel centrocytic/centroblastic) | GCB | MYC R, BCL2 R |
| U2932 | de novo DLBCL, NOS | ABC | BCL2 AMP |
| Target | Function | Ri-1 (FC) | U2904 (FC) | U2932 (FC) |
|---|---|---|---|---|
| ALDH6A1 | metabolic reprogramming | 3.750 | 2.001 | 4.112 |
| ALDH7A1 | 3.689 | 1.847 | ||
| PFKL | 3.115 | ------ | 4.105 | |
| PDK1 | 3.304 | ------ | 2.096 | |
| BCAT2 | 1.529 | ------ | ------ | |
| PFKFB4 | 4.396 | ------ | ------ | |
| HK2 | ------ | 2.015 | ------ | |
| BNIP3L | ------ | 6.746 | ------ | |
| LDHA | ------ | ------ | −9.452 | |
| PFKFB3 | ------ | ------ | 6.548 | |
| PGK1 | ------ | ------ | −5.407 | |
| ALDOC | ------ | ------ | −3.286 | |
| PDCD4 | metastasis and invasion | 1.879 | ------ | 2.370 |
| P4HA1 | ------ | 5.630 | 6.070 | |
| CXCR4 | ------ | 4.071 | ------ | |
| NPM1 | ------ | 3.611 | ------ | |
| LGALS1 | ------ | ------ | 2.468 | |
| GPI | angiogenesis | 1.590 | 4.169 | |
| HMOX1 | −3.541 | −4.303 | ||
| ZEB1 | epithelial- mesenchymal transition | 2.210 | ------ | ------ |
| VIM | EMT | 3.140 | ------ | ------ |
| EGLN1 | regulation of the hypoxic response | 2.253 | 3.048 | ------ |
| IRAK4 | apoptosis | 3.605 | ------ | ------ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duś-Szachniewicz, K.; Gdesz-Birula, K.; Zduniak, K.; Wiśniewski, J.R. Proteomic-Based Analysis of Hypoxia- and Physioxia-Responsive Proteins and Pathways in Diffuse Large B-Cell Lymphoma. Cells 2021, 10, 2025. https://doi.org/10.3390/cells10082025
Duś-Szachniewicz K, Gdesz-Birula K, Zduniak K, Wiśniewski JR. Proteomic-Based Analysis of Hypoxia- and Physioxia-Responsive Proteins and Pathways in Diffuse Large B-Cell Lymphoma. Cells. 2021; 10(8):2025. https://doi.org/10.3390/cells10082025
Chicago/Turabian StyleDuś-Szachniewicz, Kamila, Katarzyna Gdesz-Birula, Krzysztof Zduniak, and Jacek R. Wiśniewski. 2021. "Proteomic-Based Analysis of Hypoxia- and Physioxia-Responsive Proteins and Pathways in Diffuse Large B-Cell Lymphoma" Cells 10, no. 8: 2025. https://doi.org/10.3390/cells10082025
APA StyleDuś-Szachniewicz, K., Gdesz-Birula, K., Zduniak, K., & Wiśniewski, J. R. (2021). Proteomic-Based Analysis of Hypoxia- and Physioxia-Responsive Proteins and Pathways in Diffuse Large B-Cell Lymphoma. Cells, 10(8), 2025. https://doi.org/10.3390/cells10082025





