Abstract
Small double-strand RNA (dsRNA) molecules can activate endogenous genes via an RNA-based promoter targeting mechanism. RNA activation (RNAa) is an evolutionarily conserved mechanism present in diverse eukaryotic organisms ranging from nematodes to humans. Small activating RNAs (saRNAs) involved in RNAa have been successfully used to activate gene expression in cultured cells, and thereby this emergent technique might allow us to develop various biotechnological applications, without the need to synthesize hazardous construct systems harboring exogenous DNA sequences. Accordingly, this thematic issue aims to provide insights into how RNAa cellular machinery can be harnessed to activate gene expression leading to a more effective clinical treatment of various diseases.
1. Introduction
Small interfering RNAs (siRNAs) and microRNAs (miRNAs), key regulators of gene expression, are recognized as small double-stranded RNA (dsRNA) molecules [1,2,3]. They are loaded onto Ago proteins generating an active Ago–RNA complex, which regulates gene expression at transcription and post-transcription levels [4,5]. Several studies have demonstrated that dsRNAs mostly inhibit gene expression either by chromatin modifications or by translation inhibition [6,7,8,9]. A new kind of small dsRNA, small activating RNAs (saRNAs), are able to induce rather than inhibit gene expression by targeting promoter sequences of some genes, which is termed “RNA activation” (RNAa) [10,11,12,13]. saRNAs are 21 nucleotides in length and act in an Ago2-dependent manner in mammals similar to RNAi [11]. Although the first reports showed that gene expression can be activated by synthetic dsRNAs targeting promoter regions, more recent studies confirmed that RNAa is an evolutionarily conserved mechanism present in diverse eukaryotic organisms ranging from nematodes to humans [14,15]. Like small double-stranded activating RNAs, it has been evidenced that some miRNAs activate gene expression through targeting both promoter sequence and/or AU-rich elements in UTRs [11,16,17]. Several independent studies have shown that a new group of activating RNAs could be generated from natural antisense transcripts (NATs) [14,15,16]. Antisense transcripts, known as long non-coding RNAs, are transcribed from the opposite DNA strand of a protein-coding gene locus, which is complementary to the corresponding coding RNA. They are functional elements, expressed in a tissue-specific manner, and generally low in abundance [17]. Recent reports demonstrate that antisense transcripts regulate their sense (protein-coding) partners through diverse transcriptional and post-transcriptional mechanisms [18,19]. While most antisense transcripts suppress corresponding sense gene expression, they can also be regarded as targets for saRNAs [13,15]. Notably, blocking the interaction of the sense (mRNA) and antisense transcripts (forming a natural duplex) and/or destruction of antisense transcripts by siRNAs or single-stranded oligonucleotides (antagoNATs) results in locus-specific transcriptional de-repression and upregulation of the gene, possibly by activation of the RNAi machinery [14,15,16]. Apart from promoter sequences, which are known as the main target sites for saRNAs, gene activation by small RNA fragments could operate in any genomic region where antisense transcripts are presented. Despite tremendous advantages offered by saRNAs, there are still critical challenges limiting the development of these cutting-edge therapeutics. The most significant challenges include low siRNA stability, degradation and opsonization in the bloodstream, and off-target effects (Table 1). Nanoparticle-mediated delivery systems can open a new way to overcome these problems.

Table 1.
Pros and cons of using saRNAs as therapeutics.
In this review, we present recent advances and challenges in the therapeutic applications of saRNAs.
2. Small Activating RNAs Are Involved in Locus-Specific Induction of Neural Genes
saRNA-mediated locus-specific gene activation has been studied in neural cells. Based on many studies, two models of locus-specific gene activation have been reported, promoter-targeted duplex RNA activating and natural antisense disruption, leading to changes in chromatin structure [8,14,15,20]. Several studies show that endogenous saRNAs selectively activate gene expression in neurons through targeting promoter sequences [21,22]. Kuwabara et al. have reported a neural-specific dsRNA of about 20 bp containing the NRSE (neuron restrictive silencer element) sequence. This sequence, which is defined as the NRSE/RE1, is localized within promoter regions of neuron-specific genes and is recognized by neuronal restricted silencing factor/RE-1 silencing transcription factor (NRSF/REST) leading to neuron-specific gene suppression in a non-neuronal cell. During an early stage of neurogenesis, the NRSE dsRNA induces the expression of genes containing the NRSE/RE1 sequence in their promoters. Indeed, the noncoding dsRNA, as an endogenous activating RNA, interacts with NRSF/REST machinery and modulates its function. Through this process, neural stem cells can be differentiated into neuronal and glial cells [21]. Cell-mediated brain repair suffers from poor survival rate of transplanted cells and the low efficiency of differentiation into neuronal cells [23]. Diodato and his colleagues have used pre-miRNAs (as activating RNAs) to increase the expression of Emx2, a human homeobox transcription factor modulating a number of developmental mechanisms such as development of cerebral cortex. Their results showed that the transactivation of Emx2 can result in delayed differentiation, self-renewal, and decreased death of neuronally committed precursors [24]. Exogenous saRNA-mediated gene activation in the brain has been reported by Fimiani and his co-workers [22]. Synthesized saRNAs induce the Foxg1 transcription factor, a key regulator of cortico-cerebral development and function. Foxg1 allele duplication and deletion in humans results in West and Rett syndromes, respectively [25]. As a prospective RNAa therapy of Rett syndrome, Foxg1 gene expression in neural cells has been induced in vitro and in vivo by intraventricular injection of saRNA to mouse neonates [22]. NATs destruction is another alternative locus-specific gene activation mechanism, which holds great therapeutic promise. For example, Modarresi et al. have studied the regulatory role of a natural antisense transcript in the brain-derived neurotrophic factor (BDNF) locus [14]. BDNF is a member of the neurotrophin growth factor family and is essential for neuronal maturation, plasticity, memory processes, and differentiation [26,27,28,29,30,31]. Interestingly, locus-specific induction of Bdnf expression is evidenced, both in vitro and in vivo, following degradation of a natural antisense transcript by single-stranded oligonucleotides (term antagoNAT) or siRNAs [14]. Induction of endogenous Bdnf expression following Bdnf-AS repression in neurospheres induced neuronal progenitor cell differentiation. Corresponding to these in vitro experiments, in vivo Bdnf mRNA and protein expression is also induced upon intracerebroventricular injection of Bdnf-antagoNAT9 in mice. It seems that in the case of Bdnf induction, a RNAa mechanism applies through regulating epigenetic modifications, namely reduction of the H3K27me3 repressive mark at the Bdnf locus. Transient induction of neurotrophin expression using the RNAa system is suggested as a pharmacological approach for several neurological disorders, as reduced neurotrophin expression has been observed in different neurodegenerative and neurodevelopmental disorders [14].
