An Insight into the microRNAs Associated with Arteriovenous and Cavernous Malformations of the Brain
Abstract
:1. Introduction
2. Methods
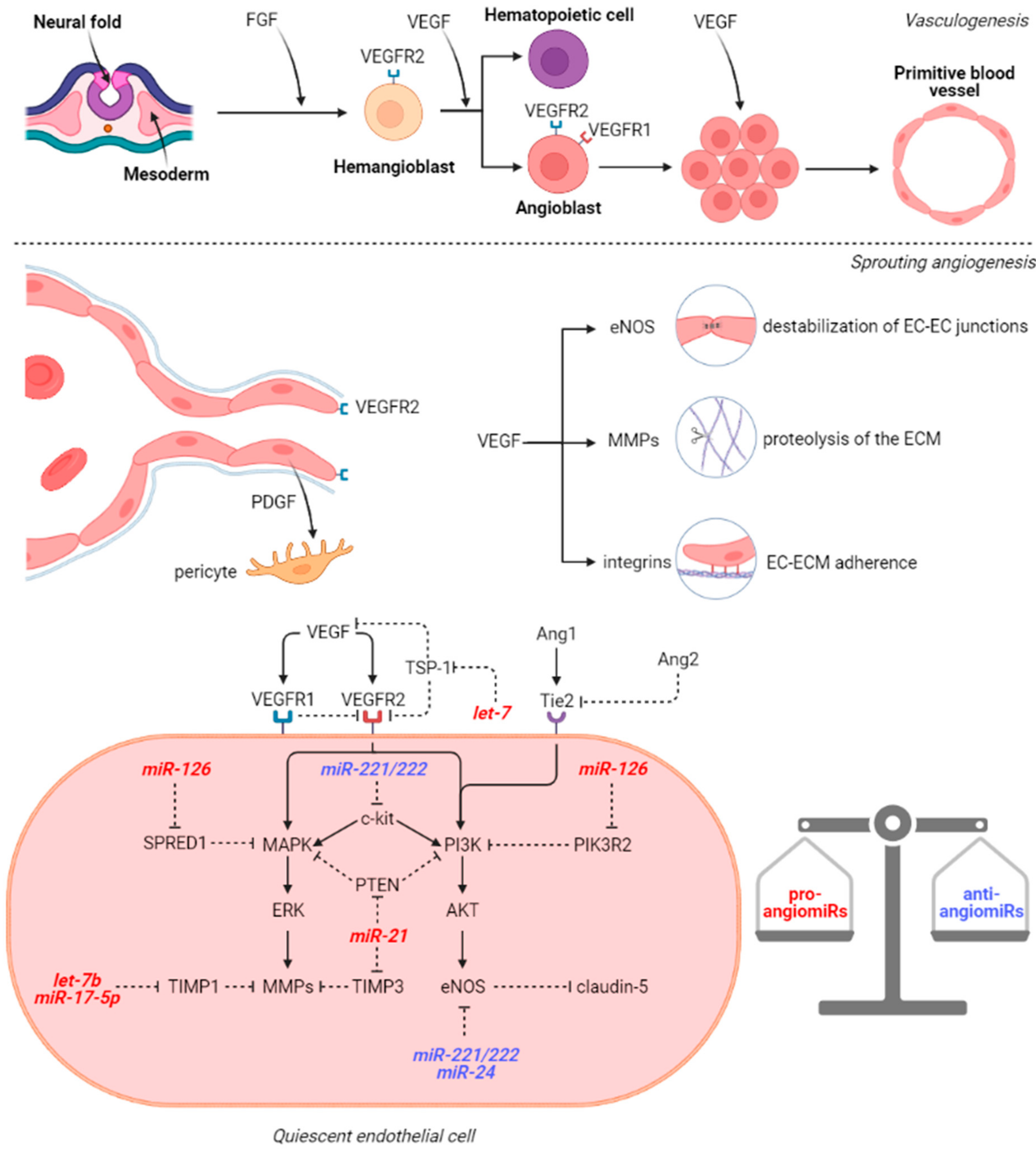
3. microRNAs Involved in Cerebral Vasculogenesis and Angiogenesis
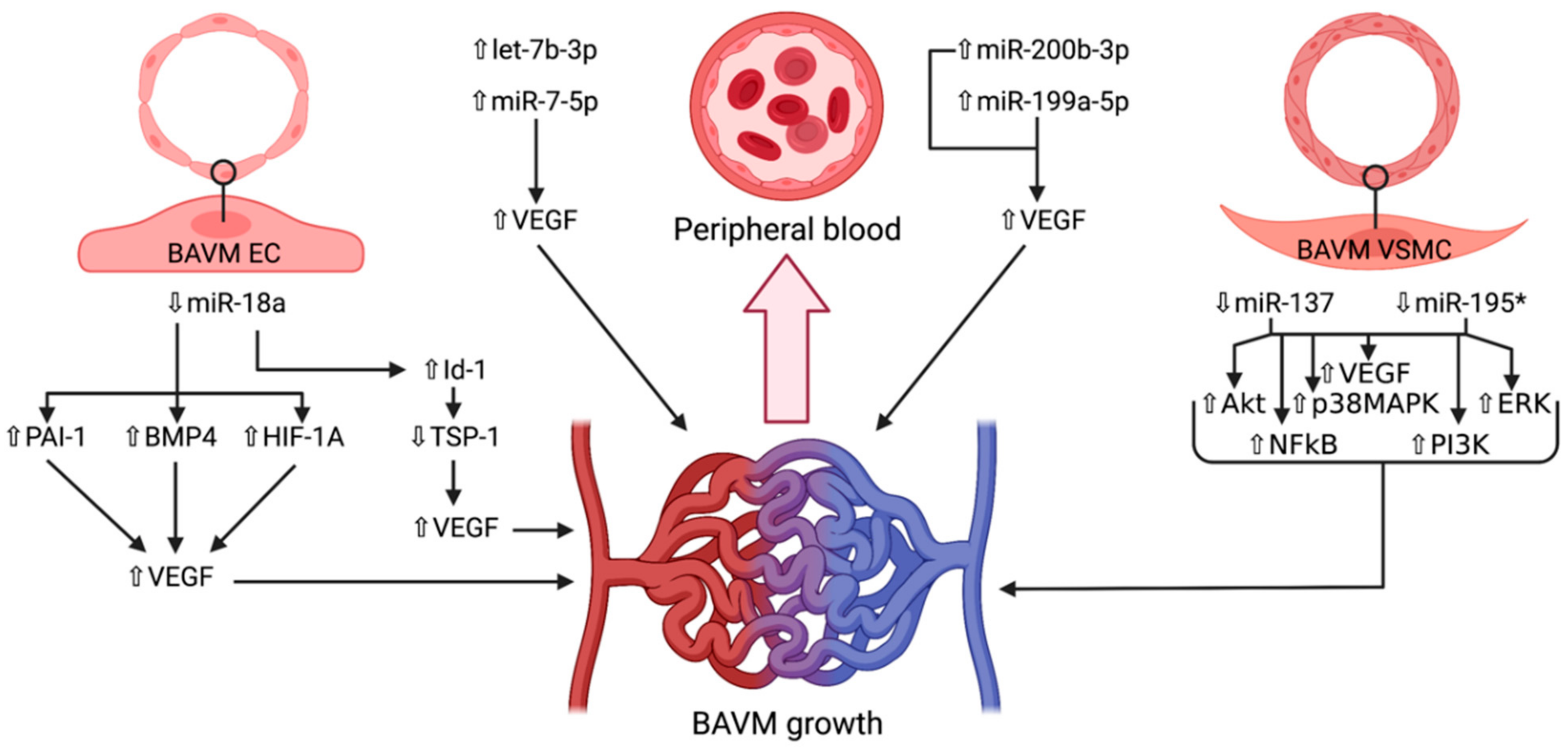
4. microRNAs in Brain Arteriovenous Malformations
4.1. miR-7-5p
4.2. miR-18a
4.3. miR-137 and miR-195*
4.4. miR-199a-5p
4.5. miR-200b-3p
4.6. let-7b-3p
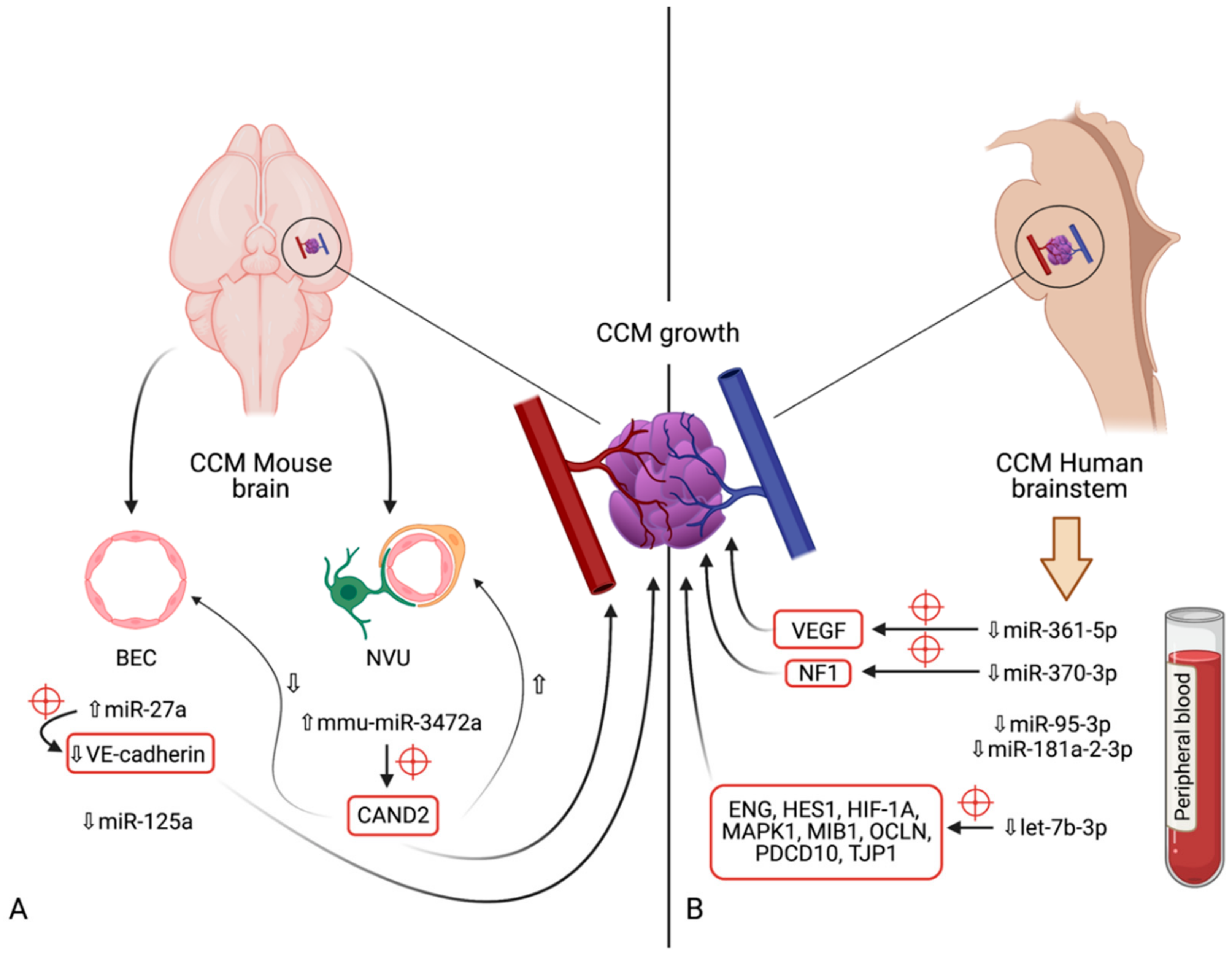
5. microRNAs in Cerebral Cavernous Malformations
5.1. miR-27a
5.2. miR-125a
5.3. mmu-miR-3472a
5.4. miR-361-5p
5.5. miR-370-3p
5.6. miR-181a-2-3p
5.7. miR-95-3p
6. Limitations
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BAVM | brain arteriovenous malformation |
| CCM | cerebral cavernous malformation |
| miRNA | microRNA |
References
- Abecassis, I.; Xu, D.; Batjer, H.; Al, E. Natural history of brain arteriovenous malformations: A systematic review. Neurosurg Focus. 2014, 37, E7. [Google Scholar] [CrossRef]
- Ion, G.; Chiriac, A.; Dobrin, N.; Poeată, I. Multimodality treatment of brain arteriovenous malformation. Case report. Rom. Neurosurg. 2016, 30, 70–76. [Google Scholar] [CrossRef]
- Kalb, S.; Gross, B.A.; Nakaji, P. Vascular Malformations (Arteriovenous Malformations and Dural Arteriovenous Fistulas). In Principles of Neurological Surgery, 4th ed.; Ellenbogen, R.G., Sekhar, L.N., Kitchen, N.D., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 20, pp. 313–324. [Google Scholar]
- Florian, I.S.; Barițchii, A.; Trifoi, S.V. Arterio-venous Mallformations. In Tratat de Chirurgie, 2nd ed.; Popescu, I., Ciuce, C., Eds.; Editura Academiei Române: București, Romania, 2014; Volume VI, pp. 402–410. (In Romanian) [Google Scholar]
- Florian, I.S.; Perju-Dumbravă, L. Therapeutic Options in Hemorrhagic Strokes; Editura Medicală Universitară, Iuliu Hațieganu: Cluj-Napoca, Romania, 2007; pp. 331–346. (In Romanian) [Google Scholar]
- Shetty, R.; Kato, Y.; Watabe, T.; Oguri, D. Unruptured Arteriovenous Malformations of Brain An overview. Rom. Neurosurg. 2010, 17, 34–45. [Google Scholar]
- Florian, I.; Beni, L.; Moisoiu, V.; Timis, T.; Florian, I.; Balașa, A.; Berindan-Neagoe, I. ‘De Novo’ Brain AVMs—Hypotheses for Development and a Systematic Review of Reported Cases. Medicina 2021, 57, 201. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Koh, E.J.; Lee, E.J.; Cheon, J.-E.; Kim, S.-K. An acquired cerebral arteriovenous malformation after brain abscess treatment: Case report and a review of the literature. Child’s Nerv. Syst. 2021, 1–4. [Google Scholar] [CrossRef]
- Bertalanffy, H.; Florian, I.A.; Timiș, T.L. Cavernous Malformations of the Pineal Region: Overview, Management, and Controversies. In Pineal Region Lesions; Florian, I.S., Ed.; Springer: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Choudhri, O.; Chen, R.P.; Bulsara, K. Cavernous Malformations of the Brain and Spinal Cord, 4th ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Iliescu, B.F.; Poeata, I. Cerebral Cavernomas. In Tratat de Chirurgie, 2nd ed.; Popescu, I., Ciuce, C., Eds.; Editura Academiei Române: București, Romania, 2014; Volume VI, pp. 411–414. (In Romanian) [Google Scholar]
- Bozinov, O.; Hatano, T.; Sarnthein, J.; Burkhardt, J.-K.; Bertalanffy, H. Current clinical management of brainstem cavernomas. Swiss Med. Wkly. 2010, 140, w13120. [Google Scholar] [CrossRef] [Green Version]
