Autotetraploid Emergence via Somatic Embryogenesis in Vitis vinifera Induces Marked Morphological Changes in Shoots, Mature Leaves, and Stomata
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Media and Culture Conditions and Plant Acclimatization
2.3. Flow Cytometry Analysis
2.4. Assessment of Genetic Stability in Regenerants by RAPD, ISSR, and SSR Markers
2.5. Ampelographic Analysis
2.6. Stomatal Characteristics
2.7. Statistical Analysis
3. Results
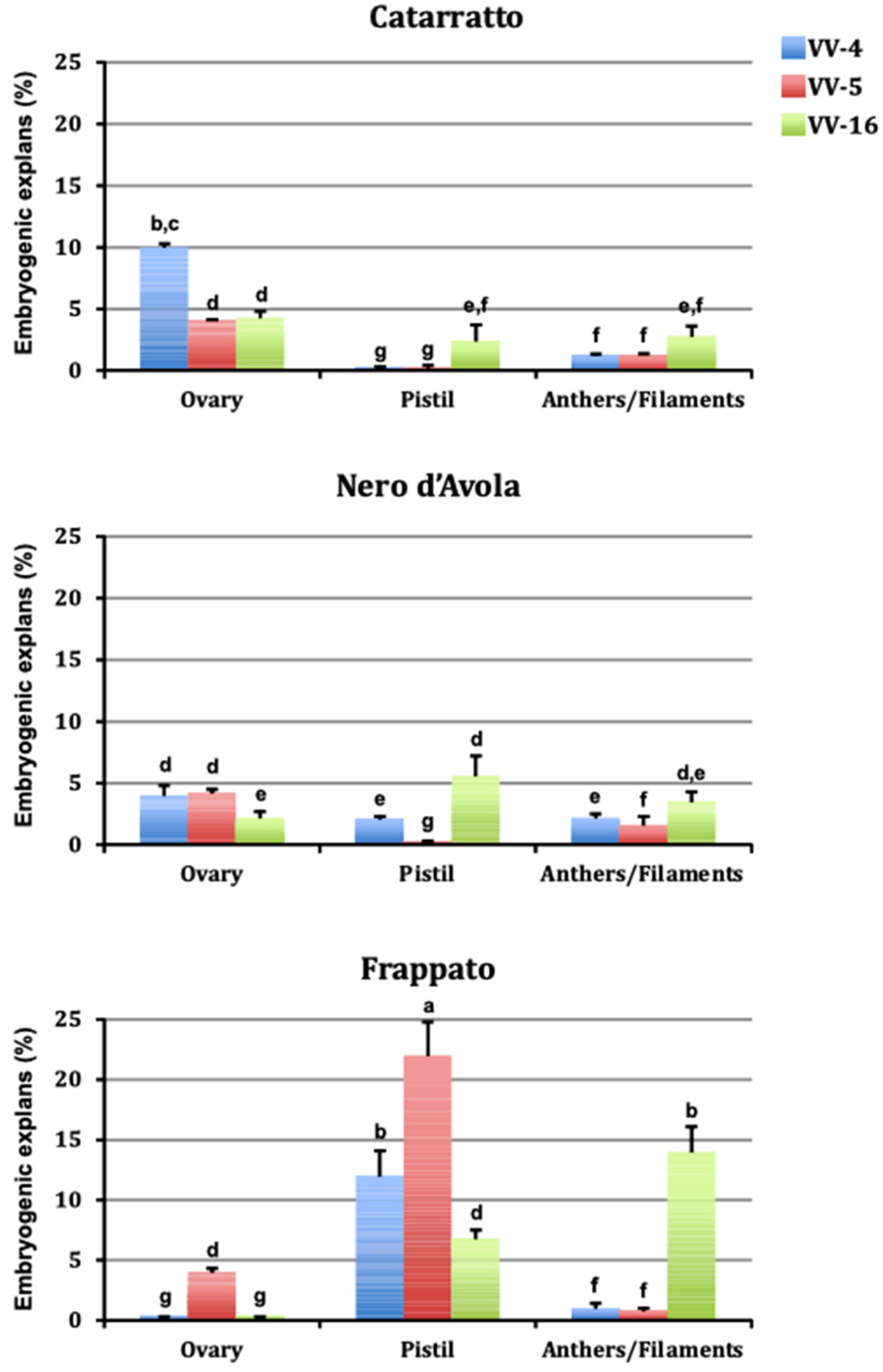
3.1. Somatic Embryogenesis
3.2. Ploidy Analysis
3.3. Assessment of Genetic Stability in Regenerants by RAPD, ISSR, and SSR Markers
3.4. Ampelographic Analysis
3.5. Leaf Morphological Characteristics and Stomata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wendel, J.F. Genome Evolution in Polyploids. In Plant Molecular Evolution; Doyle, J.J., Gaut, B.S., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 225–249. [Google Scholar] [CrossRef]
- Salman Rutland, C.A.; Hall, N.D.; McElroy, J.S. The Impact of Polyploidization on the Evolution of Weed Species: Historical Understanding and Current Limitations. Front. Agron. 2021, 3, 5. [Google Scholar] [CrossRef]
- Salman-Minkov, A.; Sabath, N.; Mayrose, I. Whole-genome duplication as a key factor in crop domestication. Nat. Plants 2016, 2, 16115. [Google Scholar] [CrossRef]
- Borrill, P.; Harrington, S.A.; Uauy, C. Genome-wide sequence and expression analysis of the NAC transcription factor family in polyploid wheat. G3 Genes Genomes Genet. 2017, 7, 3019–3029. [Google Scholar] [CrossRef]
- Wei, T.; Wang, Y.; Xie, Z.; Guo, D.; Chen, C.; Fan, Q.; Deng, X.; Liu, J.-H. Enhanced ROS scavenging and sugar accumulation contribute to drought tolerance of naturally occurring autotetraploids in Poncirus trifoliata. Plant Biotechnol. J. 2019, 17, 1394–1407. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. The role of genetic and genomic attributes in the success of polyploids. PNAS 2000, 97, 7051–7057. [Google Scholar] [CrossRef]
- Iannicelli, J.; Guariniello, J.; Tossi, V.E.; Regalado, J.J.; Di Ciaccio, L.; van Baren, C.M.; Pitta Álvarez, S.I.; Escandon, A.S. The ‘polyploid effect’ in the breeding of aromatic and medicinal species. Sci. Hortic. 2020, 260, 108854. [Google Scholar] [CrossRef]
- De Schepper, S.; Leus, L.; Eeckhaut, T.; Van Bockstaele, E.; Debergh, P.; De Loose, M. Somatic polyploid petals: Regeneration offers new roads for breeding Belgian pot azaleas. Plant Cell Tiss. Org. Cult. 2004, 76, 183–188. [Google Scholar] [CrossRef]
- Leitch, A.R.; Leitch, I.J. Genomic plasticity and the diversity of polyploid plants. Science 2008, 320, 481–483. [Google Scholar] [CrossRef]
- Sun, Q.; Sun, H.; Li, L.; Bell, R.L. In vitro colchicine-induced polyploid plantlet production and regeneration from leaf explants of the diploid pear (Pyrus communis L.) cultivar, ‘Fertility’. J. Hortic. Sci. Biotechnol. 2009, 84, 548–552. [Google Scholar] [CrossRef]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Tadeo, F.; Froelicher, Y.; Talon, M.; Navarro, L.; Ollitrault, P.; Morillon, R. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J. Exp. Bot. 2011, 62, 2507–2519. [Google Scholar] [CrossRef]
