The Arabidopsis Root Tip (Phospho)Proteomes at Growth-Promoting versus Growth-Repressing Conditions Reveal Novel Root Growth Regulators
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Primary Root Length Analyses
2.3. Scanner Growth Assay
2.4. Data Analysis
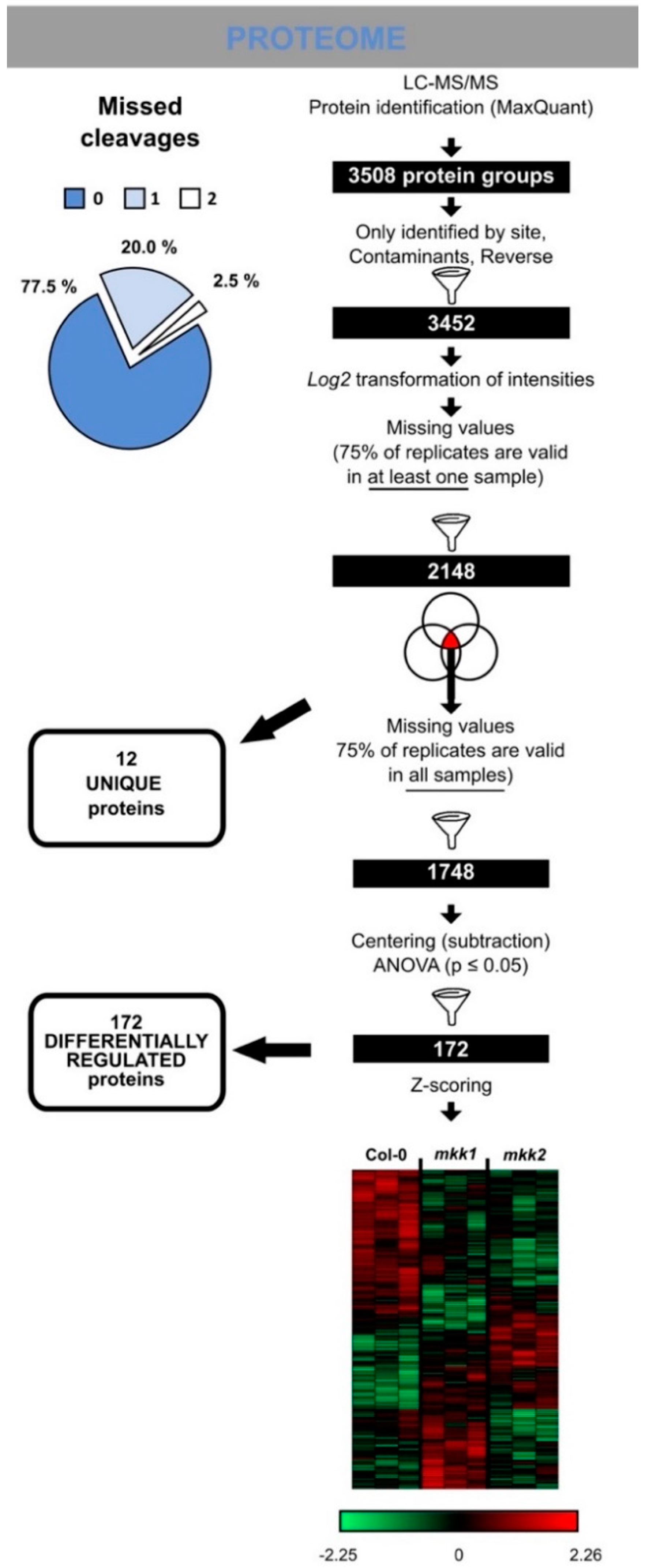
2.5. (Phospho)Proteome Analysis
2.6. Sequence Alignment
2.7. Accession Numbers
3. Results and Discussion
3.1. Establishment of Optimum NAA Levels for Repression and Promotion of Primary Root Growth
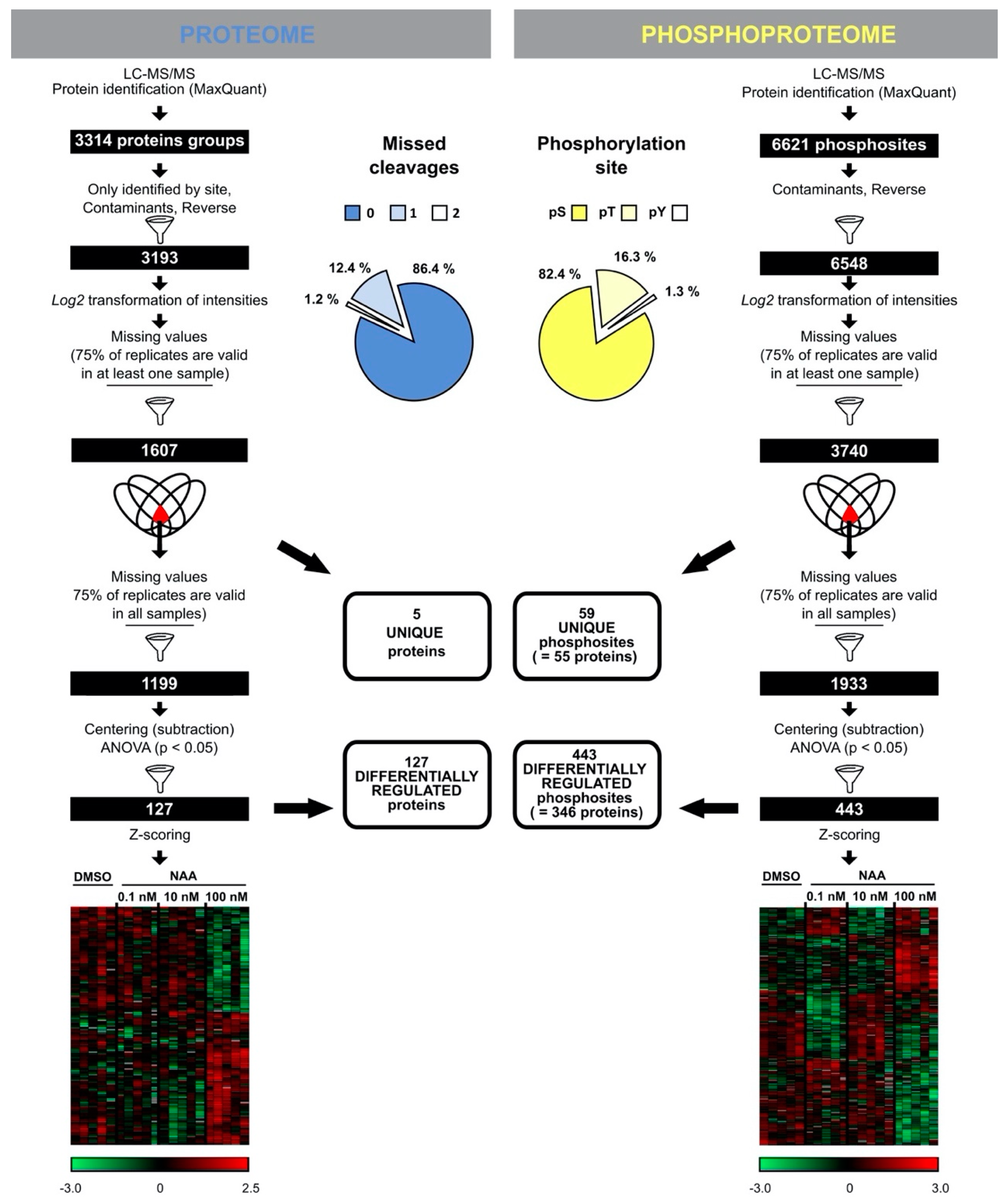
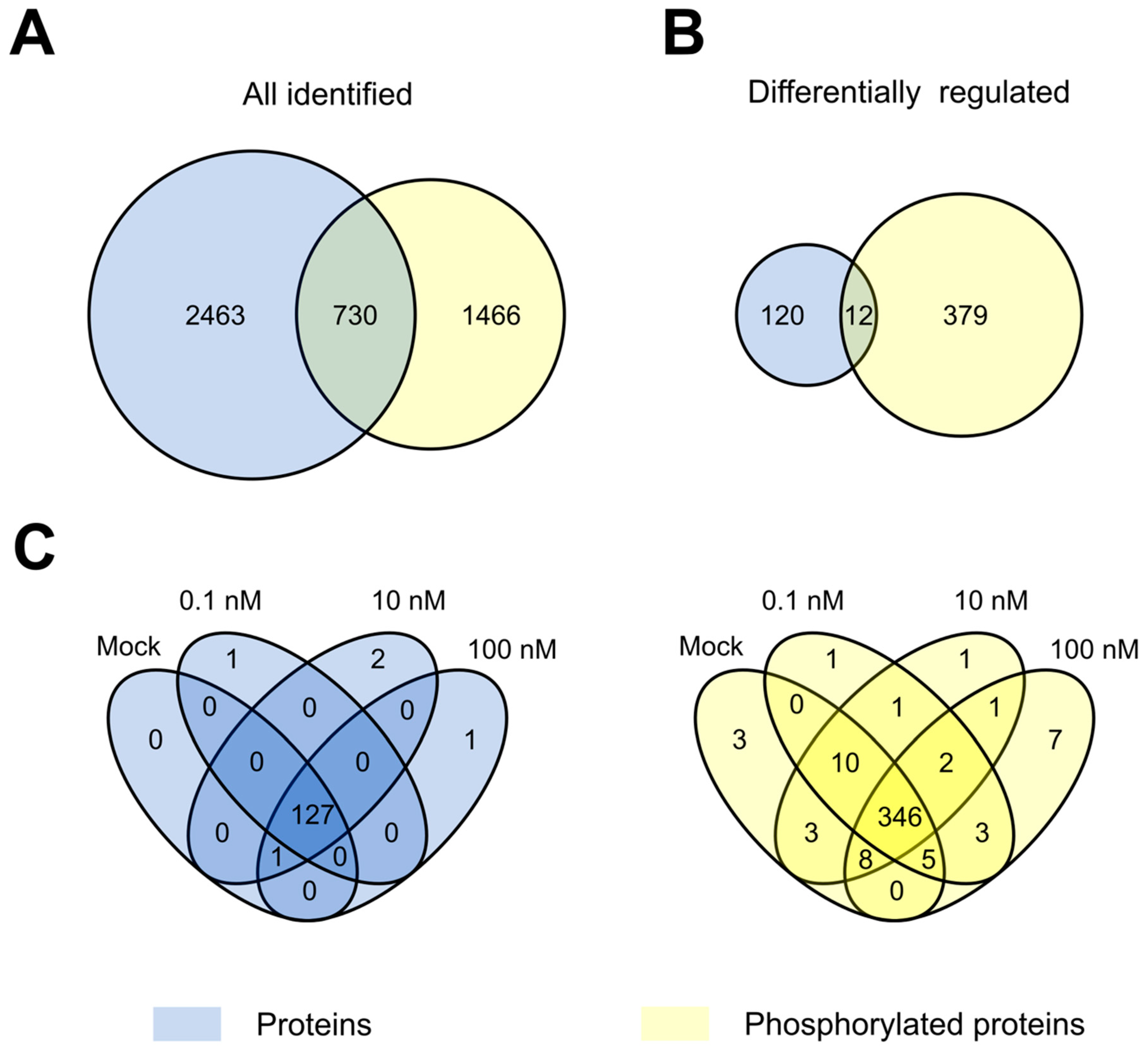
3.2. Proteome and Phosphoproteome Profiling to Unravel Concentration-Dependent Root Growth NAA Response
3.3. The Arabidopsis Root Tip Proteome at Growth-Promoting and -Repressing Conditions
3.4. The Arabidopsis Root Tip Phosphoproteome at Growth-Promoting and -Repressing Conditions
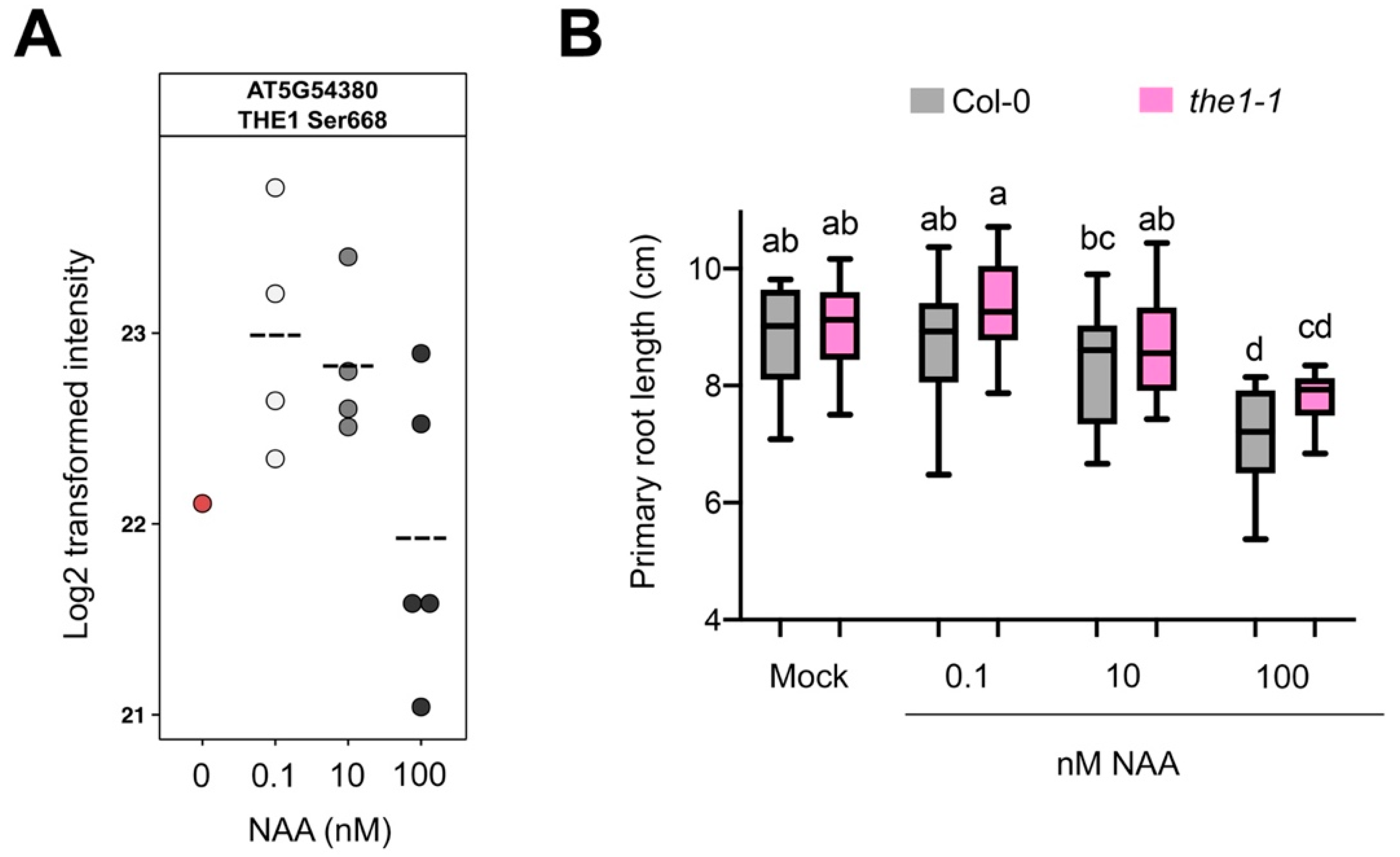
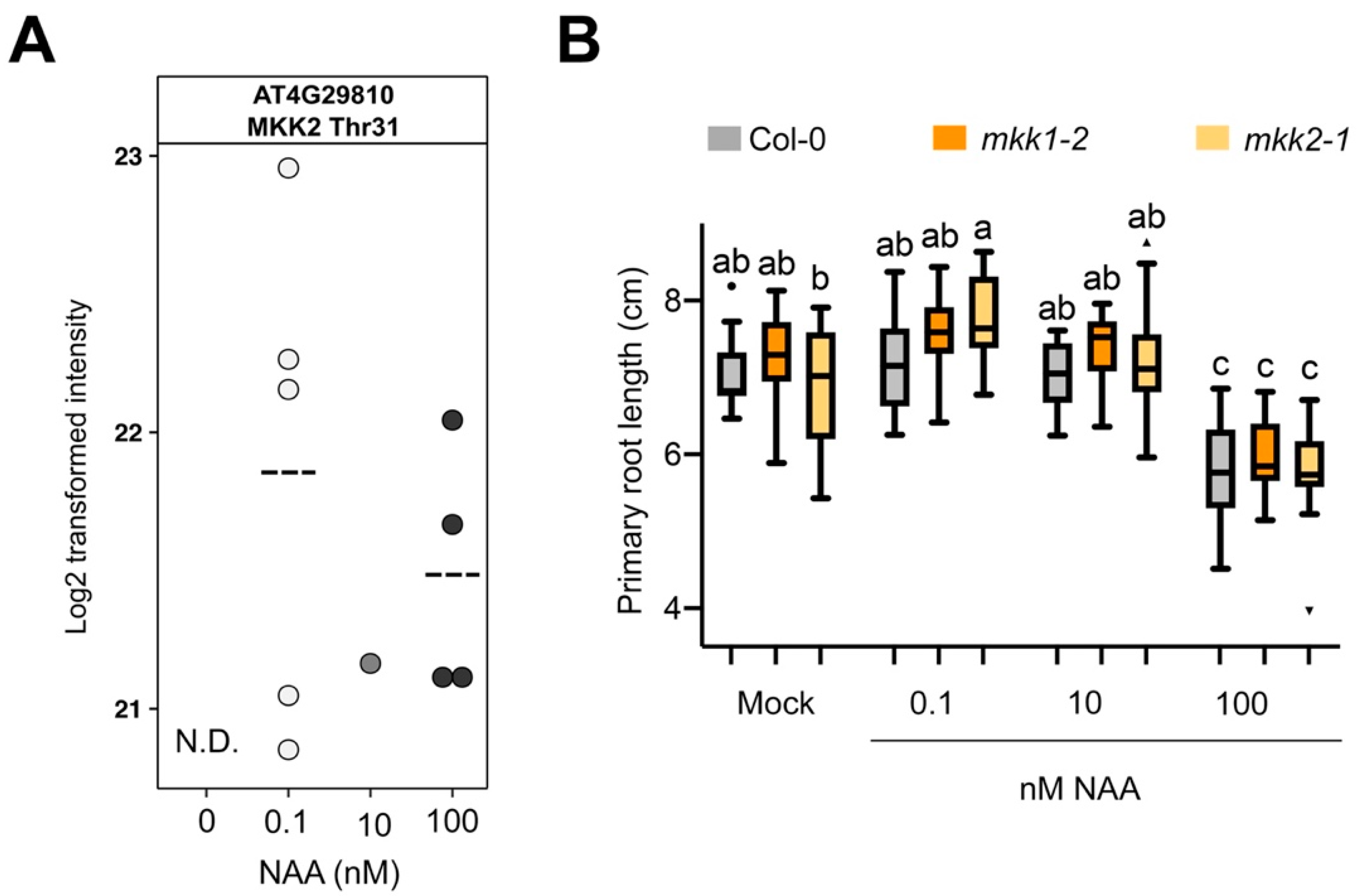
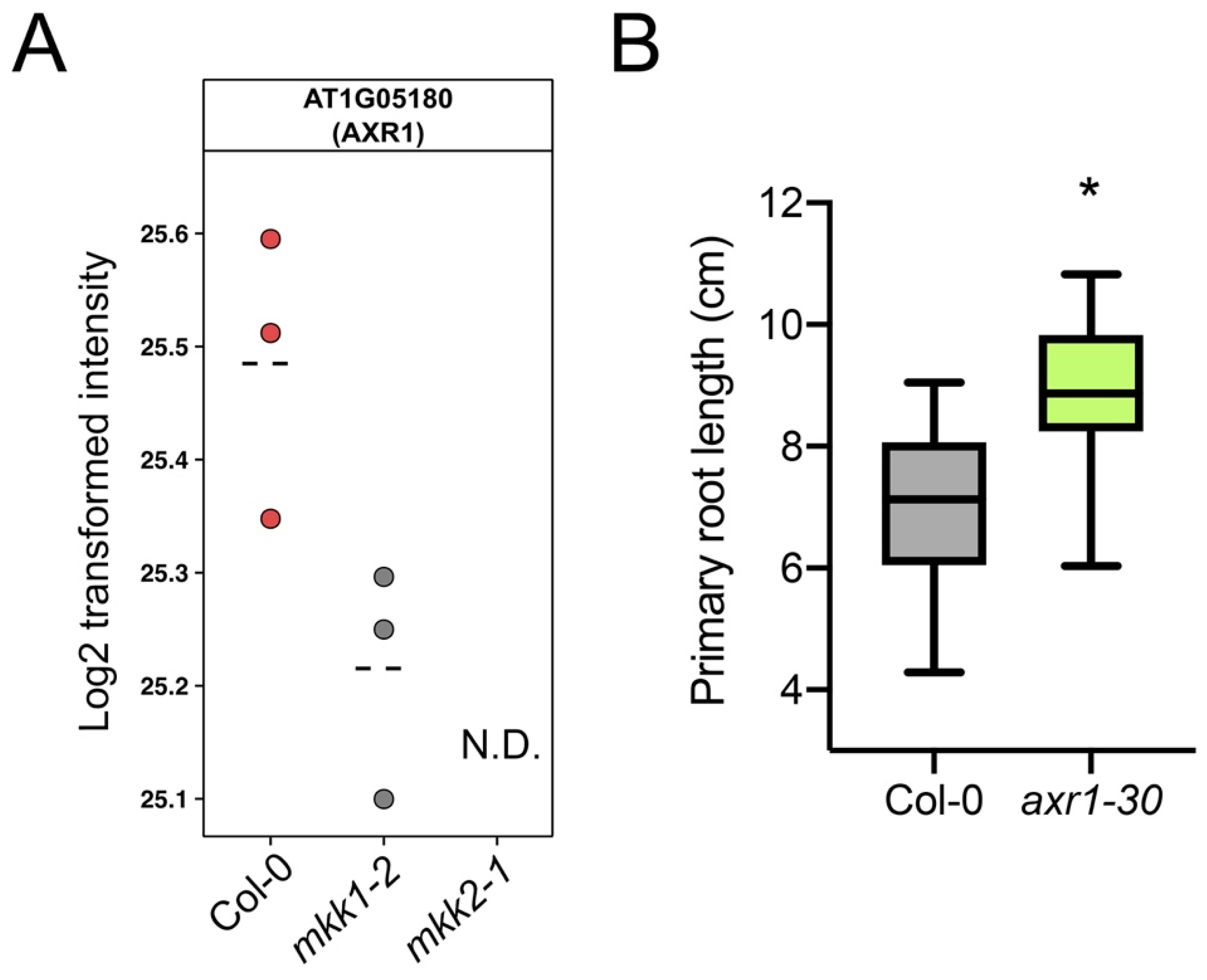
3.5. Validation of Selected Candidates
3.6. The THE1 Ligand RALF34 Controls Auxin-Dependent Primary Root Growth
3.7. Proteome Profiling Identifies Molecular Changes Downstream of MKK1 and MKK2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis Root Development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satbhai, S.B.; Ristova, D.; Busch, W. Underground Tuning: Quantitative Regulation of Root Growth. J. Exp. Bot. 2015, 66, 1099–1112. [Google Scholar] [CrossRef] [Green Version]
- Benková, E.; Hejátko, J. Hormone Interactions at the Root Apical Meristem. Plant Mol. Biol. 2009, 69, 383–396. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, C.; Weisbeek, P.; Scheres, B. Cell Fate and Cell Differentiation Status in the Arabidopsis Root. Planta 1998, 205, 483–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolan, L.; Janmaat, K.; Willemsen, V.; Linstead, P.; Poethig, S.; Roberts, K.; Scheres, B. Cellular Organisation of the Arabidopsis Thaliana Root. Development (Camb. Engl.) 1993, 119, 71–84. [Google Scholar] [CrossRef]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A Signaling Module Controlling the Stem Cell Niche in Arabidopsis Root Meristems. Curr. Biol. CB 2009, 19, 909–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, X.; Tian, H.; Yu, Q.; Zhang, F.; Wang, R.; Gao, S.; Xu, W.; Liu, J.; Shani, E.; Fu, C.; et al. PHB3 Maintains Root Stem Cell Niche Identity through ROS-Responsive AP2/ERF Transcription Factors in Arabidopsis. Cell Rep. 2018, 22, 1350–1363. [Google Scholar] [CrossRef] [Green Version]