- Akers, A.; Salman, R.A.-S.; Awad, I.A.; Dahlem, K.; Flemming, K.; Hart, B.; Kim, H.; Jusue-Torres, I.; Kondziolka, D.; Lee, C.; et al. Synopsis of Guidelines for the Clinical Management of Cerebral Cavernous Malformations: Consensus Recommendations Based on Systematic Literature Review by the Angioma Alliance Scientific Advisory Board Clinical Experts Panel. Neurosurgery 2017, 80, 665–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A Winkler, E.; Rutledge, C.; Ward, M.; Tihan, T.; Sneed, P.K.; Barbaro, N.; Garcia, P.; McDermott, M.; Chang, E.F. Radiation-induced Cavernous Malformation as a Late Sequelae of Stereotactic Radiosurgery for Epilepsy. Cureus 2018, 10, e2308. [Google Scholar] [CrossRef] [Green Version]
- Hui, X.; Wang, Q.; Zhang, S. Cavernous malformation induced by stereotactic radiosurgery: A report and literature review. Neurol. India 2018, 66, 515. [Google Scholar] [CrossRef]
- Yu, Z.; Huang, B.; Liang, R. Radiation-induced cavernous malformation after stereotactic radiosurgery for cavernous sinus meningioma: A case report. BMC Neurol. 2020, 20, 422. [Google Scholar] [CrossRef]
- Araldi, E.; Suárez, Y. MicroRNAs as regulators of endothelial cell functions in cardiometabolic diseases. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2016, 1861, 2094–2103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Li, Z.; Shi, Y.; Huang, G.; Chen, L.; Tan, H.; Wang, Z.; Yin, C.; Hu, J. Deep Sequencing of Small RNAs in Blood of Patients with Brain Arteriovenous Malformations. World Neurosurg. 2018, 115, e570–e579. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Santos, T.; Amar, A.; Tahara, S.M.; Chen, T.C.; Giannotta, S.L.; Hofman, F.M. MicroRNA-18a improves human cerebral arteriovenous malformation endothelial cell function. Stroke 2014, 45, 293–297. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Song, J.; Qu, M.; Wang, Y.; An, Q.; Song, Y.; Yan, W.; Wang, B.; Wang, X.; Zhang, S.; et al. MicroRNA-137 and -195* inhibit vasculogenesis in brain arteriovenous malformations. Ann. Neurol. 2017, 82, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dai, Y.-X.; Wang, S.-Q.; Qiu, M.-K.; Quan, Z.-W.; Liu, Y.-B.; Ou, J.-M. miR-199a-5p inhibits proliferation and induces apoptosis in hemangioma cells through targeting HIF1A. Int. J. Immunopathol. Pharmacol. 2018, 31, 0394632017749357. [Google Scholar] [CrossRef] [Green Version]
- Zammar, S.G.; El Tecle, N.E.; El Ahmadieh, T.Y.; McClendon, J.; Comair, Y.G.; Bendok, B.R. A Biological Approach to Treating Brain Arteriovenous Malformations. Neurosurgery 2014, 74, N15–N17. [Google Scholar] [CrossRef] [Green Version]
- Fong, G.-H.; Rossant, J.; Gertsenstein, M.; Breitman, M.L. Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature 1995, 376, 66–70. [Google Scholar] [CrossRef]
- Shalaby, F.; Rossant, J.; Yamaguchi, T.P.; Gertsenstein, M.; Wu, X.-F.; Breitman, M.L.; Schuh, A.C. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 1995, 376, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Yang, D.D.; Na, S.; Sandusky, G.E.; Zhang, Q.; Zhao, G. Dicer Is Required for Embryonic Angiogenesis during Mouse Development. J. Biol. Chem. 2005, 280, 9330–9335. [Google Scholar] [CrossRef] [Green Version]
- Thomas, K.A. Angiogenesis. In Encyclopedia of Cell Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 102–116. Available online: https://linkinghub.elsevier.com/retrieve/pii/B9780123944474400192 (accessed on 30 April 2021).