- Van Laere, K.; França, S.C.; Vansteenkiste, H.; Van Huylenbroeck, J.; Steppe, K.; Van Labeke, M.C. Influence of ploidy level on morphology, growth and drought susceptibility in Spathiphyllum wallisii. Acta. Physiol. Plant. 2011, 33, 1149–1156. [Google Scholar] [CrossRef]
- Tan, F.Q.; Tu, H.; Liang, W.J.; Long, J.M.; Wu, X.M.; Zhang, H.Y.; Guo, W.W. Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biol. 2015, 15, 89. [Google Scholar] [CrossRef]
- Doyle, J.J.; Coate, J.E. Polyploidy, the nucleotype, and novelty: The impact of genome doubling on the biology of the cell. Int. J. Plant Sci. 2019, 180, 1–52. [Google Scholar] [CrossRef]
- Kim, E.D.; Chen, Z.J. Unstable transcripts in Arabidopsis allotetraploids are associated with non additive gene expression in response to abiotic and biotic stresses. PLoS ONE 2011, 6, e24251. [Google Scholar] [CrossRef]
- Riddle, N.C.; Kato, A.; Birchler, J.A. Genetic variation for the response to ploidy change in Zea mays L. Theory Appl. Genet. 2006, 114, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.C.; Li, F.M.; Zhang, T. Performance of wheat crops with different chromosome ploidy: Root-sourced signals, drought tolerance, and yield performance. Planta 2006, 224, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Zhang, F.; Zhang, Z.H.; Fu, J.F.; Wang, F.; Zhang, B.; Ma, Y. Differences in salt tolerance between diploid and autotetraploid apple seedlings exposed to salt stress. Sci. Hortic. 2015, 190, 24–30. [Google Scholar] [CrossRef]
- Zhang, F.; Xue, H.; Lu, X.; Zhang, B.; Wang, F.; Ma, Y.; Zhang, Z. Autotetraploidization enhances drought stress tolerance in two apple cultivars. Trees 2015, 29, 1773–1780. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Wei, T.; Wang, Y.; Liu, J.H. Comparative transcriptome analysis reveals synergistic and disparate defense pathways in the leaves and roots of trifoliate orange (Poncirus trifoliata) autotetraploids with enhanced salt tolerance. Hortic. Res. 2020, 7, 1–14. [Google Scholar] [CrossRef]
- Marques, I.; Fernandes, I.; Paulo, O.S.; Lidon, F.C.; DaMatta, F.M.; Ramalho, J.C.; Ribeiro-Barros, A.I. A Transcriptomic Approach to Understanding the Combined Impacts of Supra-Optimal Temperatures and CO2 Revealed Different Responses in the Polyploid Coffea arabica and Its Diploid Progenitor, C. canephora. Int. J. Mol. Sci. 2021, 22, 3125. [Google Scholar] [CrossRef] [PubMed]
- Carimi, F.; Pathirana, R.; Carra, A. Somatic Embryogenesis and Agrobacterium Mediated Genetic Transformation in Vitis. In Somatic Embryogenesis and Genetic Transformation in Plants; Aslam, J., Srivastava, P.S., Sharma, M.P., Eds.; Narosa Publishing House: New Delhi, India, 2013; pp. 179–218. [Google Scholar]