Spinal muscular atrophy (SMA) is another example of a neuromuscular disorder [32]. Insufficient expression of functional survival motor neuron protein (SMN), which is correlated with disease severity, leads to muscle weakness after birth [33,34,35]. Several therapeutic efforts have focused on increasing SMN expression [32]. d’Ydewalle et al. have identified a natural antisense transcript in SMN (SMN-AS) locus, which transcriptionally suppresses SMN expression through epigenetic modifications. Importantly, knockdown of the antisense transcript induces SMN transcriptional activity either in patient-derived cells or in the central nervous system of a SMA mouse model in vivo, improving survival of the mice and indicating a novel therapeutic target for SMA [36].
We believe that the next step for clinical translation of RNAa therapeutics for the treatment of various neurodegenerative and neurodevelopmental disorders is the development of novel drug delivery systems.
3. Small Activating RNAs Are Involved in Locus-Specific Induction of Cardiac Genes
RNAa-mediated locus-specific activation of gene transcription has been reported in cardiovascular cells [37,38,39]. In all reported studies, targeting of promoter regions by small dsRNA or small hairpin RNA (shRNA) leads to transcriptional activation of cardiovascular genes, which could open the way for therapeutic strategies. For instance, targeting the promoter region of vascular endothelial growth factor (VEGF) by shRNA results in transcriptional activation, suggesting a therapeutic strategy for myocardial infarction [37]. Targeting the antisense transcript has been recently demonstrated as an alternative RNA activating system in the cyclin-dependent kinase 9 (Cdk9) locus, a key player in cardiac development [15,16]. Cdk9 is associated with specific cyclins to form a heterodimer, Cyclin T/Cdk9, which is also known as the positive transcription elongation factor-b (P-TEFb) [40,41,42]. P-TEFb activates the polymerase II transcription machinery via phosphorylation of the carboxyl-terminal domain (CTD) [43,44]. Therefore, Cdk9 is mainly involved in transcriptional regulation and plays a critical role in several differentiation pathways. Furthermore, Cdk9 regulates cardiac-specific genes including Nkx2.5, Anf, and ß-Myh via interactions with the p300/GATA4 complex, particularly involved in cardiac differentiation [45]. Moreover, we have shown recently that Cdk9 regulates apoptosis in cardiomyocytes by modulating miRNA-1 expression, a critical microRNA for cardiac differentiation [46,47,48]. It is therefore possible that both synthesis and activity of Cdk9 are tightly regulated at the transcriptional and post-transcriptional levels. In this regard, at least three non-coding RNAs are involved in Cdk9 regulation [16]. In the context of normal human cardiomyocytes, Cdk9 activity is suppressed at the protein level via interaction with 7SK non-coding RNA and at the translational level through muscle-specific microRNAs, specifically miR-1 and miR-133 [46,49,50]. We have recently reported a third mode of RNA control in the Cdk9 locus [15]. Small non-coding RNA molecules of 22bp with sequences homologous to the transcript result in transcriptional activation of Cdk9. Interestingly, NATs complementary to the most 3′ and 5′ regions of the gene were identified. Indeed, hybridization of the short single-stranded cognate transcript fragments with antisense transcripts provides the signal for transcriptional activation. The requirement of Argonaute proteins and endogenous antisense transcripts for transcriptional activation indicates that the activating single-stranded small RNAs are processed by the RNAi machinery [51,52]. Similar to siRNA knockdown, antisense transcript distraction following the sense oligoribonucleotide electroporation could represent a secondary phenomenon of the activation of the RNAi machinery. This activation may then result in a change in epigenetic modifications at the locus, leading to the induction of Cdk9 transcription as described for several genes [1,53]. As a functional consequence of RNA activation in the Cdk9 locus, an increased cardiac differentiation potential is observed in ES cells when electroporated with the sense oligoribonucleotide. Interestingly, injection into wild-type blastocysts of RNA-programmed ES cells contributes specifically to heart development in vivo, indicating that a transient RNA activation system is sufficient to create a cardiac differentiation “memory” in cells and may represent a novel tool for RNA–cell reprogramming applied in regenerative medicine.
4. Small Activating RNAs: New Insights into Cancer Therapy
Recently, RNA-based therapeutics have gained more attention in cancer therapy due to their enormous potential to selectively target previously undruggable genes and gene expression modulators. Unlike RNAi, which mostly targets sense transcripts, RNAa, as an alternative and promising new therapeutic strategy, can activate gene expression in a natural manner by targeting antisense transcripts or promoter regions [13,54,55,56]. Tumor suppressor genes, which are mostly suppressed in cancers, could be targeted by saRNA to enhance transcriptional activation and restore a normal cell phenotype.
saRNAs can exert their anticancer effects through induction of cell cycle arrest, cellular senescence, proliferation inhibition, apoptosis induction, metastasis suppression, and multidrug resistance reversal (Figure 1). Although several tumor suppressor genes including E-cadherin, NKX3-1, Wt1, and P53 have been induced by this method, p21 is the most investigated tumor suppressor gene for RNAa-mediated gene activation in several tumors and cell lines [10,57,58,59,60,61,62]. P21 is a negative regulator of the cell cycle and is rarely mutated in cancers, and therefore represents a key target for small RNA activation cancer therapy. In this regard, re-activation of the p21 gene by targeting promoter regions inhibits cell viability and proliferation rates, while it induces apoptotic cell death and sensitizes lung cancer cells to chemotherapeutic agents, providing a new approach in cancer therapy [59,60,63].