- Taddei, A.; Giampietro, C.; Conti, A.; Orsenigo, F.; Breviario, F.; Pirazzoli, V.; Potente, M.; Daly, C.; Dimmeler, S.; Dejana, E. Endothelial adherens junctions control tight junctions by VE-cadherin-mediated upregulation of claudin-5. Nat. Cell Biol. 2008, 10, 923–934. [Google Scholar] [CrossRef]
- Hogan, K.A.; Ambler, C.A.; Chapman, D.; Bautch, V.L. The neural tube patterns vessels developmentally using the VEGF signaling pathway. Development 2004, 131, 1503–1513. [Google Scholar] [CrossRef] [Green Version]
- Rundhaug, J.E. Matrix metalloproteinases and angiogenesis. J. Cell. Mol. Med. 2005, 9, 267–285. [Google Scholar] [CrossRef]
- Sato, T.N.; Tozawa, Y.; Deutsch, U.; Wolburg-Buchholz, K.; Fujiwara, Y.; Gendron-Maguire, M.; Gridley, T.; Wolburg, H.; Risau, W.; Qin, Y. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature 1995, 376, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Seegar, T.C.; Eller, B.; Tzvetkova-Robev, D.; Kolev, M.V.; Henderson, S.C.; Nikolov, D.B.; Barton, W.A. Tie1-Tie2 Interactions Mediate Functional Differences between Angiopoietin Ligands. Mol. Cell 2010, 37, 643–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Olson, E.N. AngiomiRs-Key regulators of angiogenesis. Curr. Opin. Genet. Dev. 2009, 19, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawler, P.; Lawler, J. Molecular Basis for the Regulation of Angiogenesis by Thrombospondin-1 and -2. Cold Spring Harb. Perspect. Med. 2012, 2, a006627. [Google Scholar] [CrossRef] [PubMed]
- Kuehbacher, A.; Urbich, C.; Zeiher, A.M.; Dimmeler, S. Role of Dicer and Drosha for Endothelial MicroRNA Expression and Angiogenesis. Circ. Res. 2007, 101, 59–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otsuka, M.; Zheng, M.; Hayashi, M.; Lee, J.D.; Yoshino, O.; Lin, S.; Han, J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J. Clin. Investig. 2008, 118, 1944–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006, 38, 1060–1065. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.-Z.; Li, C.; Chen, Q.; Jing, Y.; Carpenter, R.; Jiang, Y.; Kung, H.-F.; Lai, L.; Jiang, B.-H. MiR-21 Induced Angiogenesis through AKT and ERK Activation and HIF-1α Expression. PLoS ONE 2011, 6, e19139. [Google Scholar]
- Hu, J.; Ni, S.; Cao, Y.; Zhang, T.; Wu, T.; Yin, X.; Lang, Y.; Lu, H. The Angiogenic Effect of microRNA-21 Targeting TIMP3 through the Regulation of MMP2 and MMP9. PLoS ONE 2016, 11, e0149537. [Google Scholar] [CrossRef] [Green Version]
- Chistiakov, D.A.; Sobenin, I.A.; Orekhov, A.N.; Bobryshev, Y.V. Human miR-221/222 in Physiological and Atherosclerotic Vascular Remodeling. BioMed Res. Int. 2015, 2015, 354517. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; Meloni, M.; Anwar, M.; Al-Haj-Zen, A.; Sala-Newby, G.; Slater, S.; Ford, K.; Caporali, A.; Emanueli, C. MicroRNA-24-3p Targets Notch and Other Vascular Morphogens to Regulate Post-ischemic Microvascular Responses in Limb Muscles. Int. J. Mol. Sci. 2020, 21, 1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fasanaro, P.; D’Alessandra, Y.; Di Stefano, V.; Melchionna, R.; Romani, S.; Pompilio, G.; Capogrossi, M.C.; Martelli, F. MicroRNA-210 Modulates Endothelial Cell Response to Hypoxia and Inhibits the Receptor Tyrosine Kinase Ligand Ephrin-A3. J. Biol. Chem. 2008, 283, 15878–15883. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Lai, T.-C.; Jan, Y.-H.; Lin, F.-M.; Wang, W.-C.; Xiao, H.; Wang, Y.-T.; Sun, W.; Cui, X.; Li, Y.-S.; et al. Hypoxia-responsive miRNAs target argonaute 1 to promote angiogenesis. J. Clin. Investig. 2013, 123, 1057–1067. [Google Scholar] [CrossRef] [Green Version]
- Jeyapalan, Z.; Deng, Z.; Shatseva, T.; Fang, L.; He, C.; Yang, B.B. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011, 39, 3026–3041. [Google Scholar] [CrossRef] [Green Version]
- Suárez, Y.; Fernández-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Li, Y.; Han, C.; Wang, X.; She, L.; Zhang, H. miRNA microarray reveals specific expression in the peripheral blood of glioblastoma patients. Int. J. Oncol. 2014, 45, 746–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Huang, M.; Cai, Y.; Yang, Y.; Sun, X.; Ke, Y. Circ-U2AF1 promotes human glioma via derepressing neuro-oncological ventral antigen 2 by sponging hsa-miR-7-5p. J. Cell. Physiol. 2019, 234, 9144–9155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Niu, W.; Mu, M.; Hu, S.; Niu, C. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J. Exp. Clin. Cancer Res. 2020, 39, 1–20. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Y.; Li, L.; Xu, Z.; Bi, B.; Wang, Y.; Li, J.Y. MiR-7-5p is frequently downregulated in glioblastoma microvasculature and inhibits vascular endothelial cell proliferation by targeting RAF1. Tumor Biol. 2014, 35, 10177–10184. [Google Scholar] [CrossRef]
- Xu, H.; Nie, B.; Liu, L.; Zhang, C.; Zhang, Z.; Xu, M.; Mei, Y. Curcumin Prevents Brain Damage and Cognitive Dysfunction During Ischemic-reperfusion Through the Regulation of miR-7-5p. Curr. Neurovascular Res. 2020, 16, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, B. MiR-7-5p Enhances Cerebral Ischemia–Reperfusion Injury by Degrading sirt1 mRNA. J. Cardiovasc. Pharmacol. 2020, 76, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Deng, S.; Lei, Q.; He, Q.; Ren, Y.; Zhang, Y.; Nie, J.; Lu, W. miR-7-5p Affects Brain Edema After Intracerebral Hemorrhage and Its Possible Mechanism. Front. Cell Dev. Biol. 2020, 8, 598020. [Google Scholar] [CrossRef]
- Xu, M.; Xu, H.; Qin, Z.; Zhang, J.; Yang, X.; Xu, F. Increased Expression of Angiogenic Factors in Cultured Human Brain Arteriovenous Malformation Endothelial Cells. Cell Biophys. 2014, 70, 443–447. [Google Scholar] [CrossRef]
- Rangel-Castilla, L.; Russin, J.J.; Martinez-Del-Campo, E.; Soriano-Baron, H.; Spetzler, R.F.; Nakaji, P. Molecular and cellular biology of cerebral arteriovenous malformations: A review of current concepts and future trends in treatment. Neurosurg. Focus 2014, 37, E1. [Google Scholar] [CrossRef]
- Kolenda, T.; Guglas, K.; Kopczyńska, M.; Sobocińska, J.; Teresiak, A.; Bliźniak, R.; Lamperska, K. Good or not good: Role of miR-18a in cancer biology. Rep. Pract. Oncol. Radiother. 2020, 25, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, M.; Deng, D. c-Fos/microRNA-18a feedback loop modulates the tumor growth via HMBOX1 in human gliomas. Biomed. Pharmacother. 2018, 107, 1705–1711. [Google Scholar] [CrossRef]
- Gruszka, R.; Zakrzewski, K.; Liberski, P.P.; Zakrzewska, M. mRNA and miRNA Expression Analyses of the MYC/E2F/miR-17-92 Network in the Most Common Pediatric Brain Tumors. Int. J. Mol. Sci. 2021, 22, 543. [Google Scholar] [CrossRef]