- Larkin, P.J.; Scowcroft, W.R. Somaclonal variation—Anovel source of variability from cell cultures for plant improvement. Theory Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, A.M.; Linacero, R. Stress and Somaclonal Variation. In Plant Developmental Biology: Biotechnological Perspectives; Pua, E.-C., Davey, M.R., Eds.; Springer: Heidelberg, The Netherlands, 2010; pp. 45–64. [Google Scholar] [CrossRef]
- Endemann, M.; Hristoforoglu, K.; Stauber, T.; Wilhelm, E. Assessment of age-related polyploidy in Quercus robur L. somatic embryos and regenerated plants using DNA flow cytometry. Biol. Plant. 2002, 44, 339–345. [Google Scholar] [CrossRef]
- Cassells, A.C. Contamination Detection and Elimination. In Encyclopedia of Plant Cell Biology; Spier, R.E., Ed.; Wiley: Chichester, UK, 2000; pp. 577–586. [Google Scholar]
- Carimi, F.; Pathirana, R.; Carra, A. Biotechnologies for Grapevine Germplasm Management and Improvement. In Grapevines: Varieties, Cultivation and Management; Szabo, P.V., Shojania, J., Eds.; Nova Science Publishers: New York, NY, USA, 2012; pp. 199–249. [Google Scholar]
- Martinelli, L.; Gribaudo, I. Strategies for Effective Somatic Embryogenesis in Grapevine: An Appraisal. In Grapevine Molecular Physiology & Biotechnology, 2nd ed.; Roubelakis-Angelakis, K.A., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 461–493. [Google Scholar]
- Carimi, F.; Barizza, E.; Gardiman, M.; Lo Schiavo, F. Somatic embryogenesis from stigmas and styles of grapevine. Cell Dev. Biol. Plant 2005, 41, 249–252. [Google Scholar] [CrossRef]
- Perrin, M.; Gertz, C.; Masson, J.E. High efficiency initiation of regenerable embryonic callus from anther filaments of 19-grapevine genotypes grown worldwide. Plant Sci. 2004, 167, 1343–1349. [Google Scholar] [CrossRef]
- Gambino, G.; Ruffa, P.; Vallania, R.; Gribaudo, I. Somatic embryogenesis from whole flowers, anthers and ovaries of grapevine (Vitis spp). Plant Cell Tiss. Org. Cult. 2007, 90, 79–83. [Google Scholar] [CrossRef]
- Maillot, P.; Kieffer, F.; Walter, B. Somatic embryogenesis from stem nodal sections of grapevine. Vitis 2006, 45, 185–189. [Google Scholar] [CrossRef]
- Schellenbaum, P.; Mohler, V.; Wenzel, G.; Walter, B. Variation in DNA methylation patterns of grapevine somaclones (Vitis vinifera L.). BMC Plant Biol. 2008, 8, 78. [Google Scholar] [CrossRef]
- Torregrosa, L.; Fernandez, L.; Bouquet, A.; Boursiquot, J.M.; Pelsy, F.; Martínez-Zapater, J.M. Origins and Consequences of Somatic Variation in Grapevine. In Genetics, Genomics, and Breeding of Grapes; Kole, C., Ed.; Science Publishers: Enfield, UK, 2011; pp. 68–92. [Google Scholar]
- Leal, F.; Loureiro, J.; Rodriguez, E.; Pais, M.S.; Santos, C.; Pinto-Carnide, O. Nuclear DNA content of Vitis vinifera cultivars and ploidy level analyses of somatic embryo-derived plants obtained from anther culture. Plant Cell Rep. 2006, 25, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Acanda, Y.; Prado, M.J.; González, M.V.; Rey, M. Somatic embryogenesis from stamen filaments in grapevine (Vitis vinifera L. cv. Mencía): Changes in ploidy level and nuclear DNA content. Cell Dev. Biol. Plant 2013, 49, 276–284. [Google Scholar] [CrossRef]
