RETRACTED: EFA6 in Axon Regeneration, as a Microtubule Regulator and as a Guanine Nucleotide Exchange Factor
Abstract
1. Introduction
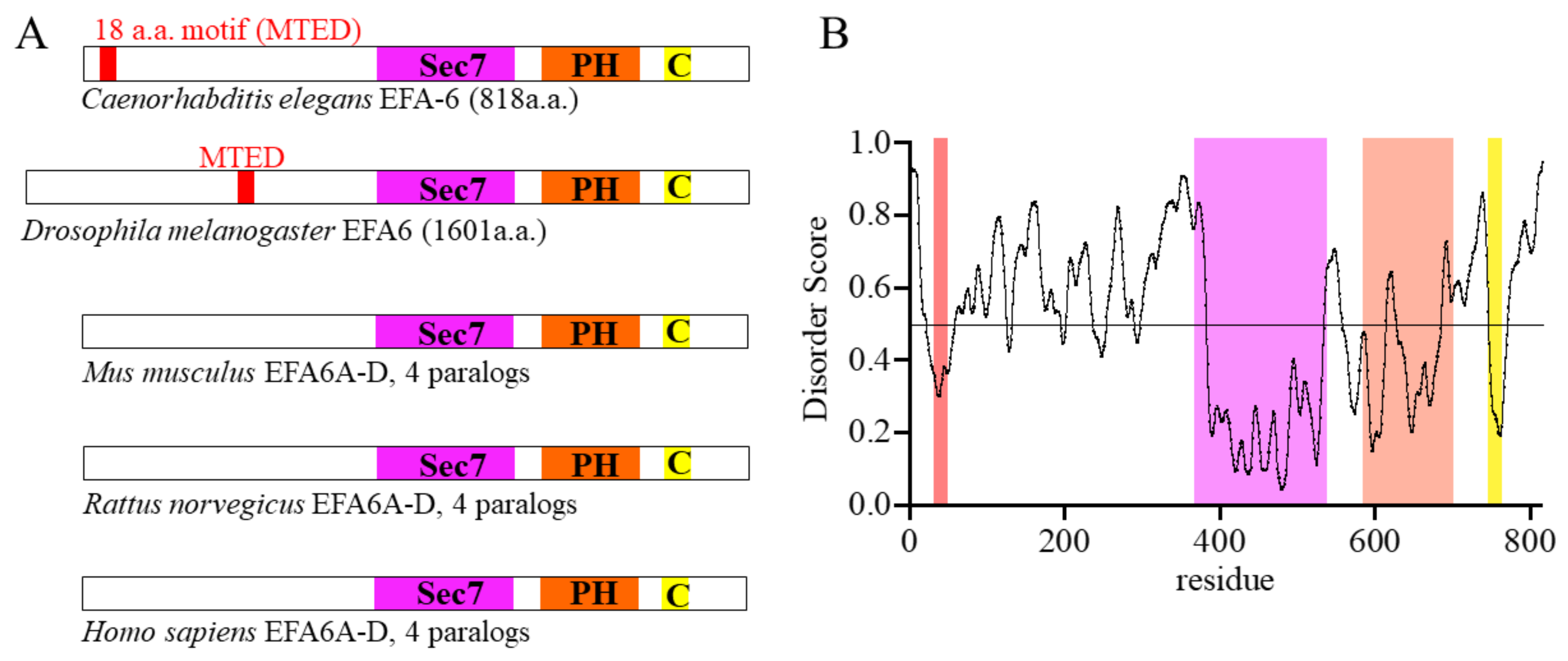
2. EFA6 Is a GEF for ARF6 and a Microtubule Regulator
3. Microtubule Dynamics in Axon Regeneration
3.1. Microtubules in Intact Axons
3.2. Microtubules in Injured and Regenerating Axons
4. ARF6, Integrins, and Axon Regeneration
5. EFA6 as an Inhibitor of Axon Regeneration
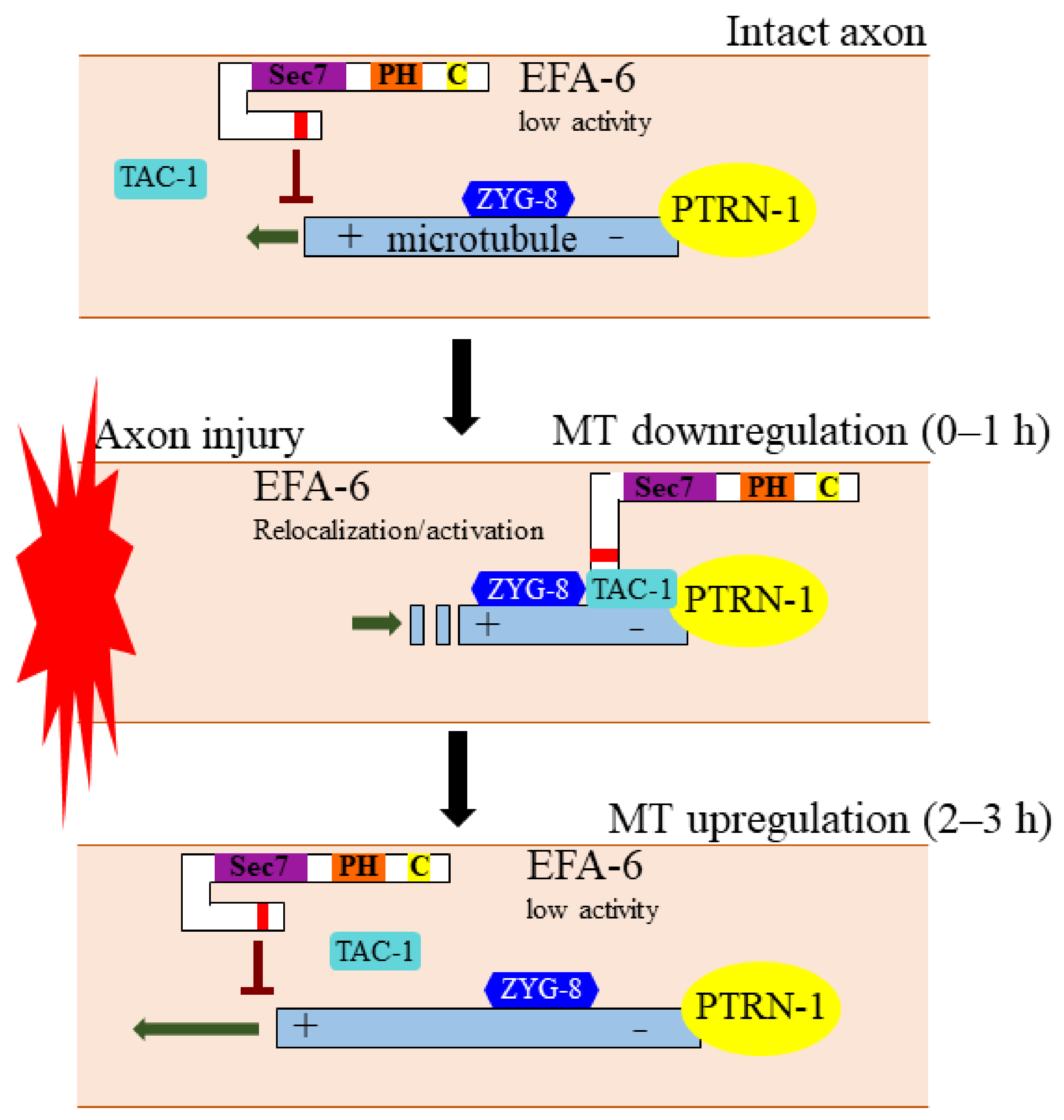
5.1. EFA6 Inhibits Axon Regeneration through Regulating Microtubule Dynamics
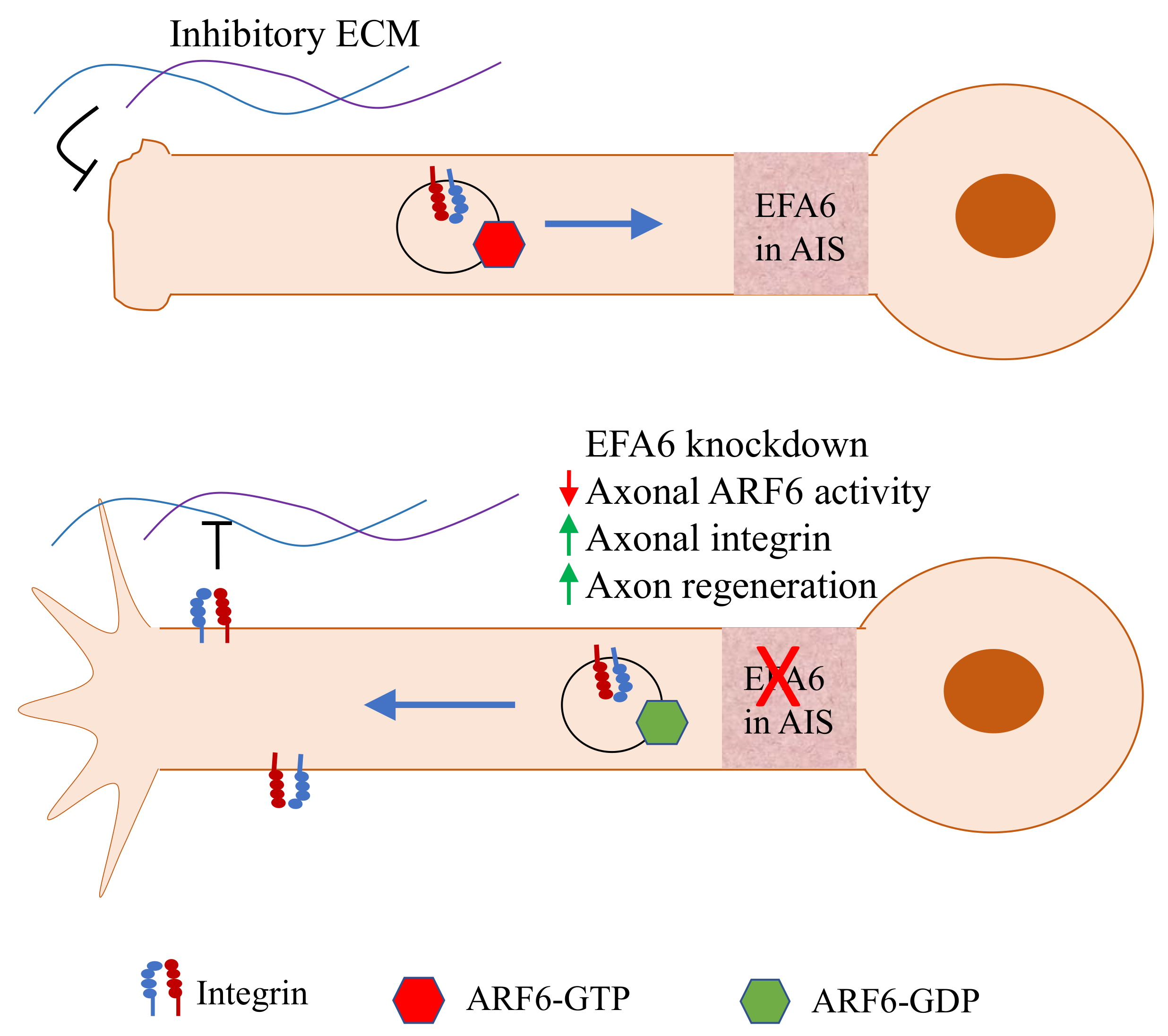
5.2. EFA6 Inhibits Axon Regeneration as a GEF for ARF6
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Tedeschi, A.; Bradke, F. Spatial and temporal arrangement of neuronal intrinsic and extrinsic mechanisms controlling axon regeneration. Curr. Opin. Neurobiol. 2017, 42, 118–127. [Google Scholar] [CrossRef]
- He, Z.; Jin, Y. Intrinsic Control of Axon Regeneration. Neuron 2016, 90, 437–451. [Google Scholar] [CrossRef]
- Aguayo, A.J.; David, S.; Bray, G.M. Influences of the glial environment on the elongation of axons after injury: Transplantation studies in adult rodents. J. Exp. Biol. 1981, 95, 231–240. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Aguayo, A. Axonal elongation into peripheral nervous system “bridges” after central nervous system injury in adult rats. Science 1981, 214, 931–933. [Google Scholar] [CrossRef]
- Geoffroy, C.G.; Hilton, B.J.; Tetzlaff, W.; Zheng, B. Evidence for an Age-Dependent Decline in Axon Regeneration in the Adult Mammalian Central Nervous System. Cell Rep. 2016, 15, 238–246. [Google Scholar] [CrossRef]
- Cafferty, W.B.J.; Duffy, P.; Huebner, E.; Strittmatter, S.M. MAG and OMgp Synergize with Nogo-A to Restrict Axonal Growth and Neurological Recovery after Spinal Cord Trauma. J. Neurosci. 2010, 30, 6825–6837. [Google Scholar] [CrossRef]
- Lee, J.K.; Chow, R.; Xie, F.; Chow, S.Y.; Tolentino, K.E.; Zheng, B. Combined Genetic Attenuation of Myelin and Semaphorin-Mediated Growth Inhibition Is Insufficient to Promote Serotonergic Axon Regeneration. J. Neurosci. 2010, 30, 10899–10904. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Geoffroy, C.G.; Chan, A.F.; Tolentino, K.E.; Crawford, M.J.; Leal, M.A.; Kang, B.; Zheng, B. Assessing Spinal Axon Regeneration and Sprouting in Nogo-, MAG-, and OMgp-Deficient Mice. Neuron 2010, 66, 663–670. [Google Scholar] [CrossRef]
- Mahar, M.; Cavalli, V. Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 2018, 19, 323–337. [Google Scholar] [CrossRef]
- Neumann, S.; Woolf, C.J. Regeneration of Dorsal Column Fibers into and beyond the Lesion Site following Adult Spinal Cord Injury. Neuron 1999, 23, 83–91. [Google Scholar] [CrossRef]
- McQuarrie, I.G.; Grafstein, B. Axon Outgrowth Enhanced by a Previous Nerve Injury. Arch. Neurol. 1973, 29, 53–55. [Google Scholar] [CrossRef]
- Rishal, I.; Fainzilber, M. Axon-soma communication in neuronal injury. Nat. Rev. Neurosci. 2014, 15, 32–42. [Google Scholar] [CrossRef]
- Ghosh-Roy, A.; Wu, Z.; Goncharov, A.; Jin, Y.; Chisholm, A.D. Calcium and Cyclic AMP Promote Axonal Regeneration in Caenorhabditis elegans and Require DLK-1 Kinase. J. Neurosci. 2010, 30, 3175–3183. [Google Scholar] [CrossRef]