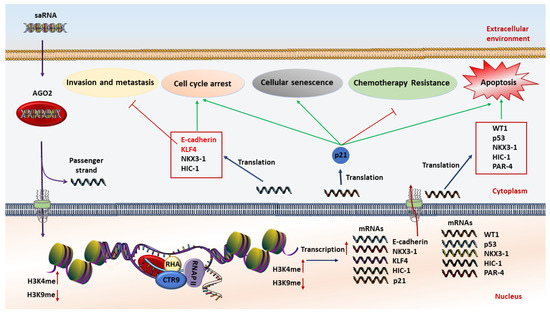
Figure 1.
Schematic illustration of the anticancer mechanism of saRNA (small activating RNA)-based therapeutics. At first, saRNAs are loaded on the AGO2 protein. Then AGO2 separates the passenger strand. After that, the complex of saRNA guide strand and AGO2 cross the nuclear and interact with promoter sequences of interested genes to increase transcription by methylation of H3K4 and/or demethylation of H3K9. The expression level of tumor suppressor genes is restored, resulting in induction of apoptosis, chemoresistance reversal, inhibition of invasion and metastasis, cell cycle arrest, and cellular senescence.
5. Towards the Development of New Therapeutic Agents
In the past few years, a steadily increasing number of clinical and preclinical studies have been performed, using various saRNA-based therapies for the treatment of a multitude of different diseases (Table 2 and Supplementary Table S1).

Table 2.
Significant preclinical and clinical studies on the features of saRNA-based therapeutics.
Due to their potential involvement in human disorders, strong efforts have been undertaken to develop new therapeutic agents applying a RNAa strategy. For instance, two companies in the USA, RNAa Therapeutics and OPKO-CURNA, are investigating therapeutic approaches based on RNA activation. Of note, mipomersen, an oligonucleotide targeting apolipoprotein B, was approved by the FDA in January 2013, but received a negative opinion from a European Medicines Agency panel [90]. RNA-activating molecules as promising drugs are under active investigation due to their high potency and specificity, locus-specific manner functions, targeting of the correct cells, small molecular size, and low toxicity [10,91]. Traditionally, an exogenous DNA construct is often required for ectopic gene expression. Apart from exogenous DNA constructs excluding regulatory elements, such systems have been problematic in the clinic because of requiring viral-based vectors for gene delivery. Several limitations and undesirable side effects of viral-based gene therapy including host genome integrity, the tedious process of construction, and various negative immunological responses which have been previously reported [92,93,94,95]. Upon delivery as mature moieties, an RNAa system offers a more natural approach and safer gene therapy method by targeting promoter regions or through natural antisense destruction, which activates endogenous targeted gene expression in a locus-specific manner in the absence of exogenous DNA. Moreover, RNAa offers an endogenous induction in a natural cellular niche leading to the correctly processed proteins with their appropriate modifications. Notably, in the case of gene activation through natural antisense destruction, short oligoribonucleotides derived from a sense transcript (mRNA) can function as an activator RNA [15], which are safer, natural, and without activating immune responses.
To improve the medicinal properties and potential in vivo applications of saRNAs, two steps must be taken. First, novel nanoparticle-based drug delivery systems must be developed to increase the drug accumulation in the targeted tissue and also address the aforementioned challenges in the therapeutic application of saRNAs. To date, different delivery systems have been developed for the delivery of saRNAs. Recently, lipid-based nanoparticles (LNPs) have attracted worldwide attention. LNP-based delivery systems face serious limitations such as toxicity, low thermodynamic stability, poor efficiency of encapsulation, and leaking challenges [96]. To overcome these limitations, various strategies have been proposed. The surface modification of delivery systems with flexible, non-ionic, and hydrophilic polymers, such as PEG, has been proposed as a robust strategy to address serum protein opsonization issues [97,98]. Furthermore, surface modification with biodegradable nano-polymers such as PEG can be used to decrease the toxicity of nanoparticles [99]. To increase the expression of P21, 2′-fluoro-modified P21 saRNA (dsP21-322-2′F) was delivered into an orthotopic bladder cancer mouse model by using a novel PEG-modified lipid nanoparticle. Results showed a significant increase in urothelium uptake and high tumor shrinkage [80]. Aptamers, nucleic acid ligands, can be used for targeted drug delivery as they can form specific three-dimensional structures based on their sequences. Yoon and his colleagues have synthesized PDAC specific 2′-Fluropyrimidine RNA-aptamers (2′F-RNA)- P19 and P1 for targeted delivery of saRNA into both PANC-1 and AsPC-1 engrafted mice. After intravenous injections of the aptamer–C/EBPα saRNA in both tumor mice models, tumor growth was significantly suppressed in comparison with mice treated with gemcitabine [84].
The second step for the development of effective saRNA therapeutics relies on tailoring site-specific chemical modifications of saRNAs. For instance, blocking the 5′-OH of the passenger strand and modifications to the 2′ backbone (i.e., 2′-OMe, 2′-Fluoro, and locked nucleic acid) has been demonstrated to reduce its off-target potential and increase endonuclease resistance and serum stability, respectively [55,58,100]. With single-stranded therapeutic oligonucleotides, designated antagoNAT, chemical modifications not only promote metabolic stability and target specificity but also minimize the length of the oligonucleotide to improve cellular uptake. In this case, 16-mer antagoNAT oligonucleotides, also known as a gapmer [101], with three locked nucleic acid (LNA) substitutions at each end and phosphorothioate-modified backbones, have been used in in vivo studies [14,102,103,104,105]. Along with chemical modifications to improve the stability and specificity, targeted and efficient in vivo delivery of oligonucleotides is also critical for RNA-based therapies. Among the currently investigated approaches [106,107,108,109], lipid-based formulations are the most promising delivery agents for systemic or localized saRNA delivery [100,110,111]. Several clinical trials that are testing RNAa-based drugs are commonly using lipid carriers [112,113]. Considering the increasing knowledge regarding RNAa-based locus-specific gene activation as a new strategy with promising therapeutic perspectives, it is clear that there is a growing scientific as well as a commercial interest to develop new therapeutic agents and clinical treatments based on this innovative approach. Lipid nanoparticle-formulated nucleoside-modified RNAs have been recently introduced as the Covid-19 vaccine, supporting the notion of efficient lipid-based delivery, safety, and efficiency of RNA therapies [114,115].