- Heidstra, R.; Sabatini, S. Plant and Animal Stem Cells: Similar yet Different. Nat. Rev. Mol. Cell Biol. 2014, 15, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Verbelen, J.-P.; De Cnodder, T.; Le, J.; Vissenberg, K.; Baluska, F. The Root Apex of Arabidopsis Thaliana Consists of Four Distinct Zones of Growth Activities: Meristematic Zone, Transition Zone, Fast Elongation Zone and Growth Terminating Zone. Plant Signal. Behav. 2006, 1, 296–304. [Google Scholar] [CrossRef] [Green Version]
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant Hormone Cross-Talk: The Pivot of Root Growth. J. Exp. Bot. 2015, 66, 1113–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sozzani, R.; Iyer-Pascuzzi, A. Postembryonic Control of Root Meristem Growth and Development. Curr. Opin. Plant Biol. 2014, 17, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Di Mambro, R.; De Ruvo, M.; Pacifici, E.; Salvi, E.; Sozzani, R.; Benfey, P.N.; Busch, W.; Novak, O.; Ljung, K.; Di Paola, L.; et al. Auxin Minimum Triggers the Developmental Switch from Cell Division to Cell Differentiation in the Arabidopsis Root. Proc. Natl. Acad. Sci. USA 2017, 114, E7641–E7649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, A.; Bannigan, A.; Sulaman, W.; Pechter, P.; Blancaflor, E.B.; Baskin, T.I. Auxin, Actin and Growth of the Arabidopsis Thaliana Primary Root. Plant J. Cell Mol. Biol. 2007, 50, 514–528. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ruan, Y.-L. Regulation of Cell Division and Expansion by Sugar and Auxin Signaling. Front. Plant Sci. 2013, 4, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrot-Rechenmann, C. Cellular Responses to Auxin: Division versus Expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef]
- Rayle, D.L.; Evans, M.L.; Hertel, R. Action of Auxin on Cell Elongation. Proc. Natl. Acad. Sci. USA 1970, 65, 184–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulkey, T.J.; Kuzmanoff, K.M.; Evans, M.L. Promotion of Growth and Shift in the Auxin Dose/Response Relationship in Maize Roots Treated with the Ethylene Biosynthesis Inhibitors Aminoethoxyvinylglycine and Cobalt. Plant Sci. Lett. 1982, 25, 43–48. [Google Scholar] [CrossRef]
- Barbez, E.; Dünser, K.; Gaidora, A.; Lendl, T.; Busch, W. Auxin Steers Root Cell Expansion via Apoplastic PH Regulation in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, E4884–E4893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, M.L.; Ishikawa, H.; Estelle, M.A. Responses of Arabidopsis Roots to Auxin Studied with High Temporal Resolution: Comparison of Wild Type and Auxin-Response Mutants. Planta 1994, 194, 215–222. [Google Scholar] [CrossRef]
- Pacheco-Villalobos, D.; Sankar, M.; Ljung, K.; Hardtke, C.S. Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-Ethylene Crosstalk in Brachypodium Distachyon Seminal Roots. PLoS Genet. 2013, 9, e1003564. [Google Scholar] [CrossRef] [Green Version]