- Blanquie, O.; Bradke, F. Cytoskeleton dynamics in axon regeneration. Curr. Opin. Neurobiol. 2018, 51, 60–69. [Google Scholar] [CrossRef]
- Friedli, L.; Rosenzweig, E.S.; Barraud, Q.; Schubert, M.; Dominici, N.; Awai, L.; Nielson, J.L.; Musienko, P.; Nout-Lomas, Y.; Zhong, H.; et al. Pronounced species divergence in corticospinal tract reorganization and functional recovery after lateralized spinal cord injury favors primates. Sci. Transl. Med. 2015, 7, 302ra134. [Google Scholar] [CrossRef]
- Rosenzweig, E.S.; Courtine, G.; Jindrich, D.L.; Brock, J.H.; Ferguson, A.R.; Strand, S.C.; Nout, Y.S.; Roy, R.R.; Miller, D.M.; Beattie, M.S.; et al. Extensive spontaneous plasticity of corticospinal projections after primate spinal cord injury. Nat. Neurosci. 2010, 13, 1505–1510. [Google Scholar] [CrossRef]
- Yanik, M.F.; Cinar, H.; Cinar, H.N.; Chisholm, A.D.; Jin, Y.; Ben-Yakar, A. Neurosurgery: Functional regeneration after laser axotomy. Nature 2004, 432, 822. [Google Scholar] [CrossRef]
- Wu, Z.; Ghosh-Roy, A.; Yanik, M.F.; Zhang, J.Z.; Jin, Y.; Chisholm, A.D. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc. Natl. Acad. Sci. USA 2007, 104, 15132–15137. [Google Scholar] [CrossRef]
- Canty, A.J.; Huang, L.; Jackson, J.S.; Little, G.E.; Knott, G.W.; Maco, B.; De Paola, V. In-Vivo single neuron axotomy triggers axon regeneration to restore synaptic density in specific cortical circuits. Nat. Commun. 2013, 4, 2038. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Z.; Ghosh-Roy, A.; Hubert, T.; Yan, D.; O’Rourke, S.; Bowerman, B.; Wu, Z.; Jin, Y.; Chisholm, A.D. Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron 2011, 71, 1043–1057. [Google Scholar] [CrossRef]
- Kim, K.W.; Tang, N.H.; Piggott, C.A.; Andrusiak, M.G.; Park, S.; Zhu, M.; Kurup, N.; Cherra, S.J., 3rd; Wu, Z.; Chisholm, A.D.; et al. Expanded genetic screening in Caenorhabditis elegans identifies new regulators and an inhibitory role for NAD(+) in axon regeneration. eLife 2018, 7, e39756. [Google Scholar] [CrossRef]
- Samara, C.; Rohde, C.B.; Gilleland, C.L.; Norton, S.; Haggarty, S.J.; Yanik, M.F. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc. Natl. Acad. Sci. USA 2010, 107, 18342–18347. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, J.G.; Jackson, C.L. ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 2011, 12, 362–375. [Google Scholar] [CrossRef]
- Gillingham, A.K.; Munro, S. The Small G Proteins of the Arf Family and Their Regulators. Annu. Rev. Cell Dev. Biol. 2007, 23, 579–611. [Google Scholar] [CrossRef] [PubMed]
- Tagliatti, E.; Fadda, M.; Falace, A.; Benfenati, F.; Fassio, A. Arf6 regulates the cycling and the readily releasable pool of synaptic vesicles at hippocampal synapse. eLife 2016, 5, e10116. [Google Scholar] [CrossRef]
- Eva, R.; Crisp, S.; Marland, J.R.K.; Norman, J.C.; Kanamarlapudi, V.; Ffrench-Constant, C.; Fawcett, J.W. ARF6 Directs Axon Transport and Traffic of Integrins and Regulates Axon Growth in Adult DRG Neurons. J. Neurosci. 2012, 32, 10352–10364. [Google Scholar] [CrossRef] [PubMed]