- Organisation Internationale de la Vigne et du Vin (OIV). Descriptor List for Grape Varieties and Vitis Species, 2nd ed.; Organisation Internationale de la Vigne et du Vin: Paris, France, 2009; Available online: http://www.oiv.int/oiv/info/enplubicationoiv (accessed on 15 January 2021).
- Prado, M.J.; Rodriguez, E.; Rey, L.; González, M.V.; Santos, C.; Rey, M. Detection of somaclonal variants in somatic embryogenesis-regenerated plants of Vitis vinifera by flow cytometry and microsatellite markers. Plant Cell Tiss. Org. Cult. 2010, 103, 49–59. [Google Scholar] [CrossRef]
- Alvarez, M.E.; Nota, F.; Cabiagno, D.A. Epigenetic control of plant immunity. Mol. Plant Pathol. 2010, 11, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Raji, M.R.; Lotfi, M.; Tohidfar, M.; Zahedi, B.; Carra, A.; Abbate, L.; Carimi, F. Somatic embryogenesis of muskmelon (Cucumis melo L.) and genetic stability assessment of regenerants using flow cytometry and ISSR markers. Protoplasma 2018, 255, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J. Applications of flow cytometry for the study of plant genomes. J. Appl. Genet. 1997, 3, 285–302. [Google Scholar] [CrossRef]
- Loureiro, J.; Pinto, G.; Lopes, T.; Doležel, J.; Santos, C. Assessment of ploidy stability of the somatic embryogenesis process in Quercus suber L. using flow cytometry. Planta 2005, 221, 815–822. [Google Scholar] [CrossRef]
- Doležel, J. Flow cytometric analysis of nuclear DNA content in higher plants. Phytochem. Anal. 1991, 2, 143–154. [Google Scholar] [CrossRef]
- Carimi, F.; Mercati, F.; Abbate, L.; Sunseri, F. Microsatellite analyses for evaluation of genetic diversity among Sicilian grapevine cultivars. Genet. Resour. Crop Evol. 2010, 57, 703–719. [Google Scholar] [CrossRef]
- Carimi, F.; Mercati, F.; De Michele, R.; Fiore, M.C.; Riccardi, P.; Sunseri, F. Intravarietal genetic diversity of the grapevine (Vitis vinifera L.) cultivar ‘Nero d’Avola’ as revealed by microsatellite markers. Genet. Resour. Crop Evol. 2011, 58, 967–975. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–479. [Google Scholar] [CrossRef]
- Carra, A.; Sajeva, M.; Abbate, L.; Siragusa, M.; Pathirana, R.; Carimi, F. Factors affecting somatic embryogenesis in eight Italian grapevine cultivars and the genetic stability of embryo-derived regenerants as assessed by molecular markers. Sci. Hortic. 2016, 204, 123–127. [Google Scholar] [CrossRef]
- Oddo, E.; Abbate, L.; Inzerillo, S.; Carimi, F.; Motisi, A.; Sajeva, M.; Nardini, A. Water relations of two Sicilian grapevine cultivars in response to potassium availability and drought stress. Plant Physiol. Biochem. 2020, 148, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Carra, A.; Sajeva, M.; Abbate, L.; Siragusa, M.; Sottile, F.; Carimi, F. In vitro plant regeneration of caper (Capparis spinosa L.) from floral explants and genetic stability of regenerants. Plant Cell Tis. Org. Cult. 2012, 109, 373–381. [Google Scholar] [CrossRef]