- Pacheco-Villalobos, D.; Díaz-Moreno, S.M.; van der Schuren, A.; Tamaki, T.; Kang, Y.H.; Gujas, B.; Novak, O.; Jaspert, N.; Li, Z.; Wolf, S.; et al. The Effects of High Steady State Auxin Levels on Root Cell Elongation in Brachypodium. Plant Cell 2016, 28, 1009–1024. [Google Scholar] [CrossRef]
- Takahashi, K.; Hayashi, K.; Kinoshita, T. Auxin Activates the Plasma Membrane H+-ATPase by Phosphorylation during Hypocotyl Elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hager, A. Role of the Plasma Membrane H+-ATPase in Auxin-Induced Elongation Growth: Historical and New Aspects. J. Plant Res. 2003, 116, 483–505. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid Auxin-Mediated Cell Expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Grandont, L.; Li, H.; Hauschild, R.; Paque, S.; Abuzeineh, A.; Rakusová, H.; Benkova, E.; Perrot-Rechenmann, C.; Friml, J. Inhibition of Cell Expansion by Rapid ABP1-Mediated Auxin Effect on Microtubules. Nature 2014, 516, 90–93. [Google Scholar] [CrossRef]
- Scheuring, D.; Löfke, C.; Krüger, F.; Kittelmann, M.; Eisa, A.; Hughes, L.; Smith, R.S.; Hawes, C.; Schumacher, K.; Kleine-Vehn, J. Actin-Dependent Vacuolar Occupancy of the Cell Determines Auxin-Induced Growth Repression. Proc. Natl. Acad. Sci. USA 2016, 113, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Löfke, C.; Dünser, K.; Scheuring, D.; Kleine-Vehn, J. Auxin Regulates SNARE-Dependent Vacuolar Morphology Restricting Cell Size. eLife 2015, 4. [Google Scholar] [CrossRef] [Green Version]
- Abel, S.; Nguyen, M.D.; Chow, W.; Theologis, A. ACS4, a Primary Indoleacetic Acid-Responsive Gene Encoding 1-Aminocyclopropane-1-Carboxylate Synthase in Arabidopsis Thaliana. Structural Characterization, Expression in Escherichia Coli, and Expression Characteristics in Response to Auxin [Corrected]. J. Biol. Chem. 1995, 270, 19093–19099. [Google Scholar] [CrossRef] [Green Version]
- Woeste, K.E.; Ye, C.; Kieber, J.J. Two Arabidopsis Mutants That Overproduce Ethylene Are Affected in the Posttranscriptional Regulation of 1-Aminocyclopropane-1-Carboxylic Acid Synthase. Plant Physiol. 1999, 119, 521–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staal, M.; De Cnodder, T.; Simon, D.; Vandenbussche, F.; Van Der Straeten, D.; Verbelen, J.-P.; Elzenga, T.; Vissenberg, K. Apoplastic Alkalinization Is Instrumental for the Inhibition of Cell Elongation in the Arabidopsis Root by the Ethylene Precursor 1-Aminocyclopropane-1-Carboxylic Acid. Plant Physiol. 2011, 155, 2049–2055. [Google Scholar] [CrossRef] [Green Version]