- Raemaekers, T.; Peric, A.; Baatsen, P.; Sannerud, R.; Declerck, I.; Baert, V.; Michiels, C.; Annaert, W. ARF6-mediated endosomal transport of Telencephalin affects dendritic filopodia-to-spine maturation. EMBO J. 2012, 31, 3252–3269. [Google Scholar] [CrossRef]
- Miyazaki, H.; Yamazaki, M.; Watanabe, H.; Maehama, T.; Yokozeki, T.; Kanaho, Y. The small GTPase ADP-ribosylation factor 6 negatively regulates dendritic spine formation. FEBS Lett. 2005, 579, 6834–6838. [Google Scholar] [CrossRef]
- Hernández-Deviez, D.J.; Roth, M.G.; Casanova, J.E.; Wilson, J.M. ARNO and ARF6 Regulate Axonal Elongation and Branching through Downstream Activation of Phosphatidylinositol 4-Phosphate 5-Kinase α. Mol. Biol. Cell 2004, 15, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Hara, Y.; Fukaya, M.; Hayashi, K.; Kawauchi, T.; Nakajima, K.; Sakagami, H. ADP Ribosylation Factor 6 Regulates Neuronal Migration in the Developing Cerebral Cortex through FIP3/Arfophilin-1-dependent Endosomal Trafficking of N-cadherin. eNeuro 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Falace, A.; Buhler, E.; Fadda, M.; Watrin, F.; Lippiello, P.; Pallesi-Pocachard, E.; Baldelli, P.; Benfenati, F.; Zara, F.; Represa, A.; et al. TBC1D24 regulates neuronal migration and maturation through modulation of the ARF6-dependent pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, D.N.; Béhar, A.; Tryoen-Tóth, P.; Bush, J.O.; Jungas, T.; Vitale, N.; Davy, A. Ephrin B1 maintains apical adhesion of neural progenitors. Development 2013, 140, 2082–2092. [Google Scholar] [CrossRef]
- Padovani, D.; Folly-Klan, M.; Labarde, A.; Boulakirba, S.; Campanacci, V.; Franco, M.; Zeghouf, M.; Cherfils, J. EFA6 controls Arf1 and Arf6 activation through a negative feedback loop. Proc. Natl. Acad. Sci. USA 2014, 111, 12378–12383. [Google Scholar] [CrossRef]
- Sakagami, H.; Suzuki, H.; Kamata, A.; Owada, Y.; Fukunaga, K.; Mayanagi, H.; Kondo, H. Distinct spatiotemporal expression of EFA6D, a guanine nucleotide exchange factor for ARF6, among the EFA6 family in mouse brain. Brain Res. 2006, 1093, 1–11. [Google Scholar] [CrossRef]
- Matsuya, S.; Sakagami, H.; Tohgo, A.; Owada, Y.; Shin, H.W.; Takeshima, H.; Nakayama, K.; Kokubun, S.; Kondo, H. Cellular and subcellular localization of EFA6C, a third member of the EFA6 family, in adult mouse Purkinje cells. J. Neurochem. 2005, 93, 674–685. [Google Scholar] [CrossRef]
- Derrien, V.; Couillault, C.; Franco, M.; Martineau, S.; Montcourrier, P.; Houlgatte, R.; Chavrier, P. A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J. Cell Sci. 2002, 115, 2867–2879. [Google Scholar] [CrossRef] [PubMed]
- Sakagami, H.; Honma, T.; Sukegawa, J.; Owada, Y.; Yanagisawa, T.; Kondo, H. Somatodendritic localization of EFA6A, a guanine nucleotide exchange factor for ADP-ribosylation factor 6, and its possible interaction with α-actinin in dendritic spines. Eur. J. Neurosci. 2007, 25, 618–628. [Google Scholar] [CrossRef]
- Franco, M.; Peters, P.J.; Boretto, J.; Van Donselaar, E.; Neri, A.; D’Souza-Schorey, C.; Chavrier, P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999, 18, 1480–1491. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ng, S.S.-M.; Wang, J.; Lai, L.; Leung, S.Y.; Franco, M.; Peng, Y.; He, M.-L.; Kung, H.-F.; Lin, M.C.-M. EFA6A Enhances Glioma Cell Invasion through ADP Ribosylation Factor 6/Extracellular Signal–Regulated Kinase Signaling. Cancer Res. 2006, 66, 1583–1590. [Google Scholar] [CrossRef] [PubMed]