- Otto, F.J. Preparation and Staining of Cells for High-Resolution DNA Analysis. In A Flow Cytometry and Cell Sorting; Radbruch, A., Ed.; Springer: Berlin/Heidelberg, Germany, 1992; pp. 65–68. [Google Scholar]
- Praça-Fontes, M.M.; Carvalho, C.R.; Clarindo, W.R.; Cruz, C.D. Revisiting the DNA C-values of the genome size-standards used in plant flow cytometry to choose the ‘best primary standards’. Plant Cell Rep. 2011, 30, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Gristina, A.S.; Fici, S.; Siragusa, M.; Fontana, I.; Garfì, G.; Carimi, F. Hybridization in Capparis spinosa L.: Molecular and morphological evidence from a Mediterranean island complex. Flora 2014, 209, 733–741. [Google Scholar] [CrossRef]
- Siragusa, M.; Carra, A.; Salvia, L.; Puglia, A.M.; De Pasquale, F.; Carimi, F. Genetic instability in calamondin (Citrus madurensis Lour.) plants derived from somatic embryogenesis induced by diphenylurea derivatives. Plant Cell Rep. 2007, 26, 1289–1296. [Google Scholar] [CrossRef]
- Meziane, M.; Frasheri, D.; Carra, A.; Boudjeniba, M.; D’Onghia, A.M.; Mercati, F.; Djelouah, K.; Carimi, F. Attempts to eradicate graft-transmissible infections through somatic embryogenesis in Citrus ssp. and analysis of genetic stability of regenerated plants. Eur. J. Plant Pathol. 2016, 10, 1–11. [Google Scholar] [CrossRef]
- Haddad, B.; Carra, A.; Saadi, A.; Haddad, N.; Mercati, F.; Gristina, A.S.; Boukhalfa, S.; Djillali, D.; Carimi, F. In vitro propagation of the relict Laperinne’s olive (Olea europaea L. subsp. laperrinei). Plant Biosyst. 2018, 152, 621–630. [Google Scholar] [CrossRef]
- De Michele, R.; La Bella, F.; Gristina, A.S.; Fontana, I.; Pacifico, D.; Garfi, G.; Motisi, A.; Crucitti, D.; Abbate, L.; Carimi, F. Phylogenetic Relationship Among Wild and Cultivated Grapevine in Sicily: A Hotspot in the Middle of the Mediterranean Basin. Front. Plant Sci. 2019, 10, 1506. [Google Scholar] [CrossRef]
- Salisbury, E.J. On the causes and ecological significance of stomatal frequency, with special reference to the woodland flora. Philos. Trans. R. Soc. Lond. Ser. B 1928, 216, 1–65. [Google Scholar] [CrossRef]
- Dhooghe, E.; Van Laere, K.; Eeckhaut, T.; Leus, L.; Van Huylenbroeck, J. Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tiss. Org. Cult. 2011, 104, 359–373. [Google Scholar] [CrossRef]
- Levin, D.A. Polyploidy and novelty in flowering plants. Am. Nat. 1983, 122, 1–25. [Google Scholar] [CrossRef]
- Estilai, A.; Shannon, M.C. Salt Tolerance in Relation to Ploidy Level in Guayule. In New Crops; Janick, J., Simon, J.E., Eds.; Wiley: New York, NY, USA, 1993; pp. 349–351. [Google Scholar]
- Remotti, P.C. Primary and secondary embryogenesis from cell suspension cultures of Gladiolus. Plant Sci. 1995, 107, 205–214. [Google Scholar] [CrossRef]