- Miao, Y.-S.; Zhao, Y.-Y.; Zhao, L.-N.; Wang, P.; Liu, Y.-H.; Ma, J.; Xue, Y.-X. MiR-18a increased the permeability of BTB via RUNX1 mediated down-regulation of ZO-1, occludin and claudin-5. Cell. Signal. 2015, 27, 156–167. [Google Scholar] [CrossRef]
- Song, Y.; Wang, P.; Zhao, W.; Yao, Y.; Liu, X.; Ma, J.; Xue, Y.; Liu, Y. MiR-18a regulates the proliferation, migration and invasion of human glioblastoma cell by targeting neogenin. Exp. Cell Res. 2014, 324, 54–64. [Google Scholar] [CrossRef]
- Wilson, N.H.; Key, B. Neogenin: One receptor, many functions. Int. J. Biochem. Cell Biol. 2007, 39, 874–878. [Google Scholar] [CrossRef]
- Cole, S.J.; Bradford, D.; Cooper, H.M. Neogenin: A multi-functional receptor regulating diverse developmental processes. Int. J. Biochem. Cell Biol. 2007, 39, 1569–1575. [Google Scholar] [CrossRef]
- De Vries, M.; Cooper, H.M. Emerging roles for neogenin and its ligands in CNS development. J. Neurochem. 2008, 106, 1483–1492. [Google Scholar] [CrossRef]
- Rodrigues, S.P.; De Wever, O.; Bruyneel, E.; Rooney, R.J.; Gespach, C. Opposing roles of netrin-1 and the dependence receptor DCC in cancer cell invasion, tumor growth and metastasis. Oncogene 2007, 26, 5615–5625. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tian, Y.; Li, Z.; Zheng, Z.; Zhu, L. miR-92 regulates the proliferation, migration, invasion and apoptosis of glioma cells by targeting neogenin. Open Med. 2020, 15, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Li, Y.; Wan, X.; Kayira, T.M.; Cao, R.; Ju, X.; Zhu, X.; Zhao, G. Down-Regulation of Neogenin Accelerated Glioma Progression through Promoter Methylation and Its Overexpression in SHG-44 Induced Apoptosis. PLoS ONE 2012, 7, e38074. [Google Scholar] [CrossRef] [Green Version]
- Sesen, J.; Driscoll, J.; Shah, N.; Moses-Gardner, A.; Luiselli, G.; Alexandrescu, S.; Zurakowski, D.; Baxter, P.A.; Su, J.M.; Fehnel, K.P.; et al. Neogenin is highly expressed in diffuse intrinsic pontine glioma and influences tumor invasion. Brain Res. 2021, 1762, 147348. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Yao, L.-L.; Pan, J.-X.; Zhang, J.-S.; Mei, L.; Wang, Y.-G.; Xiong, W.-C. Linking cortical astrocytic neogenin deficiency to the development of Moyamoya disease–like vasculopathy. Neurobiol. Dis. 2021, 154, 105339. [Google Scholar] [CrossRef]
- Yao, L.-L.; Hu, J.-X.; Li, Q.; Lee, D.; Ren, X.; Zhang, J.-S.; Sun, D.; Zhang, H.-S.; Wang, Y.-G.; Mei, L.; et al. Astrocytic neogenin/netrin-1 pathway promotes blood vessel homeostasis and function in mouse cortex. J. Clin. Investig. 2020, 130, 6490–6509. [Google Scholar] [CrossRef] [PubMed]
- Marín-Ramos, N.I.; Thein, T.Z.; Ghaghada, K.B.; Chen, T.C.; Giannotta, S.L.; Hofman, F.M. miR-18a Inhibits BMP4 and HIF-1α Normalizing Brain Arteriovenous Malformations. Circ. Res. 2020, 127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, Q.; Lin, S.; Chen, B.; Jia, B.; Ye, R.; Weygant, N.; Chu, J.; Peng, J. Qingda granule exerts neuroprotective effects against ischemia/reperfusion-induced cerebral injury via lncRNA GAS5/miR-137 signaling pathway. Int. J. Med. Sci. 2021, 18, 1687–1698. [Google Scholar] [CrossRef]
- Zhang, M.; Ge, D.; Su, Z.; Qi, B. miR-137 alleviates focal cerebral ischemic injury in rats by regulating JAK1/STAT1 signaling pathway. Hum. Exp. Toxicol. 2020, 39, 816–827. [Google Scholar] [CrossRef]
- Tian, R.; Wu, B.; Fu, C.; Guo, K. miR-137 prevents inflammatory response, oxidative stress, neuronal injury and cognitive impairment via blockade of Src-mediated MAPK signaling pathway in ischemic stroke. Aging 2020, 12, 10873–10895. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Berger, R.; Freedman, R.; Law, A.J. A VNTR Regulates miR-137 Expression Through Novel Alternative Splicing and Contributes to Risk for Schizophrenia. Sci. Rep. 2019, 9, 11793. [Google Scholar] [CrossRef] [Green Version]
- Arakawa, Y.; Yokoyama, K.; Tasaki, S.; Kato, J.; Nakashima, K.; Takeyama, M.; Nakatani, A.; Suzuki, M. Transgenic mice overexpressing miR-137 in the brain show schizophrenia-associated behavioral deficits and transcriptome profiles. PLoS ONE 2019, 14, e0220389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandratsenka, H.; Nestsiarovich, A.; Goloenko, I.; Danilenko, N.; Makarevich, A.; Obyedkov, V.; Davydenko, O.; Waszkiewicz, N. Association of MIR137 With Symptom Severity and Cognitive Functioning in Belarusian Schizophrenia Patients. Front. Psychiatry 2018, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shang, S.; Wang, J.; Zhang, T.; Nie, F.; Song, X.; Zhao, H.; Zhu, C.; Zhang, R.; Hao, D. Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia. Psychiatry Res. 2018, 265, 70–76. [Google Scholar] [CrossRef]
- Sudesh, R.; Thalamuthu, A.; John, S.; Thara, R.; Mowry, B.; Munirajan, A.K. Replication of GWAS identified miR-137 and its target gene polymorphisms in Schizophrenia of South Indian population and meta-analysis with Psychiatric Genomics Consortium. Schizophr. Res. 2018, 199, 189–194. [Google Scholar] [CrossRef]
- Sakamoto, K.; Crowley, J.J. A comprehensive review of the genetic and biological evidence supports a role for MicroRNA-137 in the etiology of schizophrenia. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2018, 177, 242–256. [Google Scholar] [CrossRef] [Green Version]
- Bier, A.; Giladi, N.; Kronfeld, N.; Lee, H.K.; Cazacu, S.; Finniss, S.; Xiang, C.; Poisson, L.; Decarvalho, A.C.; Slavin, S.; et al. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget 2013, 4, 665–676. [Google Scholar] [CrossRef] [Green Version]
- Liang, M.-L.; Hsieh, T.-H.; Ng, K.-H.; Tsai, Y.-N.; Tsai, C.-F.; Chao, M.-E.; Liu, D.-J.; Chu, S.-S.; Chen, W.; Liu, Y.-R.; et al. Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget 2016, 7, 19723–19737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, G.; Cao, Y.; Shi, L.; Sun, L.; Wang, Y.; Chen, C.; Wan, Z.; Fu, L.; You, Y. Overexpressed miRNA-137 Inhibits Human Glioma Cells Growth by Targeting Rac1. Cancer Biother. Radiopharm. 2013, 28, 327–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Wang, X.; Wang, H.; Li, Y.; Yan, W.; Han, L.; Zhang, K.; Zhang, J.; Wang, Y.; Feng, Y.; et al. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur. J. Cancer 2012, 48, 3104–3111. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.-W.; Yang, L.; Pang, J.C.-S.; Chan, A.K.-Y.; Zhou, L.; Mao, Y.; Wang, Y.; Lau, K.-M.; Poon, W.S.; Shi, Z.; et al. MIR-137 Suppresses Growth and Invasion, is Downregulated in Oligodendroglial Tumors and Targets CSE1L. Brain Pathol. 2012, 23, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Mi, Y.; Guo, N.; Xu, H.; Jiang, P.; Zhang, R.; Xu, L. Glioma in Schizophrenia: Is the Risk Higher or Lower? Front. Cell. Neurosci. 2018, 12, 289. [Google Scholar] [CrossRef]