- Vitaliani, R.; Mason, W.; Ances, B.; Zwerdling, T.; Jiang, Z.; Dalmau, J. Paraneoplastic encephalitis, psychiatric symptoms, and hypoventilation in ovarian teratoma. Ann. Neurol. 2005, 58, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Macia, E.; Partisani, M.; Favard, C.; Mortier, E.; Zimmermann, P.; Carlier, M.-F.; Gounon, P.; Luton, F.; Franco, M. The Pleckstrin Homology Domain of the Arf6-specific Exchange Factor EFA6 Localizes to the Plasma Membrane by Interacting with Phosphatidylinositol 4,5-Bisphosphate and F-actin. J. Biol. Chem. 2008, 283, 19836–19844. [Google Scholar] [CrossRef]
- O’Rourke, S.M.; Christensen, S.N.; Bowerman, B. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nat. Cell Biol. 2010, 12, 1235–1241. [Google Scholar] [CrossRef]
- Kozlowski, C.; Srayko, M.; Nedelec, F. Cortical Microtubule Contacts Position the Spindle in C. elegans Embryos. Cell 2007, 129, 499–510. [Google Scholar] [CrossRef]
- Severson, A.F.; Bowerman, B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J. Cell Biol. 2003, 161, 21–26. [Google Scholar] [CrossRef]
- Pecreaux, J.; Röper, J.-C.; Kruse, K.; Julicher, F.; Hyman, A.A.; Grill, S.W.; Howard, J. Spindle Oscillations during Asymmetric Cell Division Require a Threshold Number of Active Cortical Force Generators. Curr. Biol. 2006, 16, 2111–2122. [Google Scholar] [CrossRef]
- Qu, Y.; Hahn, I.; Lees, M.; Parkin, J.; Voelzmann, A.; Dorey, K.; Rathbone, A.; Friel, C.T.; Allan, V.J.; Okenve-Ramos, P.; et al. Efa6 protects axons and regulates their growth and branching by inhibiting microtubule polymerisation at the cortex. eLife 2019, 8, e50319. [Google Scholar] [CrossRef]
- Desai, A.; Mitchison, T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef] [PubMed]
- Janke, C.; Kneussel, M. Tubulin post-translational modifications: Encoding functions on the neuronal microtubule cytoskeleton. Trends Neurosci. 2010, 33, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Peris, L.; Wagenbach, M.; Lafanechère, L.; Brocard, J.; Moore, A.T.; Kozielski, F.; Job, D.; Wordeman, L.; Andrieux, A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 2009, 185, 1159–1166. [Google Scholar] [CrossRef]
- Suter, D.; Schaefer, A.W.; Forscher, P. Microtubule Dynamics Are Necessary for SRC Family Kinase-Dependent Growth Cone Steering. Curr. Biol. 2004, 14, 1194–1199. [Google Scholar] [CrossRef]
- Kapitein, L.C.; Hoogenraad, C.C. Building the Neuronal Microtubule Cytoskeleton. Neuron 2015, 87, 492–506. [Google Scholar] [CrossRef]
- Kapitein, L.C.; Hoogenraad, C.C. Which way to go? Cytoskeletal organization and polarized transport in neurons. Mol. Cell. Neurosci. 2011, 46, 9–20. [Google Scholar] [CrossRef]
- Goldberg, D.J.; Burmeister, D.W. Stages in axon formation: Observations of growth of Aplysia axons in culture using video-enhanced contrast-differential interference contrast microscopy. J. Cell Biol. 1986, 103, 1921–1931. [Google Scholar] [CrossRef]