- Brar, D.S.; Jain, S.M. Somaclonal Variation: Mechanism and Applications in Crop Improvement. In Current Plant Science and Biotechnology in Agriculture; Jain, S.M., Brar, D.S., Ahloowalia, B.S., Eds.; Springer: Dordrecht, The Netherlands, 1998; pp. 15–37. [Google Scholar] [CrossRef]
- Diaz-Sala, C.; Rey, M.; Boronat, A.; Besford, R.; Rodriguez, R. Variations in the DNA methylation and polypeptide patterns of adult hazel (Corylus avellana L.) associated with sequential in vitro subcultures. Plant Cell Rep. 1995, 15, 218–221. [Google Scholar] [CrossRef]
- Rakoczy-Trojanowska, M. The effects of growth regulators on somaclonal variation in rye (Secale cereale L.) and selection of somaclonal variants with increased agronomic traits. Cell Mol. Biol. Lett. 2002, 7, 1111–1120. [Google Scholar] [PubMed]
- Haoa, Y.J.; Wen, X.P.; Deng, X.X. Genetic and epigenetic evaluations of citrus calluses recovered from slow-growth culture. J. Plant Physiol. 2004, 16, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Jaligot, E.; Beulé, T.; Baurens, F.C.; Billotte, N.; Rival, A. Search for methylation-sensitive amplification polymorphisms associated with the ‘mantled’ variant phenotype in oil palm (Elaeis guineensis Jacq.). Genome 2004, 47, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Bryan, G.J.; Winfield, M.O.; Millam, S. Stability of potato (Solanum tuberosum L.) plants regenerated via somatic embryos, axillary bud proliferated shoots, microtubers and true potato seeds: A comparative phenotypic, cytogenetic and molecular assessment. Planta 2007, 226, 1449–1458. [Google Scholar] [CrossRef]
- Menéndez-Yuffá, A.; Barry-Etienne, D.B. , Georget, F.; Etienne, H. A comparative analysis of the development and quality of nursery plants derived from somatic embryogenesis and from seedlings for large-scale propagation of coffee (Coffea arabica L.). Plant Cell Tiss. Org. Cult. 2010, 102, 297–307. [Google Scholar] [CrossRef]
- Leva, A.R.; Petruccelli, R.; Rinaldi, L.M.R. Somaclonal Variation in Tissue Culture: A Case Study with Olive. In Recent Advances in Plant in vitro Culture; Leva, A.R., Rinaldi, L.M.R., Eds.; Intech Open Access Publisher: Croatia, Hungary, 2012; pp. 123–150. [Google Scholar]
- Manchanda, P.; Kaur, A.; Gosal, S.S. Somaclonal Variation for Sugar Cane Improvement. In Biotechnologies of Crop Improvement; Gosal, S., Wani, S., Eds.; Springer: Cham, Switzerland, 2018; pp. 299–326. [Google Scholar]
- Dalla Costa, L.; Malnoy, M.; Gribaudo, I. Breeding next generation tree fruits: Technical and legal challenges. Hortic. Res. 2017, 4, 17067. [Google Scholar] [CrossRef]
- Desperrier, J.C.; Berger, J.L.; Bessis, R.; Fournioux, J.C.; Labroche, C. Création clonale dirigée par embryogenèse somatique. Bullettin de l’O.I.V. 2003, 76, 751–765. [Google Scholar]