- Zhao, W.-J.; Zhang, H.-F.; Su, J.-Y. Downregulation of microRNA-195 promotes angiogenesis induced by cerebral infarction via targeting VEGFA. Mol. Med. Rep. 2017, 16, 5434–5440. [Google Scholar] [CrossRef] [Green Version]
- Pan, S.; Feng, W.; Li, Y.; Huang, J.; Chen, S.; Cui, Y.; Tian, B.; Tan, S.; Wang, Z.; Yao, S.; et al. The microRNA-195-BDNF pathway and cognitive deficits in schizophrenia patients with minimal antipsychotic medication exposure. Transl. Psychiatry 2021, 11, 117. [Google Scholar] [CrossRef]
- Huang, X.; Bao, C.; Lv, Q.; Zhao, J.; Wang, Y.; Lang, X.; Li, Z.; Yi, Z. Sex difference in cognitive impairment in drug-free schizophrenia: Association with miR-195 levels. Psychoneuroendocrinology 2020, 119, 104748. [Google Scholar] [CrossRef]
- Cao, J.; Huang, M.; Guo, L.; Zhu, L.; Hou, J.; Zhang, L.; Pero, A.; Ng, S.; El Gaamouch, F.; Elder, G.; et al. MicroRNA-195 rescues ApoE4-induced cognitive deficits and lysosomal defects in Alzheimer’s disease pathogenesis. Mol. Psychiatry 2020. [Google Scholar] [CrossRef]
- Zhu, H.-C.; Wang, L.-M.; Wang, M.; Song, B.; Tan, S.; Teng, J.-F.; Duan, D.-X. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res. Bull. 2012, 88, 596–601. [Google Scholar] [CrossRef]
- Ai, J.; Sun, L.-H.; Che, H.; Zhang, R.; Zhang, T.-Z.; Wu, W.-C.; Su, X.-L.; Chen, X.; Yang, G.; Li, K.; et al. MicroRNA-195 Protects Against Dementia Induced by Chronic Brain Hypoperfusion via Its Anti-Amyloidogenic Effect in Rats. J. Neurosci. 2013, 33, 3989–4001. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, X.-M.; Zhao, L.-J.; Sun, L.-L.; Yan, M.-L.; Tian, Y.; Zhang, S.; Duan, M.-J.; Zhao, H.-M.; Li, W.-R.; et al. MicroRNA-195 prevents dendritic degeneration and neuron death in rats following chronic brain hypoperfusion. Cell Death Dis. 2017, 8, e2850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, L.; Zhang, W.; Shi, S.; Peng, Y.; Wang, D.; Zhang, L.; Zhang, J. microRNA-195 attenuates neuronal apoptosis in rats with ischemic stroke through inhibiting KLF5-mediated activation of the JNK signaling pathway. Mol. Med. 2020, 26, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H.-Y.; Wang, Y.-S.; Hsu, P.-Y.; Chen, C.-Y.; Liao, Y.-C.; Juo, S.-H.H. miR-195 Has a Potential to Treat Ischemic and Hemorrhagic Stroke through Neurovascular Protection and Neurogenesis. Mol. Ther. Methods Clin. Dev. 2019, 13, 121–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.-R.; Li, D.; Weng, J.-C.; Li, C.-B.; Wang, L.; Wu, Z.; Zhang, J.-T. MicroRNA-195 Functions as a Tumor Suppressor by Directly Targeting Fatty Acid Synthase in Malignant Meningioma. World Neurosurg. 2020, 136, e355–e364. [Google Scholar] [CrossRef]
- Jia, Y.; Tian, Y.; An, S.; Yang, D. Effects of microRNA-195 on the Prognosis of Glioma Patients and the Proliferation and Apoptosis of Human Glioma Cells. Pathol. Oncol. Res. 2020, 26, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-P.; Zhang, N.-N.; Ren, X.-Q.; He, J.; Li, Y. miR-103/miR-195/miR-15b Regulate SALL4 and Inhibit Proliferation and Migration in Glioma. Molecules 2018, 23, 2938. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Cheung, H.H.; Lee, T.L.; Lu, G.; Poon, W.S.; Chan, W.Y. Molecular Mechanisms of Regulation and Action of microRNA-199a in Testicular Germ Cell Tumor and Glioblastomas. PLoS ONE 2013, 8, e83980. [Google Scholar] [CrossRef]
- Li, Z.; Gu, X.-D.; Fang, Y.; Xiang, J.; Chen, Z. microRNA expression profiles in human colorectal cancers with brain metastases. Oncol. Lett. 2012, 3, 346–350. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Li, Y.; Li, J.; Zhang, D.; Luo, C.; Zhang, B.; Sun, X. microRNA-199a-5p suppresses glioma progression by inhibiting MAGT1. J. Cell. Biochem. 2019, 120, 15248–15254. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, Q.; Zhu, J.-W.; Liu, Z.-F. MicroRNA-199a-5p regulates glioma progression via targeting MARCH8. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7482–7487. [Google Scholar] [CrossRef]
- Chi, G.; Yang, F.; Xu, D.; Liu, W. Silencing hsa_circ_PVT1 (circPVT1) suppresses the growth and metastasis of glioblastoma multiforme cells by up-regulation of miR-199a-5p. Artif. Cells Nanomed. Biotechnol. 2020, 48, 188–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, W.; Li, Y.-C.; Huang, Q.-Y.; Tang, X.-Q. lncRNA ANRIL Ameliorates Oxygen and Glucose Deprivation (OGD) Induced Injury in Neuron Cells via miR-199a-5p/CAV-1 Axis. Neurochem. Res. 2020, 45, 772–782. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Luan, L.; Liu, Q.; Liu, Y.; Lan, X.; Li, Z.; Liu, W. MiRNA-199a-5p Protects Against Cerebral Ischemic Injury by Down-Regulating DDR1 in Rats. World Neurosurg. 2019, 131, e486–e494. [Google Scholar] [CrossRef]
- Bao, N.; Fang, B.; Lv, H.; Jiang, Y.; Chen, F.; Wang, Z.; Ma, H. Upregulation of miR-199a-5p Protects Spinal Cord Against Ischemia/Reperfusion-Induced Injury via Downregulation of ECE1 in Rat. Cell. Mol. Neurobiol. 2018, 38, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, L.; Wu, Q.; Zheng, G.; Long, H.; Wu, H.; Zhou, C.; Guo, T.; Zhong, T.; Wang, L.; et al. Long noncoding RNA H19 upregulates vascular endothelial growth factor A to enhance mesenchymal stem cells survival and angiogenic capacity by inhibiting miR-199a-5p. Stem Cell Res. Ther. 2018, 9, 109. [Google Scholar] [CrossRef]
- Heuslein, J.L.; Gorick, C.M.; McDonnell, S.P.; Song, J.; Annex, B.H.; Price, R.J. Exposure of Endothelium to Biomimetic Flow Waveforms Yields Identification of miR-199a-5p as a Potent Regulator of Arteriogenesis. Mol. Ther. Nucleic Acids 2018, 12, 829–844. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Yang, L.; Meng, L.; Cui, H. Inhibition of miR-200b-3p alleviates hypoxia-ischemic brain damage via targeting Slit2 in neonatal rats. Biochem. Biophys. Res. Commun. 2020, 523, 931–938. [Google Scholar] [CrossRef]
- Visani, M.; Marucci, G.; De Biase, D.; Giangaspero, F.; Buttarelli, F.R.; Brandes, A.A.; Franceschi, E.; Acquaviva, G.; Ciarrocchi, A.; Rhoden, K.J.; et al. miR-196B-5P and miR-200B-3P Are Differentially Expressed in Medulloblastomas of Adults and Children. Diagnostics 2020, 10, 265. [Google Scholar] [CrossRef]
- Minn, Y.-K.; Lee, D.; Hyung, W.; Kim, J.; Choi, J.; Yang, S.-H.; Song, H.; Lim, B.; Kim, S.H. MicroRNA-200 family members and ZEB2 are associated with brain metastasis in gastric adenocarcinoma. Int. J. Oncol. 2014, 45, 2403–2410. [Google Scholar] [CrossRef] [Green Version]