- Dent, E.W.; Gertler, F.B. Cytoskeletal Dynamics and Transport in Growth Cone Motility and Axon Guidance. Neuron 2003, 40, 209–227. [Google Scholar] [CrossRef]
- Letourneau, P.C. Differences in the organization of actin in the growth cones compared with the neurites of cultured neurons from chick embryos. J. Cell Biol. 1983, 97, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Flynn, K.C.; Hellal, F.; Neukirchen, D.; Jacob, S.; Tahirovic, S.; Dupraz, S.; Stern, S.; Garvalov, B.K.; Gurniak, C.; Shaw, A.E.; et al. ADF/Cofilin-Mediated Actin Retrograde Flow Directs Neurite Formation in the Developing Brain. Neuron 2012, 76, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Mallavarapu, A.; Mitchison, T. Regulated Actin Cytoskeleton Assembly at Filopodium Tips Controls Their Extension and Retraction. J. Cell Biol. 1999, 146, 1097–1106. [Google Scholar] [CrossRef]
- Mitchison, T.; Kirschner, M. Cytoskeletal dynamics and nerve growth. Neuron 1988, 1, 761–772. [Google Scholar] [CrossRef]
- Witte, H.; Neukirchen, D.; Bradke, F. Microtubule stabilization specifies initial neuronal polarization. J. Cell Biol. 2008, 180, 619–632. [Google Scholar] [CrossRef]
- Tang, N.H.; Chisholm, A.D. Regulation of Microtubule Dynamics in Axon Regeneration: Insights from C. elegans. F1000Research 2016, 5, 764. [Google Scholar] [CrossRef] [PubMed]
- Baas, P.W.; Rao, A.N.; Matamoros, A.J.; Leo, L. Stability properties of neuronal microtubules. Cytoskeleton 2016, 73, 442–460. [Google Scholar] [CrossRef]
- Forscher, P.; Smith, S.J. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 1988, 107, 1505–1516. [Google Scholar] [CrossRef]
- Biswas, S.; Kalil, K. The Microtubule-Associated Protein Tau Mediates the Organization of Microtubules and Their Dynamic Exploration of Actin-Rich Lamellipodia and Filopodia of Cortical Growth Cones. J. Neurosci. 2018, 38, 291–307. [Google Scholar] [CrossRef]
- Schaefer, A.W.; Kabir, N.; Forscher, P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 2002, 158, 139–152. [Google Scholar] [CrossRef]
- Hanz, S.; Perlson, E.; Willis, D.; Zheng, J.-Q.; Massarwa, R.; Huerta, J.J.; Koltzenburg, M.; Kohler, M.; Van-Minnen, J.; Twiss, J.L.; et al. Axoplasmic Importins Enable Retrograde Injury Signaling in Lesioned Nerve. Neuron 2003, 40, 1095–1104. [Google Scholar] [CrossRef]
- Hill, C.E.; Beattie, M.S.; Bresnahan, J.C. Degeneration and Sprouting of Identified Descending Supraspinal Axons after Contusive Spinal Cord Injury in the Rat. Exp. Neurol. 2001, 171, 153–169. [Google Scholar] [CrossRef]
- Bradke, F.; Fawcett, J.W.; Spira, M.E. Assembly of a new growth cone after axotomy: The precursor to axon regeneration. Nat. Rev. Neurosci. 2012, 13, 183–193. [Google Scholar] [CrossRef]
- Sahly, I.; Khoutorsky, A.; Erez, H.; Spira, M.E.; Prager-Khoutorsky, M. On-line confocal imaging of the events leading to structural dedifferentiation of an axonal segment into a growth cone after axotomy. J. Comp. Neurol. 2005, 494, 705–720. [Google Scholar] [CrossRef]
- Erez, H.; Malkinson, G.; Prager-Khoutorsky, M.; De Zeeuw, C.I.; Hoogenraad, C.C.; Spira, M.E. Formation of microtubule-based traps controls the sorting and concentration of vesicles to restricted sites of regenerating neurons after axotomy. J. Cell Biol. 2007, 176, 497–507. [Google Scholar] [CrossRef]