- Boso, S.; Alonso-Villaverde, V.; Santiago, J.L.; Gago, P.; Dürrenberger, M.; Düggelin, M.; Kassemeyer, H.H.; Martinez, M.C. Macro- and microscopic leaf characteristics of six grapevine genotypes (Vitis spp.) with different susceptibilities to grapevine downy mildew. Vitis 2010, 49, 43–50. [Google Scholar] [CrossRef]
- Muganu, M.; Paolocci, M. Adaptation of Local Grapevine Germplasm: Exploitation of Natural Defence Mechanisms to Biotic Stresses. In The Mediterranean Genetic Code—Grapevine and Olive; Poljuha, D., Sladonja, B., Eds.; Intech Open Access Publisher: London, UK, 2013; pp. 221–246. [Google Scholar]
- Leroy, X.J.; Leon, K.; Branchard, M. IRSS and somaclonal variation: A new molecular technique for an important in vitro phenomenon. Electron. J. Biotech. 2000, 3, 1–2. [Google Scholar]
- Yang, X.M.; Cao, Z.Y.; An, L.Z.; Wang, Y.M.; Fang, X.W. In vitro tetraploid induction via colchicine treatment from diploid somatic embryos in grapevine (Vitis vinifera L.). Euphytica 2006, 152, 217. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M.; Nuez, F. Embryogenesis induction, callogenesis, and plant regeneration by in vitro culture of tomato isolated microspores and whole anthers. J. Exp. Bot. 2007, 58, 1119–1132. [Google Scholar] [CrossRef] [PubMed]



| OIV Code | OIV Descriptor | Levels | 2n (E51) | 4n (E33) | 4n (E34) |
|---|---|---|---|---|---|
| OIV 001 | Young shoot: opening of the shoot tip | (1) Closed; (3) half open; (5) fully open | 1 | 5 | 5 |
| OIV 007 | Shoot: color of the dorsal side of internodes | (1) Green; (2) green and red; (3) red | 2 | 2 | 2 |
| OIV 008 | Shoot: color of the ventral side of internodes | (1) Green; (2) green and red; (3) red | 2 | 2 | 2 |
| OIV 065 | Mature leaf: size of blade | (1) Very small; (3) small; (5) medium; (7) large; (9) very large | 3 | 5 * | 7 * |
| OIV 067 | Mature leaf: shape of blade | (1) Cordate; (2) wedge-shaped; (3) pentagonal; (4) circular; (5) kidney-shaped | 3 | 3 | 3 |
| OIV 068 | Mature leaf: number of lobes | (1) One (entire leaf); (2) three; (3) five; (4) seven; (5) more than seven | 3 | 2 | 2 |
| OIV 072 | Mature leaf: goffering of blade | (1) Absent or very weak; (3) weak; (5) medium; (7) strong; (9) very strong | 1 | 1 * | 3 * |
| OIV 074 | Mature leaf: profile of blade in cross section | (1) Flat; (2) V-shaped; (3) involute; 4) revolute; (5) twisted | 1 | 2 | 2 |
| OIV 075 | Mature leaf: blistering of upper side of blade | (1) Absent or very weak; (3) weak; (5) medium; (7) strong; (9) very strong | 1 | 3 | 3 |
| OIV 076 | Mature leaf: shape of teeth | (1) Both sides concave; (2) both sides straight; (3) both sides convex; (4) one side concave, one side convex; (5) mixture between, both sides straight and both sides convex | 4 | 5 * | 4 * |
| OIV 078 | Mature leaf: length of teeth compared with their width | (1) Very short; (3) short; (5) medium; (7) long; (9) very long | 3 | 5 * | 7 * |
| OIV 079 | Mature leaf: degree of opening/overlapping of petiole sinuses | (1) Very wide open; (3) open; (5) closed; (7) overlapped; (9) strongly overlapped | 3 | 7 | 7 |