- Markakis, M.N.; De Cnodder, T.; Lewandowski, M.; Simon, D.; Boron, A.; Balcerowicz, D.; Doubbo, T.; Taconnat, L.; Renou, J.-P.; Höfte, H.; et al. Identification of Genes Involved in the ACC-Mediated Control of Root Cell Elongation in Arabidopsis Thaliana. BMC Plant Biol. 2012, 12, 208. [Google Scholar] [CrossRef] [Green Version]
- Slade, W.O.; Ray, W.K.; Hildreth, S.B.; Winkel, B.S.J.; Helm, R.F. Exogenous Auxin Elicits Changes in the Arabidopsis Thaliana Root Proteome in a Time-Dependent Manner. Proteomes 2017, 5, 16. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of Auxin-Regulated Gene Expression in Plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of Auxin Perception by the TIR1 Ubiquitin Ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef]
- Fendrych, M.; Akhmanova, M.; Merrin, J.; Glanc, M.; Hagihara, S.; Takahashi, K.; Uchida, N.; Torii, K.U.; Friml, J. Rapid and Reversible Root Growth Inhibition by TIR1 Auxin Signalling. Nat. Plants 2018, 4, 453–459. [Google Scholar] [CrossRef]
- Mattei, B.; Sabatini, S.; Schininà, M.E. Proteomics in Deciphering the Auxin Commitment in the Arabidopsis Thaliana Root Growth. J. Proteome Res. 2013, 12, 4685–4701. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, H.; Berke, L.; Heck, A.J.R.; Mohammed, S.; Scheres, B.; Menke, F.L.H. Quantitative Phosphoproteomics after Auxin-Stimulated Lateral Root Induction Identifies an SNX1 Protein Phosphorylation Site Required for Growth. Mol. Cell. Proteom. MCP 2013, 12, 1158–1169. [Google Scholar] [CrossRef] [Green Version]
- Cao, M.; Chen, R.; Li, P.; Yu, Y.; Zheng, R.; Ge, D.; Zheng, W.; Wang, X.; Gu, Y.; Gelová, Z.; et al. TMK1-Mediated Auxin Signalling Regulates Differential Growth of the Apical Hook. Nature 2019, 568, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Yu, Q.; Liu, J.; Wen, X.; Yan, Z.; Hu, K.; Li, H.; Kong, X.; Li, C.; Tian, H.; et al. Non-Canonical AUX/IAA Protein IAA33 Competes with Canonical AUX/IAA Repressor IAA5 to Negatively Regulate Auxin Signaling. EMBO J. 2020, 39, e101515. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zheng, R.; He, J.; Zhou, Z.; Wang, J.; Xiong, Y.; Xu, T. Noncanonical Auxin Signaling Regulates Cell Division Pattern during Lateral Root Development. Proc. Natl. Acad. Sci. USA 2019, 116, 21285–21290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, Y.; Walley, J.W.; Shen, Z.; Lang, M.G.; Briggs, S.P.; Estelle, M.; Kelley, D.R. Quantitative Early Auxin Root Proteomics Identifies GAUT10, a Galacturonosyltransferase, as a Novel Regulator of Root Meristem Maintenance. Mol. Cell. Proteom. 2019, 18. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Liu, J.; Bi, D.; Zhang, Z.; Cheng, F.; Chen, S.; Zhang, Y. MEKK1, MKK1/MKK2 and MPK4 Function Together in a Mitogen-Activated Protein Kinase Cascade to Regulate Innate Immunity in Plants. Cell Res. 2008, 18, 1190–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hématy, K.; Sado, P.