- Zakharenko, S.; Popov, S. Dynamics of Axonal Microtubules Regulate the Topology of New Membrane Insertion into the Growing Neurites. J. Cell Biol. 1998, 143, 1077–1086. [Google Scholar] [CrossRef]
- Song, Y.; Ori-McKenney, K.M.; Zheng, Y.; Han, C.; Jan, L.Y.; Jan, Y.N. Regeneration of Drosophila sensory neuron axons and dendrites is regulated by the Akt pathway involving Pten and microRNA bantam. Genes Dev. 2012, 26, 1612–1625. [Google Scholar] [CrossRef]
- Stone, M.C.; Nguyen, M.M.; Tao, J.; Allender, D.L.; Rolls, M.M. Global Up-Regulation of Microtubule Dynamics and Polarity Reversal during Regeneration of an Axon from a Dendrite. Mol. Biol. Cell 2010, 21, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Ghosh-Roy, A.; Goncharov, A.; Jin, Y.; Chisholm, A.D. Kinesin-13 and Tubulin Posttranslational Modifications Regulate Microtubule Growth in Axon Regeneration. Dev. Cell 2012, 23, 716–728. [Google Scholar] [CrossRef]
- Chen, L.; Chuang, M.; Koorman, T.; Boxem, M.; Jin, Y.; Chisholm, A.D. Axon injury triggers EFA-6 mediated destabilization of axonal microtubules via TACC and doublecortin like kinase. eLife 2015, 4, e08695. [Google Scholar] [CrossRef]
- Li, Y.; Raisman, G. Sprouts from Cut Corticospinal Axons Persist in the Presence of Astrocytic Scarring in Long-Term Lesions of the Adult Rat Spinal Cord. Exp. Neurol. 1995, 134, 102–111. [Google Scholar] [CrossRef]
- Tom, V.J.; Steinmetz, M.P.; Miller, J.H.; Doller, C.M.; Silver, J. Studies on the Development and Behavior of the Dystrophic Growth Cone, the Hallmark of Regeneration Failure, in an In Vitro Model of the Glial Scar and after Spinal Cord Injury. J. Neurosci. 2004, 24, 6531–6539. [Google Scholar] [CrossRef]
- Ertürk, A.; Hellal, F.; Enes, J.; Bradke, F. Disorganized Microtubules Underlie the Formation of Retraction Bulbs and the Failure of Axonal Regeneration. J. Neurosci. 2007, 27, 9169–9180. [Google Scholar] [CrossRef]
- Steinmetz, M.P.; Horn, K.P.; Tom, V.J.; Miller, J.H.; Busch, S.A.; Nair, D.; Silver, D.J.; Silver, J. Chronic Enhancement of the Intrinsic Growth Capacity of Sensory Neurons Combined with the Degradation of Inhibitory Proteoglycans Allows Functional Regeneration of Sensory Axons through the Dorsal Root Entry Zone in the Mammalian Spinal Cord. J. Neurosci. 2005, 25, 8066–8076. [Google Scholar] [CrossRef]
- Ruschel, J.; Hellal, F.; Flynn, K.C.; Dupraz, S.; Elliott, D.A.; Tedeschi, A.; Bates, M.; Sliwinski, C.; Brook, G.; Dobrindt, K.; et al. Systemic administration of epothilone B promotes axon regeneration after spinal cord injury. Science 2015, 348, 347–352. [Google Scholar] [CrossRef]
- Sengottuvel, V.; Leibinger, M.; Pfreimer, M.; Andreadaki, A.; Fischer, D. Taxol Facilitates Axon Regeneration in the Mature CNS. J. Neurosci. 2011, 31, 2688–2699. [Google Scholar] [CrossRef] [PubMed]
- Hellal, F.; Hurtado, A.; Ruschel, J.; Flynn, K.C.; Laskowski, C.J.; Umlauf, M.; Kapitein, L.C.; Strikis, D.; Lemmon, V.; Bixby, J.; et al. Microtubule Stabilization Reduces Scarring and Causes Axon Regeneration after Spinal Cord Injury. Science 2011, 331, 928–931. [Google Scholar] [CrossRef]