| OIV 082 | Mature leaf: degree of opening/overlapping of upper lateral sinuses | (1) Open; (2) closed; (3) slightly overlapped; (4) strongly overlapped; (5) absence of sinus | 1 | 5 | 5 |
| OIV 093 | Mature leaf: length of petiole compared to length of middle vein | (1) Much shorter; (3) slightly shorter; (5) equal; (7) slightly longer; (9) much longer | 3 (0.72) | 1 (0.56) | 1 (0.59) |
| OIV 094 | Mature leaf: depth of upper lateral sinuses | (1) Absent or very shallow; (3) shallow; (5) medium; (7) deep; (9) very deep | 5 | 1 | 1 |
| Characteristics | 2n (E51) | 4n (E33) | 4n (E34) |
|---|---|---|---|
| Leaf length (cm) | 8.4 ± 0.7 a | 9.5 ± 1 a | 9.5 ± 0.5 a |
| Leaf width (cm) | 9.4 ± 1.1 a | 13 ± 0.5 b | 11.8 ± 0.3 b |
| Leaf area (cm2) | 43.3 ± 8.7 a | 79.8 ± 6.1 b | 82.6 ± 3.1 b |
| Stomatal length (μm) | 18.3 ± 0.5 a | 27.1 ± 0.5 b | 24.1 ± 0.5 b |
| Stomatal width (μm) | 13.4 ± 0.3 a | 16.3 ± 0.3 b | 15.5 ± 0.6 b |
| Number of chloroplastsper guard cell pair | 44.3 ± 2.8 a | 47.1 ± 2.6 a | 46.1 ± 2.6 a |
| SI | 10.7 ± 0.3 a | 10.1 ± 1.1 b | 9.0 ± 0.5 b |
| Genotype | Embryogenic Explants (%) |
|---|---|
| ‘Catarratto’ | 2.9 ± 0.2 b |
| ‘Nero d’Avola’ | 2.8 ± 0.6 b |
| ‘Frappato’ | 6.7 ± 0.7 a |
| Media | Embryogenic Explants (%) | Explant Type | Embryogenic Explants (%) |
|---|---|---|---|
| VV-4 (5 µM CPPU + 5 µM 2,4-D) | 3.6 + 0.2 a | Ovary | 3.6 + 0.2 b |
| VV-5 (20 µM NOA + 4 µM TDZ) | 4.2 + 0.3 a | Pistil | 5.7 + 0.6 a |
| VV-16 (10 µM NOA + 4.4 µM BA) | 4.6 + 0.1 a | Anther/filament | 3.1 + 0.2 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catalano, C.; Abbate, L.; Motisi, A.; Crucitti, D.; Cangelosi, V.; Pisciotta, A.; Di Lorenzo, R.; Carimi, F.; Carra, A. Autotetraploid Emergence via Somatic Embryogenesis in Vitis vinifera Induces Marked Morphological Changes in Shoots, Mature Leaves, and Stomata. Cells 2021, 10, 1336. https://doi.org/10.3390/cells10061336
Catalano C, Abbate L, Motisi A, Crucitti D, Cangelosi V, Pisciotta A, Di Lorenzo R, Carimi F, Carra A. Autotetraploid Emergence via Somatic Embryogenesis in Vitis vinifera Induces Marked Morphological Changes in Shoots, Mature Leaves, and Stomata. Cells. 2021; 10(6):1336. https://doi.org/10.3390/cells10061336
Chicago/Turabian StyleCatalano, Caterina, Loredana Abbate, Antonio Motisi, Dalila Crucitti, Vincenzo Cangelosi, Antonino Pisciotta, Rosario Di Lorenzo, Francesco Carimi, and Angela Carra. 2021. "Autotetraploid Emergence via Somatic Embryogenesis in Vitis vinifera Induces Marked Morphological Changes in Shoots, Mature Leaves, and Stomata" Cells 10, no. 6: 1336. https://doi.org/10.3390/cells10061336
APA StyleCatalano, C., Abbate, L., Motisi, A., Crucitti, D., Cangelosi, V., Pisciotta, A., Di Lorenzo, R., Carimi, F., & Carra, A. (2021). Autotetraploid Emergence via Somatic Embryogenesis in Vitis vinifera Induces Marked Morphological Changes in Shoots, Mature Leaves, and Stomata. Cells, 10(6), 1336. https://doi.org/10.3390/cells10061336