-E.; Van Tuinen, A.; Rochange, S.; Desnos, T.; Balzergue, S.; Pelletier, S.; Renou, J.-P.; Höfte, H. A Receptor-like Kinase Mediates the Response of Arabidopsis Cells to the Inhibition of Cellulose Synthesis. Curr. Biol. CB 2007, 17, 922–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, E.; Vu, L.D.; Van den Broeck, L.; Lin, Z.; Ramakrishna, P.; van de Cotte, B.; Gaudinier, A.; Goh, T.; Slane, D.; Beeckman, T.; et al. RALFL34 Regulates Formative Cell Divisions in Arabidopsis Pericycle during Lateral Root Initiation. J. Exp. Bot. 2016, 67, 4863–4875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoenaers, S.; Balcerowicz, D.; Breen, G.; Hill, K.; Zdanio, M.; Mouille, G.; Holman, T.J.; Oh, J.; Wilson, M.H.; Nikonorova, N.; et al. The Auxin-Regulated CrRLK1L Kinase ERULUS Controls Cell Wall Composition during Root Hair Tip Growth. Curr. Biol. CB 2018, 28, 722–732.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wymer, C.L.; Bibikova, T.N.; Gilroy, S. Cytoplasmic Free Calcium Distributions during the Development of Root Hairs of Arabidopsis Thaliana. Plant J. Cell Mol. Biol. 1997, 12, 427–439. [Google Scholar] [CrossRef]
- Van Aken, O.; Pecenková, T.; van de Cotte, B.; De Rycke, R.; Eeckhout, D.; Fromm, H.; De Jaeger, G.; Witters, E.; Beemster, G.T.S.; Inzé, D.; et al. Mitochondrial Type-I Prohibitins of Arabidopsis Thaliana Are Required for Supporting Proficient Meristem Development. Plant J. Cell Mol. Biol. 2007, 52, 850–864. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Krens, S.F.G.; Fendrych, M.; Friml, J. Real-Time Analysis of Auxin Response, Cell Wall PH and Elongation in Arabidopsis Thaliana Hypocotyls. Bio-Protocol 2018, 8, e2685. [Google Scholar] [CrossRef]
- Nikonorova, N.; Van den Broeck, L.; Zhu, S.; van de Cotte, B.; Dubois, M.; Gevaert, K.; Inzé, D.; De Smet, I. Early Mannitol-Triggered Changes in the Arabidopsis Leaf (Phospho)Proteome Reveal Growth Regulators. J. Exp. Bot. 2018, 69, 4591–4607. [Google Scholar] [CrossRef]
- Vu, L.D.; Stes, E.; Van Bel, M.; Nelissen, H.; Maddelein, D.; Inzé, D.; Coppens, F.; Martens, L.; Gevaert, K.; De Smet, I. Up-to-Date Workflow for Plant (Phospho)Proteomics Identifies Differential Drought-Responsive Phosphorylation Events in Maize Leaves. J. Proteome Res. 2016, 15, 4304–4317. [Google Scholar] [CrossRef] [Green Version]