- Comijn, J.; Berx, G.; Vermassen, P.; Verschueren, K.; van Grunsven, L.; Bruyneel, E.; Mareel, M.; Huylebroeck, D.; van Roy, F. The Two-Handed E Box Binding Zinc Finger Protein SIP1 Downregulates E-Cadherin and Induces Invasion. Mol. Cell 2001, 7, 1267–1278. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yuan, J.; Yuan, X.; Zhao, J.; Zhang, Z.; Weng, L.; Liu, J. MicroRNA-200b inhibits the growth and metastasis of glioma cells via targeting ZEB2. Int. J. Oncol. 2016, 48, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cui, H.; Zhu, Z.; Wang, L. MicroRNA-200b-3p suppresses epithelial-mesenchymal transition and inhibits tumor growth of glioma through down-regulation of ERK5. Biochem. Biophys. Res. Commun. 2016, 478, 1158–1164. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Jiang, Q.; Luo, D.; Zhao, L.; Fu, X.; Chen, Y.; Song, X.; Li, L.; Zhao, H.; He, Y.; et al. miR-200b is a key regulator of tumor progression and metabolism targeting lactate dehydrogenase A in human malignant glioma. Oncotarget 2016, 7, 48423–48431. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Gong, S.; Yan, T.; Yang, Y. MicroRNA-200b expression level is negatively associated with pathological grading in human gliomas. Cancer Manag. Res. 2018, 10, 2825–2834. [Google Scholar] [CrossRef]
- Liu, H.; Tian, T.; Qin, S.; Li, W.; Zhang, X.; Wang, X.; Gao, Y.; Huang, G. Folic acid deficiency enhances abeta accumulation in APP/PS1 mice brain and decreases amyloid-associated miRNAs expression. J. Nutr. Biochem. 2015, 26, 1502–1508. [Google Scholar] [CrossRef]
- Higaki, S.; Muramatsu, M.; Matsuda, A.; Matsumoto, K.; Satoh, J.-I.; Michikawa, M.; Niida, S. Defensive effect of microRNA-200b/c against amyloid-beta peptide-induced toxicity in Alzheimer’s disease models. PLoS ONE 2018, 13, e0196929. [Google Scholar] [CrossRef]
- Boese, A.S.; Saba, R.; Campbell, K.; Majer, A.; Medina, S.; Burton, L.; Booth, T.F.; Chong, P.; Westmacott, G.R.; Dutta, S.M.; et al. MicroRNA abundance is altered in synaptoneurosomes during prion disease. Mol. Cell. Neurosci. 2016, 71, 13–24. [Google Scholar] [CrossRef]
- Osei, J.; Kelly, W.; Toffolo, K.; Donahue, K.; Levy, B.; Bard, J.; Wang, J.; Levy, E.; Nowak, N.; Poulsen, D. Thymosin beta 4 induces significant changes in the plasma miRNA profile following severe traumatic brain injury in the rat lateral fluid percussion injury model. Expert Opin. Biol. Ther. 2018, 18, 159–164. [Google Scholar] [CrossRef]
- Lee, K.H.; Lim, B.J.; Ferreira, V.H.; Min, S.Y.; Hong, Y.-M.; Jo, J.-H.; Han, S.H. Expression of human miR-200b-3p and -200c-3p in cytomegalovirus-infected tissues. Biosci. Rep. 2018, 38, BSR20180961. [Google Scholar] [CrossRef] [Green Version]
- Kar, S.; Bali, K.K.; Baisantry, A.; Geffers, R.; Samii, A.; Bertalanffy, H. Genome-Wide Sequencing Reveals MicroRNAs Downregulated in Cerebral Cavernous Malformations. J. Mol. Neurosci. 2017, 61, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Alsharafi, W.A.; Xiao, B.; Abuhamed, M.M.; Luo, Z. miRNAs: Biological and clinical determinants in epilepsy. Front. Mol. Neurosci. 2015, 8, 59. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, P.K.; Roncon, P.; Lukasiuk, K.; Gorter, J.A.; Aronica, E.; Pitkänen, A.; Petretto, E.; Johnson, M.R.; Simonato, M. Meta-Analysis of MicroRNAs Dysregulated in the Hippocampal Dentate Gyrus of Animal Models of Epilepsy. Eneuro 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.D.; van Vliet, E.; Chen, B.J.; Janitz, M.; Anink, J.J.; Baayen, J.C.; Idema, S.; Devore, S.; Friedman, D.; Diehl, B.; et al. Coding and non-coding transcriptome of mesial temporal lobe epilepsy: Critical role of small non-coding RNAs. Neurobiol. Dis. 2020, 134, 104612. [Google Scholar] [CrossRef]
- Li, S.; Chen, L.; Zhou, X.; Li, J.; Liu, J. miRNA-223-3p and let-7b-3p as potential blood biomarkers associated with the ischemic penumbra in rats. Acta Neurobiol. Exp. 2019, 79, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Hao, S.; Ye, M.; Zhang, A.; Nan, Y.; Wang, G.; Jia, Z.; Yuan, T.; Guo, L.; Pu, P.; et al. MicroRNAs let-7b/i suppress human glioma cell invasion and migration by targeting IKBKE directly. Biochem. Biophys. Res. Commun. 2015, 458, 307–312. [Google Scholar] [CrossRef]
- Song, H.; Zhang, Y.; Liu, N.; Zhang, D.; Wan, C.; Zhao, S.; Kong, Y.; Yuan, L. Let-7b inhibits the malignant behavior of glioma cells and glioma stem-like cells via downregulation of E2F2. J. Physiol. Biochem. 2016, 72, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, M.; Gruszka, R.; Stawiski, K.; Fendler, W.; Kordacka, J.; Grajkowska, W.; Daszkiewicz, P.; Liberski, P.P.; Zakrzewski, K. Expression-based decision tree model reveals distinct microRNA expression pattern in pediatric neuronal and mixed neuronal-glial tumors. BMC Cancer 2019, 19, 544. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Y.; Choi, J.; Ting, K.K.; Coleman, P.; Chen, J.; Cogger, V.C.; Wan, L.; Shi, Z.; Moller, T.; et al. Targeting miR-27a/VE-cadherin interactions rescues cerebral cavernous malformations in mice. PLoS Biol. 2020, 18, e3000734. [Google Scholar] [CrossRef]
- Zhou, Z.; Tang, A.T.; Wong, W.-Y.; Bamezai, S.; Goddard, L.M.; Shenkar, R.; Zhou, S.; Yang, J.; Wright, A.C.; Foley, M.; et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3–KLF2/4 signalling. Nature 2016, 532, 122–126. [Google Scholar] [CrossRef] [Green Version]
- Cuttano, R.; Rudini, N.; Bravi, L.; Corada, M.; Giampietro, C.; Papa, E.; Morini, M.F.; Maddaluno, L.; Baeyens, N.; Adams, R.H.; et al. KLF 4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO Mol. Med. 2016, 8, 6–24. [Google Scholar] [CrossRef]
- Lu, J.; Zhou, N.; Yang, P.; Deng, L.; Liu, G. MicroRNA-27a-3p Downregulation Inhibits Inflammatory Response and Hippocampal Neuronal Cell Apoptosis by Upregulating Mitogen-Activated Protein Kinase 4 (MAP2K4) Expression in Epilepsy: In Vivo and In Vitro Studies. Med. Sci. Monit. 2019, 25, 8499–8508. [Google Scholar] [CrossRef] [PubMed]
- Xi, T.; Jin, F.; Zhu, Y.; Wang, J.; Tang, L.; Wang, Y.; Liebeskind, D.S.; Scalzo, F.; He, Z. miR-27a-3p protects against blood–brain barrier disruption and brain injury after intracerebral hemorrhage by targeting endothelial aquaporin-11. J. Biol. Chem. 2018, 293, 20041–20050. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Zhao, M.; Wang, Y.; Liu, A.; Lv, M.; Li, Y.; Yang, X.; Wu, Z. Neuroprotective effects of miR-27a against traumatic brain injury via suppressing FoxO3a-mediated neuronal autophagy. Biochem. Biophys. Res. Commun. 2017, 482, 1141–1147. [Google Scholar] [CrossRef]