6. Conclusions and Future Perspectives
RNAa-based drugs have gained great attention in the past few years owing to their high potential in treating various disorders. However, delivery challenges slow down the clinical translation of this new class of therapeutic agents. Enzymatic digestion, low cellular uptake, poor endosomal escape, off-target effects, and fast clearance from the bloodstream have necessitated optimization of not only the RNAa molecules but also of the delivery systems. Many non-viral delivery systems have been designed and developed for the delivery of RNAa-based therapeutic systems. Nanoparticle-based delivery systems have several advantages including high functionalization and targeting capacity and ease of large-scale development, which make them a promising delivery system for medical applications. For instance, several studies demonstrated that PEGylation of different materials such as PEI can enhance their circulation time and reduce hemolysis and serum opsonization. Chemical modifications of RNA molecules herald a new era in the development of these cutting-edge RNA-based therapeutics. However, these modifications might be limited due to the occurrence of severe side effects and functionality reduction, which are topics that urgently need to be addressed.
Future applications of RNAa-based therapeutics will significantly advance due to the progress in the development of drug delivery vehicles, reduction of the off-targeting problem, and huge large-scale synthesis capacity that will be offered by pharmaceutical companies. The combinatorial strategy using RNAa with chemotherapeutic drugs also could be a new strong treatment modality for different types of diseases.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/10/3/591/s1, Table S1: Significant preclinical and clinical studies on the features of saRNA-based therapeutics—Extended characteristics.
Author Contributions
Conceptualization, H.G. and K.-D.W.; writing—original draft preparation, H.G. and M.E.; writing—review and editing, S.A., N.W. and K.-D.W.; supervision, K.-D.W., H.G., and N.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors acknowledge the Shahid Beheshti University of Medical Sciences and the University Cote d’Azur for support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aghamiri, S.; Mehrjardi, K.F.; Shabani, S.; Keshavarz-Fathi, M.; Kargar, S.; Rezaei, N. Nanoparticle-siRNA: A potential strategy for ovarian cancer therapy? Nanomedicine 2019, 14, 2083–2100. [Google Scholar] [CrossRef] [PubMed]
- Setten, R.L.; Rossi, J.J.; Han, S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular Mechanisms and Biological Functions of siRNA. Int. J. Biomed Sci. 2017, 13, 48–57. [Google Scholar] [PubMed]
- Sun, G.; Yeh, S.Y.; Yuan, C.W.; Chiu, M.J.; Yung, B.S.; Yen, Y. Molecular Properties, Functional Mechanisms, and Applications of Sliced siRNA. Mol. Ther. Nucleic Acids 2015, 4, e221. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Volpe, T.; Martienssen, R.A. RNA interference and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 2011, 3, a003731. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Xiao, H.; Zhang, J.; Liang, X.J.; Huang, Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019, 37, 801–825. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, M.; Kang, M.R.; Yang, G.; Li, L.-C. Targeted p21WAF1/CIP1 Activation by RNAa Inhibits Hepatocellular Carcinoma Cells. Nucleic Acid Ther. 2012, 22, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Jiang, Q.; Chang, N.; Wang, X.; Liu, C.; Xiong, J.; Cao, H.; Liang, Z. Small activating RNA binds to the genomic target site in a seed-region-dependent manner. Nucleic Acids Res. 2016, 44, 2274–2282. [Google Scholar] [CrossRef]
- Wang, J.; Place, R.F.; Portnoy, V.; Huang, V.; Kang, M.R.; Kosaka, M.; Ho, M.K.C.; Li, L.C. Inducing gene expression by targeting promoter sequences using small activating RNAs. J. Biol. Methods 2015, 2. [Google Scholar] [CrossRef]
- Portnoy, V.; Huang, V.; Place, R.F.; Li, L.C. Small RNA and transcriptional upregulation. Wiley Interdiscip. Rev. RNA 2011, 2, 748–760. [Google Scholar] [CrossRef] [PubMed]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.P.; van der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarian, H.; Wagner, N.; Michiels, J.F.; Cuzin, F.; Wagner, K.D.; Rassoulzadegan, M. Small RNA-directed epigenetic programming of embryonic stem cell cardiac differentiation. Sci. Rep. 2017, 7, 41799. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarian, H.; Grandjean, V.; Cuzin, F.; Rassoulzadegan, M. A Network of Regulations by Small Non-Coding RNAs: The P-TEFb Kinase in Development and Pathology. Front. Genet. 2011, 2, 95. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhang, L.; Luo, W.; Zhang, X. Characteristics of Antisense Transcript Promoters and the Regulation of Their Activity. Int. J. Mol. Sci. 2015, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Kocerha, J.; Modarresi, F.; Engström, P.G.; Chalk, A.M.; Brothers, S.P.; Koesema, E.; St Laurent, G.; Wahlestedt, C. RNAi screen indicates widespread biological function for human natural antisense transcripts. PLoS ONE 2010, 5, e13177. [Google Scholar] [CrossRef]
- Kim, D.H.; Saetrom, P.; Snøve, O.; Rossi, J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16230–16235. [Google Scholar] [CrossRef]