- Feng, D.F.; Doolittle, R.F. Progressive Sequence Alignment as a Prerequisite to Correct Phylogenetic Trees. J. Mol. Evol. 1987, 25, 351–360. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 Update of the PRIDE Database and Its Related Tools. Nucleic Acids Res. 2016, 44, D447-56. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.R.; Chalkley, R.J. MS-Viewer: A Web-Based Spectral Viewer for Proteomics Results. Mol. Cell. Proteom. MCP 2014, 13, 1392–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Bel, M.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Van de Peer, Y.; Coppens, F.; Vandepoele, K. PLAZA 4.0: An Integrative Resource for Functional, Evolutionary and Comparative Plant Genomics. Nucleic Acids Res. 2018, 46, D1190–D1196. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H. Rab GTPases as Coordinators of Vesicle Traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Chantarachot, T.; Bailey-Serres, J. Polysomes, Stress Granules, and Processing Bodies: A Dynamic Triumvirate Controlling Cytoplasmic MRNA Fate and Function. Plant Physiol. 2018, 176, 254–269. [Google Scholar] [CrossRef] [Green Version]
- Shigeto, J.; Kiyonaga, Y.; Fujita, K.; Kondo, R.; Tsutsumi, Y. Putative Cationic Cell-Wall-Bound Peroxidase Homologues in Arabidopsis, AtPrx2, AtPrx25, and AtPrx71, Are Involved in Lignification. J. Agric. Food Chem. 2013, 61, 3781–3788. [Google Scholar] [CrossRef]
- Shigeto, J.; Nagano, M.; Fujita, K.; Tsutsumi, Y. Catalytic Profile of Arabidopsis Peroxidases, AtPrx-2, 25 and 71, Contributing to Stem Lignification. PLoS ONE 2014, 9, e105332. [Google Scholar] [CrossRef] [Green Version]
- Remy, E.; Cabrito, T.R.; Baster, P.; Batista, R.A.; Teixeira, M.C.; Friml, J.; Sá-Correia, I.; Duque, P. A Major Facilitator Superfamily Transporter Plays a Dual Role in Polar Auxin Transport and Drought Stress Tolerance in Arabidopsis. Plant Cell 2013, 25, 901–926. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Li, L.; Ye, H.; Yu, X.; Algreen, A.; Yin, Y. Three Related Receptor-like Kinases Are Required for Optimal Cell Elongation in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 7648–7653. [Google Scholar] [CrossRef] [Green Version]
- Schoenaers, S.; Balcerowicz, D.; Costa, A.; Vissenberg, K. The Kinase ERULUS Controls Pollen Tube Targeting and Growth in Arabidopsis Thaliana. Front. Plant Sci. 2017, 8, 1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-W.; Michniewicz, M.; Bergmann, D.C.; Wang, Z.-Y. Brassinosteroid Regulates Stomatal Development by GSK3-Mediated Inhibition of a MAPK Pathway. Nature 2012, 482, 419–422. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Jwa, N.-S. The Rice MAPKK-MAPK Interactome: The Biological Significance of MAPK Components in Hormone Signal Transduction. Plant Cell Rep. 2013, 32, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, S. MAPK Cascades in Plant Disease Resistance Signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Qu, N.; Gao, M.; Zhang, Z.; Ding, X.; Yang, F.; Li, Y.; Dong, O.X.; Chen, S.; Li, X.; et al. The MEKK1-MKK1/MKK2-MPK4 Kinase Cascade Negatively Regulates Immunity Mediated by a Mitogen-Activated Protein Kinase Kinase Kinase in Arabidopsis. Plant Cell 2012, 24, 2225–2236. [Google Scholar] [CrossRef] [Green Version]