- Smith, D.S.; Skene, J.H.P. A Transcription-Dependent Switch Controls Competence of Adult Neurons for Distinct Modes of Axon Growth. J. Neurosci. 1997, 17, 646–658. [Google Scholar] [CrossRef] [PubMed]
- Kaas, J.H.; Qi, H.-X.; Burish, M.J.; Gharbawie, O.A.; Onifer, S.M.; Massey, J.M. Cortical and subcortical plasticity in the brains of humans, primates, and rats after damage to sensory afferents in the dorsal columns of the spinal cord. Exp. Neurol. 2008, 209, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2009, 339, 269–280. [Google Scholar] [CrossRef]
- Andrews, M.R.; Soleman, S.; Cheah, M.; Tumbarello, D.A.; Mason, M.R.J.; Moloney, E.; Verhaagen, J.; Bensadoun, J.-C.; Schneider, B.; Aebischer, P.; et al. Axonal Localization of Integrins in the CNS Is Neuronal Type and Age Dependent. eNeuro 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Monsul, N.T.; Geisendorfer, A.R.; Han, P.J.; Banik, R.; Pease, M.; Skolasky, R.L.; Hoffman, P.N. Intraocular injection of dibutyryl cyclic AMP promotes axon regeneration in rat optic nerve. Exp. Neurol. 2004, 186, 124–133. [Google Scholar] [CrossRef]
- Qiu, J.; Cai, D.; Dai, H.; McAtee, M.; Hoffman, P.N.; Bregman, B.S.; Filbin, M.T. Spinal Axon Regeneration Induced by Elevation of Cyclic AMP. Neuron 2002, 34, 895–903. [Google Scholar] [CrossRef]
- Leon, S.; Yin, Y.; Nguyen, J.; Irwin, N.; Benowitz, L.I. Lens Injury Stimulates Axon Regeneration in the Mature Rat Optic Nerve. J. Neurosci. 2000, 20, 4615–4626. [Google Scholar] [CrossRef]
- Richardson, P.M.; Issa, V.M.K. Peripheral injury enhances central regeneration of primary sensory neurones. Nature 1984, 309, 791–793. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, F.; Udina, E.; Navarro, X. Extracellular Matrix Components in Peripheral Nerve Regeneration. Int. Rev. Neurobiol. 2013, 108, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Alé, A.; Santos, D.; Barwig, C.; Freier, T.; Navarro, X.; Udina, E.; Gonzalez-Perez, F. Substratum preferences of motor and sensory neurons in postnatal and adult rats. Eur. J. Neurosci. 2015, 43, 431–442. [Google Scholar] [CrossRef]
- Gardiner, N.J.; Fernyhough, P.; Tomlinson, D.R.; Mayer, U.; von der Mark, H.; Streuli, C.H. α7 integrin mediates neurite outgrowth of distinct populations of adult sensory neurons. Mol. Cell. Neurosci. 2005, 28, 229–240. [Google Scholar] [CrossRef]
- Wallquist, W.; Zelano, J.; Plantman, S.; Kaufman, S.J.; Cullheim, S.; Hammarberg, H. Dorsal root ganglion neurons up-regulate the expression of laminin-associated integrins after peripheral but not central axotomy. J. Comp. Neurol. 2004, 480, 162–169. [Google Scholar] [CrossRef]
- Hammarberg, H.; Wallquist, W.; Piehl, F.; Cullheim, S. Regulation of laminin-associated integrin subunit mRNAs in rat spinal motoneurons during postnatal development and after axonal injury. J. Comp. Neurol. 2000, 428, 294–304. [Google Scholar] [CrossRef]
- Werner, A.; Willem, M.; Jones, L.L.; Kreutzberg, G.W.; Mayer, U.; Raivich, G. Impaired Axonal Regeneration in α7 Integrin-Deficient Mice. J. Neurosci. 2000, 20, 1822–1830. [Google Scholar] [CrossRef] [PubMed]