- Sabirzhanov, B.; Zhao, Z.; Stoica, B.A.; Loane, D.J.; Wu, J.; Borroto, C.; Dorsey, S.G.; Faden, A.I. Downregulation of miR-23a and miR-27a following Experimental Traumatic Brain Injury Induces Neuronal Cell Death through Activation of Proapoptotic Bcl-2 Proteins. J. Neurosci. 2014, 34, 10055–10071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raoof, R.; Bauer, S.; El Naggar, H.; Connolly, N.; Brennan, G.P.; Brindley, E.; Hill, T.; McArdle, H.; Spain, E.; Forster, R.J.; et al. Dual-center, dual-platform microRNA profiling identifies potential plasma biomarkers of adult temporal lobe epilepsy. EBioMedicine 2018, 38, 127–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasmeen, S.; Kaur, S.; Mirza, A.H.; Brodin, B.; Pociot, F.; Kruuse, C. miRNA-27a-3p and miRNA-222-3p as Novel Modulators of Phosphodiesterase 3a (PDE3A) in Cerebral Microvascular Endothelial Cells. Mol. Neurobiol. 2019, 56, 5304–5314. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Volsko, C.; Garcia, J.P.; Agirre, E.; Allan, K.C.; Tesar, P.; Trapp, B.D.; Castelo-Branco, G.; Sim, F.J.; Dutta, R. Oligodendrocyte Intrinsic miR-27a Controls Myelination and Remyelination. Cell Rep. 2019, 29, 904–919.e9. [Google Scholar] [CrossRef] [Green Version]
- Abdel, R.H.; Kholoussi, N.M.; Eissa, E.; El Nady, H.G.; Fayed, D.B.; Abdelkawy, R.F.M. MicroRNAs as Immune Regulators of Inflammation in Children with Epilepsy. Int. J. Mol. Cell Med. 2020, 9, 188–197. [Google Scholar] [CrossRef]
- Liu, D.-Z.; Tian, Y.; Ander, B.P.; Xu, H.; Stamova, B.S.; Zhan, X.; Turner, R.; Jickling, G.; Sharp, F.R. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J. Cereb. Blood Flow Metab. 2010, 30, 92–101. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, L.; Yan, G.; Zhang, W.; Huan, Z.; Li, J. MiR-125a-5p Alleviates Dysfunction and Inflammation of Pentylenetetrazol- induced Epilepsy Through Targeting Calmodulin-dependent Protein Kinase IV (CAMK4). Curr. Neurovascular Res. 2019, 16, 365–372. [Google Scholar] [CrossRef]
- Tiedt, S.; Prestel, M.; Malik, R.; Schieferdecker, N.; Duering, M.; Kautzky, V.; Stoycheva, I.; Böck, J.; Northoff, B.; Klein, M.; et al. RNA-Seq Identifies Circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as Potential Biomarkers for Acute Ischemic Stroke. Circ. Res. 2017, 121, 970–980. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Ma, C.; Wang, Y.; Wang, J.; Zheng, J.; Du, D.; Liao, X.; Chen, Y.; Chen, Y.; Bihl, J.; et al. Microvesicles-mediated communication between endothelial cells modulates, endothelial survival, and angiogenic function via transferring of miR-125a-5p. J. Cell. Biochem. 2019, 120, 3160–3172. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Liao, X.; Liu, H.; Wang, Y.; Chen, Y.; Zhao, B.; Lazartigues, E.; Yang, Y.; Ma, X. MicroRNA-125a-5p alleviates the deleterious effects of ox-LDL on multiple functions of human brain microvessel endothelial cells. Am. J. Physiol. Cell Physiol. 2017, 312, C119–C130. [Google Scholar] [CrossRef]
- Koskimäki, J.; Zhang, D.; Li, Y.; Saadat, L.; Moore, T.; Lightle, R.; Polster, S.; Carrión-Penagos, J.; Lyne, S.B.; Zeineddine, H.A.; et al. Transcriptome clarifies mechanisms of lesion genesis versus progression in models of Ccm3 cerebral cavernous malformations. Acta Neuropathol. Commun. 2019, 7, 132. [Google Scholar] [CrossRef]
- Wei, T.; Song, J.; Xu, M.; Lv, L.; Liu, C.; Shen, J.; Huang, Y. NEURL rs6584555 and CAND2 rs4642101 contribute to postoperative atrial fibrillation: A prospective study among Chinese population. Oncotarget 2016, 7, 42617–42624. [Google Scholar] [CrossRef] [Green Version]
- Shiraishi, S.; Zhou, C.; Aoki, T.; Sato, N.; Chiba, T.; Tanaka, K.; Yoshida, S.; Nabeshima, Y.; Nabeshima, Y.-I.; Tamura, T.-A. TBP-interacting Protein 120B (TIP120B)/Cullin-associated and Neddylation-dissociated 2 (CAND2) Inhibits SCF-dependent Ubiquitination of Myogenin and Accelerates Myogenic Differentiation. J. Biol. Chem. 2007, 282, 9017–9028. [Google Scholar] [CrossRef] [Green Version]
- Goldenberg, S.J.; Cascio, T.C.; Shumway, S.D.; Garbutt, K.C.; Liu, J.; Xiong, Y.; Zheng, N. Structure of the Cand1-Cul1-Roc1 Complex Reveals Regulatory Mechanisms for the Assembly of the Multisubunit Cullin-Dependent Ubiquitin Ligases. Cell 2004, 119, 517–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaue, T.; Sakakibara, I.; Uesugi, T.; Fujisaki, A.; Nakashiro, K.-I.; Hamakawa, H.; Kubota, E.; Joh, T.; Imai, Y.; Izutani, H.; et al. The CUL3-SPOP-DAXX axis is a novel regulator of VEGFR2 expression in vascular endothelial cells. Sci. Rep. 2017, 7, 42845. [Google Scholar] [CrossRef]
- Sakaue, T.; Fujisaki, A.; Nakayama, H.; Maekawa, M.; Hiyoshi, H.; Kubota, E.; Joh, T.; Izutani, H.; Higashiyama, S. Neddylated Cullin 3 is required for vascular endothelial-cadherin-mediated endothelial barrier function. Cancer Sci. 2017, 108, 208–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.-W.; Lo, H.-H.; Chiu, Y.-L.; Chang, S.-J.; Huang, P.-H.; Liao, K.-H.; Tasi, C.-F.; Wu, C.-H.; Tsai, T.-N.; Cheng, C.-C.; et al. Dysregulated miR-361-5p/VEGF Axis in the Plasma and Endothelial Progenitor Cells of Patients with Coronary Artery Disease. PLoS ONE 2014, 9, e98070. [Google Scholar] [CrossRef] [Green Version]
- Hemond, C.C.; Healy, B.C.; Tauhid, S.; Mazzola, M.A.; Quintana, F.J.; Gandhi, R.; Weiner, H.L.; Bakshi, R. MRI phenotypes in MS. Neurol. Neuroimmunol. Neuroinflamm. 2019, 6, e530. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Long, G.; Zhao, C.; Li, H.; Chaugai, S.; Wang, Y.; Chen, C.; Wang, D.W. Atherosclerosis-Related Circulating miRNAs as Novel and Sensitive Predictors for Acute Myocardial Infarction. PLoS ONE 2014, 9, e105734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, T.F.; Piergiorge, R.M.; Dos Santos, J.M.; Gusmao, J.; Pimentel, M.; Santos-Rebouças, C.B. Network Profiling of Brain-Expressed X-Chromosomal MicroRNA Genes Implicates Shared Key MicroRNAs in Intellectual Disability. J. Mol. Neurosci. 2019, 67, 295–304. [Google Scholar] [CrossRef]
- Lulli, V.; Buccarelli, M.; Ilari, R.; Castellani, G.; De Dominicis, C.; Di Giamberardino, A.; D′alessandris, Q.G.; Giannetti, S.; Martini, M.; Stumpo, V.; et al. Mir-370-3p Impairs Glioblastoma Stem-Like Cell Malignancy Regulating a Complex Interplay between HMGA2/HIF1A and the Oncogenic Long Non-Coding RNA (lncRNA) NEAT1. Int. J. Mol. Sci. 2020, 21, 3610. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wu, T.; Li, Y.; Xu, Z.; Zhang, S.; Liu, B.; Chen, Q.; Tian, D. MicroRNA-370-3p inhibits human glioma cell proliferation and induces cell cycle arrest by directly targeting β-catenin. Brain Res. 2016, 1644, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zong, J.; Zhao, C. lncRNA CTBP1-AS2 promotes proliferation and migration of glioma by modulating miR-370-3p–Wnt7a-mediated epithelial–mesenchymal transition. Biochem. Cell Biol. 2020, 98, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Visitchanakun, P.; Tangtanatakul, P.; Trithiphen, O.; Soonthornchai, W.; Wongphoom, J.; Tachaboon, S.; Srisawat, N.; Leelahavanichkul, A. Plasma miR-370-3P as a Biomarker of Sepsis-Associated Encephalopathy, the Transcriptomic Profiling Analysis of Microrna-Arrays from Mouse Brains. Shock 2019, 54, 347–357. [Google Scholar] [CrossRef]