- Kuwabara, T.; Hsieh, J.; Nakashima, K.; Taira, K.; Gage, F.H. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 2004, 116, 779–793. [Google Scholar] [CrossRef]
- Fimiani, C.; Goina, E.; Su, Q.; Gao, G.; Mallamaci, A. RNA activation of haploinsufficient Foxg1 gene in murine neocortex. Sci. Rep. 2016, 6, 39311. [Google Scholar] [CrossRef]
- Burns, T.C.; Verfaillie, C.M.; Low, W.C. Stem cells for ischemic brain injury: A critical review. J. Comp. Neurol. 2009, 515, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Diodato, A.; Pinzan, M.; Granzotto, M.; Mallamaci, A. Promotion of cortico-cerebral precursors expansion by artificial pri-miRNAs targeted against the Emx2 locus. Curr. Gene Ther. 2013, 13, 152–161. [Google Scholar] [CrossRef]
- Guerrini, R.; Parrini, E. Epilepsy in Rett syndrome, and CDKL5- and FOXG1-gene-related encephalopathies. Epilepsia 2012, 53, 2067–2078. [Google Scholar] [CrossRef] [PubMed]
- Ortega, J.A.; Alcántara, S. BDNF/MAPK/ERK-induced BMP7 expression in the developing cerebral cortex induces premature radial glia differentiation and impairs neuronal migration. Cereb. Cortex 2010, 20, 2132–2144. [Google Scholar] [CrossRef]
- Antal, A.; Chaieb, L.; Moliadze, V.; Monte-Silva, K.; Poreisz, C.; Thirugnanasambandam, N.; Nitsche, M.A.; Shoukier, M.; Ludwig, H.; Paulus, W. Brain-derived neurotrophic factor (BDNF) gene polymorphisms shape cortical plasticity in humans. Brain Stimul. 2010, 3, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.V.; Escobar, M.L. A role for MAPK and PI-3K signaling pathways in brain-derived neurotrophic factor modification of conditioned taste aversion retention. Behav. Brain Res. 2011, 217, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Nikolakopoulou, A.M.; Meynard, M.M.; Marshak, S.; Cohen-Cory, S. Synaptic maturation of the Xenopus retinotectal system: Effects of brain-derived neurotrophic factor on synapse ultrastructure. J. Comp. Neurol. 2010, 518, 972–989. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.C.; Maguschak, K.A.; Ye, K.; Jang, S.W.; Myers, K.M.; Ressler, K.J. Prelimbic cortical BDNF is required for memory of learned fear but not extinction or innate fear. Proc. Natl. Acad. Sci. USA 2010, 107, 2675–2680. [Google Scholar] [CrossRef] [PubMed]
- Chapleau, C.A.; Larimore, J.L.; Theibert, A.; Pozzo-Miller, L. Modulation of dendritic spine development and plasticity by BDNF and vesicular trafficking: Fundamental roles in neurodevelopmental disorders associated with mental retardation and autism. J. Neurodev. Disord. 2009, 1, 185–196. [Google Scholar] [CrossRef]
- d’Ydewalle, C.; Sumner, C.J. Spinal Muscular Atrophy Therapeutics: Where do we Stand? Neurotherapeutics 2015, 12, 303–316. [Google Scholar] [CrossRef]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Lorson, C.L.; Hahnen, E.; Androphy, E.J.; Wirth, B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 1999, 96, 6307–6311. [Google Scholar] [CrossRef]
- Lefebvre, S.; Burlet, P.; Liu, Q.; Bertrandy, S.; Clermont, O.; Munnich, A.; Dreyfuss, G.; Melki, J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997, 16, 265–269. [Google Scholar] [CrossRef] [PubMed]
- d’Ydewalle, C.; Ramos, D.M.; Pyles, N.J.; Ng, S.Y.; Gorz, M.; Pilato, C.M.; Ling, K.; Kong, L.; Ward, A.J.; Rubin, L.L.; et al. The Antisense Transcript SMN-AS1 Regulates SMN Expression and Is a Novel Therapeutic Target for Spinal Muscular Atrophy. Neuron 2017, 93, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Turunen, M.P.; Husso, T.; Musthafa, H.; Laidinen, S.; Dragneva, G.; Laham-Karam, N.; Honkanen, S.; Paakinaho, A.; Laakkonen, J.P.; Gao, E.; et al. Epigenetic upregulation of endogenous VEGF-A reduces myocardial infarct size in mice. PLoS ONE 2014, 9, e89979. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wang, T.; Rao, K.; Yang, J.; Zhang, S.; Wang, S.; Liu, J.; Ye, Z. Up-regulation of VEGF by small activator RNA in human corpus cavernosum smooth muscle cells. J. Sex Med. 2011, 8, 2773–2780. [Google Scholar] [CrossRef] [PubMed]
- Husso, T.; Ylä-Herttuala, S.; Turunen, M.P. A New Gene Therapy Approach for Cardiovascular Disease by Non-coding RNAs Acting in the Nucleus. Mol. Ther. Nucleic Acids 2014, 3, e197. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Taube, R.; Fujinaga, K.; Peterlin, B.M. P-TEFb containing cyclin K and Cdk9 can activate transcription via RNA. J. Biol. Chem. 2002, 277, 16873–16878. [Google Scholar] [CrossRef]
- Marshall, N.F.; Price, D.H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995, 270, 12335–12338. [Google Scholar] [CrossRef] [PubMed]
- Price, D.H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell Biol. 2000, 20, 2629–2634. [Google Scholar] [CrossRef]
- Leucci, E.; De Falco, G.; Onnis, A.; Cerino, G.; Cocco, M.; Luzzi, A.; Crupi, D.; Tigli, C.; Bellan, C.; Tosi, P.; et al. The role of the Cdk9/Cyclin T1 complex in T cell differentiation. J. Cell Physiol. 2007, 212, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Pevarello, P.; Barbacid, M.; Bischoff, J.R. CDK inhibitors in cancer therapy: What is next? Trends Pharmacol. Sci. 2008, 29, 16–21. [Google Scholar] [CrossRef]