- Teige, M.; Scheikl, E.; Eulgem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 Pathway Mediates Cold and Salt Stress Signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, S.; He, W.-D.; Shao, X.-H.; Li, C.-Y.; Wei, Y.-R.; Deng, G.-M.; Kuang, R.-B.; Hu, C.-H.; Yi, G.-J.; et al. Comparative Phosphoproteomics Reveals an Important Role of MKK2 in Banana (Musa Spp.) Cold Signal Network. Sci. Rep. 2017, 7, 40852. [Google Scholar] [CrossRef]
- Wang, F.; Jing, W.; Zhang, W. The Mitogen-Activated Protein Kinase Cascade MKK1-MPK4 Mediates Salt Signaling in Rice. Plant Sci. Int. J. Exp. Plant Biol. 2014, 227, 181–189. [Google Scholar] [CrossRef]
- Qiu, J.; Zhou, L.; Yun, B.-W.; Nielsen, H.B.; Fiil, B.K.; Petersen, K.; Mackinlay, J.; Loake, G.J.; Mundy, J.; Morris, P.C. Arabidopsis Mitogen-Activated Protein Kinase Kinases MKK1 and MKK2 Have Overlapping Functions in Defense Signaling Mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 2008, 148, 212–222. [Google Scholar] [CrossRef] [Green Version]
- Gonneau, M.; Desprez, T.; Martin, M.; Doblas, V.G.; Bacete, L.; Miart, F.; Sormani, R.; Hématy, K.; Renou, J.; Landrein, B.; et al. Receptor Kinase THESEUS1 Is a Rapid Alkalinization Factor 34 Receptor in Arabidopsis. Curr. Biol. CB 2018, 28, 2452–2458.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dressano, K.; Ceciliato, P.H.O.; Silva, A.L.; Guerrero-Abad, J.C.; Bergonci, T.; Ortiz-Morea, F.A.; Bürger, M.; Silva-Filho, M.C.; Moura, D.S. BAK1 Is Involved in AtRALF1-Induced Inhibition of Root Cell Expansion. PLoS Genet. 2017, 13, e1007053. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [Green Version]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma Membrane H(+)-ATPase Regulation in the Center of Plant Physiology. Mol. Plant 2016, 9, 323–337. [Google Scholar] [CrossRef] [Green Version]
- Stanko, V.; Giuliani, C.; Retzer, K.; Djamei, A.; Wahl, V.; Wurzinger, B.; Wilson, C.; Heberle-Bors, E.; Teige, M.; Kragler, F. Timing Is Everything: Highly Specific and Transient Expression of a MAP Kinase Determines Auxin-Induced Leaf Venation Patterns in Arabidopsis. Mol. Plant 2014, 7, 1637–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Karunarathna, N.; Jurgens, G.; Estelle, M. AXL and AXR1 Have Redundant Functions in RUB Conjugation and Growth and Development in Arabidopsis. Plant J. Cell Mol. Biol. 2007, 52, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, C.; Britton, J.H.; Estelle, M. Growth and Development of the Axr1 Mutants of Arabidopsis. Plant Cell 1990, 2, 1071–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitts, R.J.; Cernac, A.; Estelle, M. Auxin and Ethylene Promote Root Hair Elongation in Arabidopsis. Plant J. Cell Mol. Biol. 1998, 16, 553–560. [Google Scholar] [CrossRef]