- Ebrahimkhani, S.; Vafaee, F.; Young, P.E.; Hur, S.S.J.; Hawke, S.; Devenney, E.; Beadnall, H.; Barnett, M.H.; Suter, C.M.; Buckland, M.E. Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci. Rep. 2017, 7, 14293. [Google Scholar] [CrossRef] [Green Version]
- Plata-Bello, J.; Fariña-Jerónimo, H.; Betancor, I.; Salido, E. High Expression of FOXP2 Is Associated with Worse Prognosis in Glioblastoma. World Neurosurg. 2021, in press. [Google Scholar] [CrossRef]
- RahulAgrawal, R.; Pandey, P.; Jha, P.; Dwivedi, V.; Sarkar, C.; Kulshreshtha, R. Hypoxic signature of microRNAs in glioblastoma: Insights from small RNA deep sequencing. BMC Genom. 2014, 15, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Piwecka, M.; Rolle, K.; Belter, A.; Barciszewska, A.M.; Żywicki, M.; Michalak, M.; Nowak, S.; Naskręt-Barciszewska, M.Z.; Barciszewski, J. Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Mol. Oncol. 2015, 9, 1324–1340. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Jiao, B.-H.; Fan, F.-S.; Lu, S.-K.; Song, J.; Guo, C.-Y.; Yang, J.-K.; Yang, L. Downregulation of miR-95-3p inhibits proliferation, and invasion promoting apoptosis of glioma cells by targeting CELF2. Int. J. Oncol. 2015, 47, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.J.; Lee, H.W.; Kim, H.R.; Song, H.J.; Lee, D.H.; Lee, H.; Shin, C.H.; Joung, J.-G.; Kim, D.-H.; Joo, K.M.; et al. Overexpression of microRNA-95-3p suppresses brain metastasis of lung adenocarcinoma through downregulation of cyclin D1. Oncotarget 2015, 6, 20434–20448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Author | miRNA | Location | Expression in BAVM | Putative Targets (BAVM) | Known Cerebral Pathologies Associated (Expression)—Respective Putative Target(s) |
|---|---|---|---|---|---|
| Chen et al., 2018 [18] | miR-7-5p | Peripheral blood | Upregulated | VEGF | Glioma, glioblastoma (downregulated)—RAF Ischemia-reperfusion injury Hemorrhagic stroke (downregulated)—PI3K/AKT pathway |
| Ferreira et al., 2014 [19] | miR-18a | BAVM BEC | Downregulated | Id-1, TSP-1 (indirect) | Pediatric medulloblastoma, ependymoma, astrocytoma (upregulated)—RUNX1 Glioblastoma (upregulated)—neogenin Moyamoya-like vasculopathy (downregulated) |
| Marin-Ramos et al., 2020 [46] | PAI-1, BMP4, HIF-1A | ||||
| Huang et al., 2017 [20] | miR-137 | VSMC | Downregulated | Akt, p38MAPK, PI3K, ERK, VEGF, NFkB | Ischemic stroke—lncRNA GAS5//JAK1 and STAT 1//Src and MAPK Schizophrenia (upregulated) Gliomas (downregulated)—Rac1 or Cox-2 Oligodendrogliomas (downregulated)—CSE1L |
| Huang et al., 2017 [20] | miR-195* | VSMC | Downregulated | Akt, p38MAPK, PI3K, ERK, VEGF, NFkB | Ischemic stroke—VEGFA schizophrenia (upregulated)—BDNF Alzheimer’s disease (downregulated)—BACE1//KLF5 signalling//NFkB Gliomas (downregulated) Malignant meningioma (downregulated)—FASN |
| Chen et al., 2018 [18] | miR-199a-5p | Peripheral blood | Upregulated | VEGF | Glioma (upregulated)—MAGT1//MARCH8 Testicular germ cell tumors (downregulated) Ischemic stroke (upregulated)—CAV-1 mediated MEK/ERK pathway//DDR1//ECE1 Hemangioma—HIF-1A |
| Chen et al., 2018 [18] | miR-200b-3p | Peripheral blood | Upregulated | VEGF | Ischemic stroke (upregulated) Medulloblastoma (upregulated) Gastric adenocrcinoma metastasis (upregulated)—ZEB2 Glioma (downregulated)—ERK5 Alzheimer’s disease mouse models (downregulated)—APP Cytomegalus infection (upregulated) |
| Chen et al., 2018 [18] | let-7b-5p | Peripheral blood | Upregulated | NS | CCM (downregulated) Epilepsy (downregulated) Ischemic stroke models (downregulated) Glioma (downregulated)—IKBKE, E2F2, oncogenes KRAS, HMGA2, and MYC |
| Author | miRNA | Location | Expression in CCM | Putative Targets | Known Cerebral Pathologies Associated (Expression)—Respective Putative Target(s) |
|---|---|---|---|---|---|
| Li et al., 2020 [105] | miR-27a | BEC (mouse model) | Upregulated | VE-cadherin | Temporal lobe epilepsy (upregulated)—GLRA2 Neuroprotection in rat models after TBI—Bax, Noxa, and Puma//FoxO3a |
| Li et al., 2020 [105] | miR-125a | Downregulated | NS | Epilepsy—CAMK4 Ischemic stroke | |
| Koskimäki et al., 2019 [121] | mmu-miR-3472a | Mouse NVU | Dysregulated | CAND2 | NS |
| Kar et al., 2017 [97] | miR-361-5p | Brainstem CCM | Downregulated | VEGF, EGFL7 | Acute myocardial infarction Ischemic stroke Pulmonary embolism Multiple sclerosis Intellectual disability |
| Kar et al., 2017 [97] | miR-370-3p | Downregulated | NF1 | Glioma (downregulated)—HMGA2, HIF-1A, NEAT1//CTBP1-AS2 Sepsis associated encepahlopathy in mouse models | |
| Kar et al., 2017 [97] | miR-181a-2-3p | Downregulated | NS | Glioblastoma (downregulated)—FOXP2 | |
| Kar et al., 2017 [97] | miR-95-3p | Downregulated | NS | Glioma (upregulated)—CELF2 Lung cell adenocarcinoma metastasis (downregulated)—cyclin D1 | |
| Kar et al., 2017 [97] | let-7b-5p | Downregulated | ENG, HES1, HIF-1A, MAPK1, MIB1, OCLN, PDCD10, TJP1 | BAVM (upregulated) Epilepsy (downregulated) Ischemic stroke models (downregulated) Glioma (downregulated)—IKBKE, E2F2, oncogenes KRAS, HMGA2 and MYC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Florian, I.A.; Buruiana, A.; Timis, T.L.; Susman, S.; Florian, I.S.; Balasa, A.; Berindan-Neagoe, I. An Insight into the microRNAs Associated with Arteriovenous and Cavernous Malformations of the Brain. Cells 2021, 10, 1373. https://doi.org/10.3390/cells10061373
Florian IA, Buruiana A, Timis TL, Susman S, Florian IS, Balasa A, Berindan-Neagoe I. An Insight into the microRNAs Associated with Arteriovenous and Cavernous Malformations of the Brain. Cells. 2021; 10(6):1373. https://doi.org/10.3390/cells10061373
Chicago/Turabian StyleFlorian, Ioan Alexandru, Andrei Buruiana, Teodora Larisa Timis, Sergiu Susman, Ioan Stefan Florian, Adrian Balasa, and Ioana Berindan-Neagoe. 2021. "An Insight into the microRNAs Associated with Arteriovenous and Cavernous Malformations of the Brain" Cells 10, no. 6: 1373. https://doi.org/10.3390/cells10061373
APA StyleFlorian, I. A., Buruiana, A., Timis, T. L., Susman, S., Florian, I. S., Balasa, A., & Berindan-Neagoe, I. (2021). An Insight into the microRNAs Associated with Arteriovenous and Cavernous Malformations of the Brain. Cells, 10(6), 1373. https://doi.org/10.3390/cells10061373