- Kaichi, S.; Takaya, T.; Morimoto, T.; Sunagawa, Y.; Kawamura, T.; Ono, K.; Shimatsu, A.; Baba, S.; Heike, T.; Nakahata, T.; et al. Cyclin-dependent kinase 9 forms a complex with GATA4 and is involved in the differentiation of mouse ES cells into cardiomyocytes. J. Cell Physiol. 2011, 226, 248–254. [Google Scholar] [CrossRef]
- Tarhriz, V.; Wagner, K.D.; Masoumi, Z.; Molavi, O.; Hejazi, M.S.; Ghanbarian, H. CDK9 Regulates Apoptosis of Myoblast Cells by Modulation of microRNA-1 Expression. J. Cell Biochem. 2018, 119, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.D.; Wagner, N.; Ghanbarian, H.; Grandjean, V.; Gounon, P.; Cuzin, F.; Rassoulzadegan, M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev. Cell 2008, 14, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Olson, E.N.; Bassel-Duby, R. Mending broken hearts: Cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 2013, 14, 529–541. [Google Scholar] [CrossRef]
- Sayed, D.; Hong, C.; Chen, I.Y.; Lypowy, J.; Abdellatif, M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007, 100, 416–424. [Google Scholar] [CrossRef]
- Peterlin, B.M.; Brogie, J.E.; Price, D.H. 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 2012, 3, 92–103. [Google Scholar] [CrossRef]
- Mattick, J.S. The State of Long Non-Coding RNA Biology. Noncoding RNA 2018, 4, 17. [Google Scholar] [CrossRef]
- Mattick, J.S.; Rinn, J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, H.; Burnett, J.C.; Rossi, J.J. The role of antisense long noncoding RNA in small RNA-triggered gene activation. RNA 2014, 20, 1916–1928. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Okino, S.T.; Zhao, H.; Pookot, D.; Place, R.F.; Urakami, S.; Enokida, H.; Dahiya, R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA 2006, 103, 17337–17342. [Google Scholar] [CrossRef]
- Matsui, M.; Sakurai, F.; Elbashir, S.; Foster, D.J.; Manoharan, M.; Corey, D.R. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem. Biol. 2010, 17, 1344–1355. [Google Scholar] [CrossRef]
- Werner, A. Biological functions of natural antisense transcripts. BMC Biol. 2013, 11, 31. [Google Scholar] [CrossRef]
- Chen, Z.; Place, R.F.; Jia, Z.J.; Pookot, D.; Dahiya, R.; Li, L.C. Antitumor effect of dsRNA-induced p21(WAF1/CIP1) gene activation in human bladder cancer cells. Mol. Cancer Ther. 2008, 7, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Place, R.F.; Wang, J.; Noonan, E.J.; Meyers, R.; Manoharan, M.; Charisse, K.; Duncan, R.; Huang, V.; Wang, X.; Li, L.C. Formulation of Small Activating RNA Into Lipidoid Nanoparticles Inhibits Xenograft Prostate Tumor Growth by Inducing p21 Expression. Mol. Ther. Nucleic Acids 2012, 1, e15. [Google Scholar] [CrossRef]
- Wei, J.; Zhao, J.; Long, M.; Han, Y.; Wang, X.; Lin, F.; Ren, J.; He, T.; Zhang, H. p21WAF1/CIP1 gene transcriptional activation exerts cell growth inhibition and enhances chemosensitivity to cisplatin in lung carcinoma cell. BMC Cancer 2010, 10, 632. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.M.; Dai, C.; Huang, Y.; Zheng, C.F.; Dong, Q.Z.; Wang, G.; Li, X.W.; Zhang, X.F.; Li, B.; Chen, G. Anti-cancer effects of p21WAF1/CIP1 transcriptional activation induced by dsRNAs in human hepatocellular carcinoma cell lines. Acta Pharmacol. Sin. 2011, 32, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Frixen, U.H.; Behrens, J.; Sachs, M.; Eberle, G.; Voss, B.; Warda, A.; Löchner, D.; Birchmeier, W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J. Cell Biol. 1991, 113, 173–185. [Google Scholar] [CrossRef]
- Richards, F.M.; McKee, S.A.; Rajpar, M.H.; Cole, T.R.; Evans, D.G.; Jankowski, J.A.; McKeown, C.; Sanders, D.S.; Maher, E.R. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum. Mol. Genet. 1999, 8, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Zandsalimi, F.; Talaei, S.; Noormohammad Ahari, M.; Aghamiri, S.; Raee, P.; Roshanzamiri, S.; Yarian, F.; Bandehpour, M.; Zohrab Zadeh, Z. Antimicrobial peptides: A promising strategy for lung cancer drug discovery? Expert Opin. Drug Discov. 2020, 15, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Sarker, D.; Plummer, R.; Meyer, T.; Sodergren, M.H.; Basu, B.; Chee, C.E.; Huang, K.W.; Palmer, D.H.; Ma, Y.T.; Evans, T.R.J.; et al. MTL-CEBPA, a Small Activating RNA Therapeutic Upregulating C/EBP-α, in Patients with Advanced Liver Cancer: A First-in-Human, Multicenter, Open-Label, Phase I Trial. Clin. Cancer Res. 2020, 26, 3936–3946. [Google Scholar] [CrossRef]
- Kramer, E.D.; Abrams, S.I. Granulocytic Myeloid-Derived Suppressor Cells as Negative Regulators of Anticancer Immunity. Front. Immunol. 2020, 11, 1963. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Z.; Liu, X.; Wang, J.; Li, F.; Li, C.; Shan, B. Up-regulation of p21WAF1/CIP1 by small activating RNA inhibits the in vitro and in vivo growth of pancreatic cancer cells. Tumori 2012, 98, 804–811. [Google Scholar] [CrossRef]
- Yang, K.; Zheng, X.Y.; Qin, J.; Wang, Y.B.; Bai, Y.; Mao, Q.Q.; Wan, Q.; Wu, Z.M.; Xie, L.P. Up-regulation of p21WAF1/Cip1 by saRNA induces G1-phase arrest and apoptosis in T24 human bladder cancer cells. Cancer Lett. 2008, 265, 206–214. [Google Scholar] [CrossRef]
- Qin, Q.; Lin, Y.W.; Zheng, X.Y.; Chen, H.; Mao, Q.Q.; Yang, K.; Huang, S.J.; Zhao, Z.Y. RNAa-mediated overexpression of WT1 induces apoptosis in HepG2 cells. World J. Surg. Oncol. 2012, 10, 11. [Google Scholar] [CrossRef]