- Fendrych, M.; Leung, J.; Friml, J. TIR1/AFB-Aux/IAA Auxin Perception Mediates Rapid Cell Wall Acidification and Growth of Arabidopsis Hypocotyls. eLife 2016, 5, 1–19. [Google Scholar] [CrossRef]
- Shih, H.-W.; DePew, C.L.; Miller, N.D.; Monshausen, G.B. The Cyclic Nucleotide-Gated Channel CNGC14 Regulates Root Gravitropism in Arabidopsis Thaliana. Curr. Biol. CB 2015, 25, 3119–3125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monshausen, G.B.; Miller, N.D.; Murphy, A.S.; Gilroy, S. Dynamics of Auxin-Dependent Ca2+ and PH Signaling in Root Growth Revealed by Integrating High-Resolution Imaging with Automated Computer Vision-Based Analysis. Plant J. Cell Mol. Biol. 2011, 65, 309–318. [Google Scholar] [CrossRef]
- Vanneste, S.; Friml, J. Calcium: The Missing Link in Auxin Action. Plants 2013, 2, 650–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikonorova, N.; Murphy, E.; Fonseca de Lima, C.F.; Zhu, S.; van de Cotte, B.; Vu, L.D.; Balcerowicz, D.; Li, L.; Kong, X.; De Rop, G.; et al. The Arabidopsis Root Tip (Phospho)Proteomes at Growth-Promoting versus Growth-Repressing Conditions Reveal Novel Root Growth Regulators. Cells 2021, 10, 1665. https://doi.org/10.3390/cells10071665
Nikonorova N, Murphy E, Fonseca de Lima CF, Zhu S, van de Cotte B, Vu LD, Balcerowicz D, Li L, Kong X, De Rop G, et al. The Arabidopsis Root Tip (Phospho)Proteomes at Growth-Promoting versus Growth-Repressing Conditions Reveal Novel Root Growth Regulators. Cells. 2021; 10(7):1665. https://doi.org/10.3390/cells10071665
Chicago/Turabian StyleNikonorova, Natalia, Evan Murphy, Cassio Flavio Fonseca de Lima, Shanshuo Zhu, Brigitte van de Cotte, Lam Dai Vu, Daria Balcerowicz, Lanxin Li, Xiangpei Kong, Gieljan De Rop, and et al. 2021. "The Arabidopsis Root Tip (Phospho)Proteomes at Growth-Promoting versus Growth-Repressing Conditions Reveal Novel Root Growth Regulators" Cells 10, no. 7: 1665. https://doi.org/10.3390/cells10071665
APA StyleNikonorova, N., Murphy, E., Fonseca de Lima, C. F., Zhu, S., van de Cotte, B., Vu, L. D., Balcerowicz, D., Li, L., Kong, X., De Rop, G., Beeckman, T., Friml, J., Vissenberg, K., Morris, P. C., Ding, Z., & De Smet, I. (2021). The Arabidopsis Root Tip (Phospho)Proteomes at Growth-Promoting versus Growth-Repressing Conditions Reveal Novel Root Growth Regulators. Cells, 10(7), 1665. https://doi.org/10.3390/cells10071665