- Mao, Q.; Zheng, X.; Yang, K.; Qin, J.; Bai, Y.; Jia, X.; Li, Y.; Xie, L. Suppression of migration and invasion of PC3 prostate cancer cell line via activating E-cadherin expression by small activating RNA. Cancer Investig. 2010, 28, 1013–1018. [Google Scholar] [CrossRef]
- Mao, Q.; Li, Y.; Zheng, X.; Yang, K.; Shen, H.; Qin, J.; Bai, Y.; Kong, D.; Jia, X.; Xie, L. Up-regulation of E-cadherin by small activating RNA inhibits cell invasion and migration in 5637 human bladder cancer cells. Biochem. Biophys. Res. Commun. 2008, 375, 566–570. [Google Scholar] [CrossRef]
- Junxia, W.; Ping, G.; Yuan, H.; Lijun, Z.; Jihong, R.; Fang, L.; Min, L.; Xi, W.; Ting, H.; Ke, D.; et al. Double strand RNA-guided endogeneous E-cadherin up-regulation induces the apoptosis and inhibits proliferation of breast carcinoma cells in vitro and in vivo. Cancer Sci. 2010, 101, 1790–1796. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Place, R.F.; Huang, V.; Wang, X.; Noonan, E.J.; Magyar, C.E.; Huang, J.; Li, L.C. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010, 70, 10182–10191. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Meng, L.; Xu, F.; Lian, J.; Xu, Y.; Xie, X.; Wang, X.; He, H.; Wang, C.; Zhu, Y. Enhanced wild-type p53 expression by small activating RNA dsP53-285 induces cell cycle arrest and apoptosis in pheochromocytoma cell line PC12. Oncol. Rep. 2017, 38, 3160–3166. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, F.; Pan, S.; Gu, Y.; Guo, S.; Dai, Q.; Yu, Y.; Zhang, W. Small activating RNA restores the activity of the tumor suppressor HIC-1 on breast cancer. PLoS ONE 2014, 9, e86486. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Shen, J.; Xie, Y.Q.; Lin, Y.W.; Qin, J.; Mao, Q.Q.; Zheng, X.Y.; Xie, L.P. Promoter-targeted double-stranded small RNAs activate PAWR gene expression in human cancer cells. Int. J. Biochem. Cell Biol. 2013, 45, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Kang, M.R.; Wang, J.; Huang, V.; Place, R.F.; Sun, Y.; Li, L.C. Targeted induction of endogenous NKX3-1 by small activating RNA inhibits prostate tumor growth. Prostate 2013, 73, 1591–1601. [Google Scholar] [CrossRef]
- Zeng, T.; Duan, X.; Zhu, W.; Liu, Y.; Wu, W.; Zeng, G. SaRNA-mediated activation of TRPV5 reduces renal calcium oxalate deposition in rat via decreasing urinary calcium excretion. Urolithiasis 2018, 46, 271–278. [Google Scholar] [CrossRef]
- Kang, M.R.; Park, K.H.; Lee, C.W.; Lee, M.Y.; Han, S.B.; Li, L.C.; Kang, J.S. Small activating RNA induced expression of VHL gene in renal cell carcinoma. Int. J. Biochem. Cell Biol. 2018, 97, 36–42. [Google Scholar] [CrossRef]
- Xia, W.; Li, D.; Wang, G.; Ni, J.; Zhuang, J.; Ha, M.; Wang, J.; Ye, Y. Small activating RNA upregulates NIS expression: Promising potential for hepatocellular carcinoma endoradiotherapy. Cancer Gene Ther. 2016, 23, 333–340. [Google Scholar] [CrossRef]
- Kang, M.R.; Yang, G.; Place, R.F.; Charisse, K.; Epstein-Barash, H.; Manoharan, M.; Li, L.C. Intravesical delivery of small activating RNA formulated into lipid nanoparticles inhibits orthotopic bladder tumor growth. Cancer Res. 2012, 72, 5069–5079. [Google Scholar] [CrossRef]
- Reebye, V.; Sætrom, P.; Mintz, P.J.; Huang, K.W.; Swiderski, P.; Peng, L.; Liu, C.; Liu, X.; Lindkaer-Jensen, S.; Zacharoulis, D.; et al. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology 2014, 59, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Huan, H.; Wen, X.; Chen, X.; Wu, L.; Liu, W.; Habib, N.A.; Bie, P.; Xia, F. C/EBPα Short-Activating RNA Suppresses Metastasis of Hepatocellular Carcinoma through Inhibiting EGFR/β-Catenin Signaling Mediated EMT. PLoS ONE 2016, 11, e0153117. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Feng, C.L.; Zheng, W.S.; Huang, S.; Zhang, W.X.; Wu, H.N.; Zhan, Y.; Han, Y.X.; Wu, S.; Jiang, J.D. Tumor-selective lipopolyplex encapsulated small active RNA hampers colorectal cancer growth in vitro and in orthotopic murine. Biomaterials 2017, 141, 13–28. [Google Scholar] [CrossRef]
- Yoon, S.; Huang, K.W.; Reebye, V.; Mintz, P.; Tien, Y.W.; Lai, H.S.; Sætrom, P.; Reccia, I.; Swiderski, P.; Armstrong, B.; et al. Targeted Delivery of C/EBPα -saRNA by Pancreatic Ductal Adenocarcinoma-specific RNA Aptamers Inhibits Tumor Growth In Vivo. Mol. Ther. 2016, 24, 1106–1116. [Google Scholar] [CrossRef]
- Li, C.; Jiang, W.; Hu, Q.; Li, L.C.; Dong, L.; Chen, R.; Zhang, Y.; Tang, Y.; Thrasher, J.B.; Liu, C.B.; et al. Enhancing DPYSL3 gene expression via a promoter-targeted small activating RNA approach suppresses cancer cell motility and metastasis. Oncotarget 2016, 7, 22893–22910. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, M.; Yuan, H.; Zhan, Y.; Xu, H.; Wang, S.; Yang, W.; Liu, J.; Ye, Z.; Li, L.C. saRNA guided iNOS up-regulation improves erectile function of diabetic rats. J. Urol. 2013, 190, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Jiang, K.; Jiang, P.; He, H.; Chen, K.; Shao, J.; Deng, G. Mechanism of Notch1-saRNA-1480 reversing androgen sensitivity in human metastatic castration-resistant prostate cancer. Int. J. Mol. Med. 2020, 46, 265–279. [Google Scholar] [CrossRef]
- Huang, K.W.; Reebye, V.; Czysz, K.; Ciriello, S.; Dorman, S.; Reccia, I.; Lai, H.S.; Peng, L.; Kostomitsopoulos, N.; Nicholls, J.; et al. Liver Activation of Hepatocellular Nuclear Factor-4α by Small Activating RNA Rescues Dyslipidemia and Improves Metabolic Profile. Mol. Ther. Nucleic Acids 2020, 19, 361–370. [Google Scholar] [CrossRef]
- Zhu, Q.; Wu, X.; Huang, Y.; Tang, M.; Wu, L. Upregulation of FHIT gene expression in endometrial carcinoma by RNA activation. Int. J. Clin. Exp. Pathol. 2020, 13, 1372–1380. [Google Scholar] [PubMed]
- Wahlestedt, C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013, 12, 433–446. [Google Scholar] [CrossRef]
- Poller, W.; Tank, J.; Skurk, C.; Gast, M. Cardiovascular RNA interference therapy: The broadening tool and target spectrum. Circ. Res. 2013, 113, 588–602. [Google Scholar] [CrossRef]
- Hedman, M.; Hartikainen, J.; Ylä-Herttuala, S. Progress and prospects: Hurdles to cardiovascular gene therapy clinical trials. Gene Ther. 2011, 18, 743. [Google Scholar] [CrossRef]
- Cotrim, A.P.; Baum, B.J. Gene therapy: Some history, applications, problems, and prospects. Toxicol. Pathol. 2008, 36, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J. The challenge of gene therapy and DNA delivery. J. Pharm. Pharmacol. 2001, 53, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Jafarlou, M.; Baradaran, B.; Saedi, T.; Jafarlou, V.; Shanehbandi, D.; Maralani, M.; Othman, F. An overview of the history, applications, advantages, disadvantages and prospects of gene therapy. J. Biol. Regul. Homeost Agents 2016, 30, 315–321. [Google Scholar] [PubMed]
- Aldosari, B.N.; Alfagih, I.M.; Almurshedi, A.S. Lipid Nanoparticles as Delivery Systems for RNA-Based Vaccines. Pharmaceutics 2021, 13, 206. [Google Scholar] [CrossRef]
- Owens, D.E.; Peppas, N.A. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006, 307, 93–102. [Google Scholar] [CrossRef]
- Kaczmarek, J.C.; Patel, A.K.; Kauffman, K.J.; Fenton, O.S.; Webber, M.J.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem. Int. Ed. Engl. 2016, 55, 13808–13812. [Google Scholar] [CrossRef] [PubMed]
- Ickenstein, L.M.; Garidel, P. Lipid-based nanoparticle formulations for small molecules and RNA drugs. Expert Opin. Drug Deliv. 2019, 16, 1205–1226. [Google Scholar] [CrossRef]
- Watts, J.K.; Yu, D.; Charisse, K.; Montaillier, C.; Potier, P.; Manoharan, M.; Corey, D.R. Effect of chemical modifications on modulation of gene expression by duplex antigene RNAs that are complementary to non-coding transcripts at gene promoters. Nucleic Acids Res. 2010, 38, 5242–5259. [Google Scholar] [CrossRef]
- Wahlestedt, C.; Salmi, P.; Good, L.; Kela, J.; Johnsson, T.; Hökfelt, T.; Broberger, C.; Porreca, F.; Lai, J.; Ren, K.; et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. USA 2000, 97, 5633–5638. [Google Scholar] [CrossRef]
- Veedu, R.N.; Wengel, J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol. 2009, 6, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qu, Z.; Kim, S.; Shi, V.; Liao, B.; Kraft, P.; Bandaru, R.; Wu, Y.; Greenberger, L.; Horak, I. Down-modulation of cancer targets using locked nucleic acid (LNA)-based antisense oligonucleotides without transfection. Gene Ther. 2011, 18, 326. [Google Scholar] [CrossRef]
- Mook, O.; Vreijling, J.; Wengel, S.L.; Wengel, J.; Zhou, C.; Chattopadhyaya, J.; Baas, F.; Fluiter, K. In vivo efficacy and off-target effects of locked nucleic acid (LNA) and unlocked nucleic acid (UNA) modified siRNA and small internally segmented interfering RNA (sisiRNA) in mice bearing human tumor xenografts. Artif. DNA PNA XNA 2010, 1, 36–44. [Google Scholar] [CrossRef]
- Fluiter, K.; ten Asbroek, A.L.; de Wissel, M.B.; Jakobs, M.E.; Wissenbach, M.; Olsson, H.; Olsen, O.; Oerum, H.; Baas, F. In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res. 2003, 31, 953–962. [Google Scholar] [CrossRef] [PubMed]
- Dirin, M.; Winkler, J. Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther. 2013, 13, 875–888. [Google Scholar] [CrossRef]
- Bennett, C.F.; Swayze, E.E. RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. [Google Scholar] [CrossRef]
- Shim, M.S.; Kwon, Y.J. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010, 277, 4814–4827. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liu, Y. In vivo delivery of RNAi with lipid-based nanoparticles. Annu. Rev. Biomed. Eng. 2011, 13, 507–530. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.; Carver, K.; Cao, C.; Ming, X. Receptors, endocytosis, and trafficking: The biological basis of targeted delivery of antisense and siRNA oligonucleotides. J. Drug Target. 2013, 21, 27–43. [Google Scholar] [CrossRef]
- Aghamiri, S.; Talaei, S.; Roshanzamiri, S.; Zandsalimi, F.; Fazeli, E.; Aliyu, M.; Kheiry Avarvand, O.; Ebrahimi, Z.; Keshavarz-Fathi, M.; Ghanbarian, H. Delivery of genome editing tools: A promising strategy for HPV-related cervical malignancy therapy. Expert Opin. Drug Deliv. 2020, 17, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Yee, F.; Ericson, H.; Reis, D.J.; Wahlestedt, C. Cellular uptake of intracerebroventricularly administered biotin- or digoxigenin-labeled antisense oligodeoxynucleotides in the rat. Cell Mol. Neurobiol. 1994, 14, 475–486. [Google Scholar] [CrossRef]
- Rigo, F.; Hua, Y.; Krainer, A.R.; Bennett, C.F. Antisense-based therapy for the treatment of spinal muscular atrophy. J. Cell Biol. 2012, 199, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Güler, A.; et al. Immunogenic BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and T. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).