- Kloss, C.U.; Werner, A.; Klein, M.A.; Shen, J.; Menuz, K.; Probst, J.C.; Kreutzberg, G.W.; Raivich, G. Integrin family of cell adhesion molecules in the injured brain: Regulation and cellular localization in the normal and regenerating mouse facial motor nucleus. J. Comp. Neurol. 1999, 411, 162–178. [Google Scholar] [CrossRef]
- Ekstrom, P.A.; Mayer, U.; Panjwani, A.; Pountney, D.; Pizzey, J.; Tonge, D.A. Involvement of α7β1 integrin in the conditioning-lesion effect on sensory axon regeneration. Mol. Cell. Neurosci. 2003, 22, 383–395. [Google Scholar] [CrossRef]
- Bentley, M.; Banker, G. The cellular mechanisms that maintain neuronal polarity. Nat. Rev. Neurosci. 2016, 17, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Britt, D.J.; Farías, G.G.; Guardia, C.M.; Bonifacino, J.S. Mechanisms of Polarized Organelle Distribution in Neurons. Front. Cell. Neurosci. 2016, 10, 88. [Google Scholar] [CrossRef] [PubMed]
- Lasiecka, Z.M.; Winckler, B. Mechanisms of polarized membrane trafficking in neurons—Focusing in on endosomes. Mol. Cell. Neurosci. 2011, 48, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Franssen, E.H.P.; Zhao, R.-R.; Koseki, H.; Kanamarlapudi, V.; Hoogenraad, C.C.; Eva, R.; Fawcett, J.W. Exclusion of Integrins from CNS Axons Is Regulated by Arf6 Activation and the AIS. J. Neurosci. 2015, 35, 8359–8375. [Google Scholar] [CrossRef]
- Bi, X.; Lynch, G.; Zhou, J.; Gall, C.M. Polarized distribution of α5 integrin in dendrites of hippocampal and cortical neurons. J. Comp. Neurol. 2001, 435, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Powelka, A.M.; Sun, J.; Li, J.; Gao, M.; Shaw, L.M.; Sonnenberg, A.; Hsu, V.W. Stimulation-dependent recycling of integrin β1 regulated by ARF6 and Rab11. Traffic 2004, 5, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Eva, R.; Dassie, E.; Caswell, P.T.; Dick, G.; Ffrench-Constant, C.; Norman, J.C.; Fawcett, J.W. Rab11 and its effector Rab coupling protein contribute to the trafficking of β1 integrins during axon growth in adult dorsal root ganglion neurons and PC12 cells. J. Neurosci. 2010, 30, 11654–11669. [Google Scholar] [CrossRef] [PubMed]
- Caswell, P.T.; Chan, M.; Lindsay, A.J.; McCaffrey, M.W.; Boettiger, D.; Norman, J.C. Rab-coupling protein coordinates recycling of α5β1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 2008, 183, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuis, B.; Haenzi, B.; Andrews, M.R.; Verhaagen, J.; Fawcett, J.W. Integrins promote axonal regeneration after injury of the nervous system. Biol. Rev. 2018, 93, 1339–1362. [Google Scholar] [CrossRef]
- Weisenberg, R.C. Microtubule Formation in vitro in Solutions Containing Low Calcium Concentrations. Science 1972, 177, 1104–1105. [Google Scholar] [CrossRef] [PubMed]
- Karr, T.L.; Kristofferson, D.; Purich, D.L. Calcium ion induces endwise depolymerization of bovine brain microtubules. J. Biol. Chem. 1980, 255, 11853–11856. [Google Scholar] [CrossRef]
- Marcum, J.M.; Dedman, J.R.; Brinkley, B.R.; Means, A.R. Control of microtubule assembly-disassembly by calcium-dependent regulator protein. Proc. Natl. Acad. Sci. USA 1978, 75, 3771–3775. [Google Scholar] [CrossRef]
- Nishida, E.; Sakai, H. Calcium-sensitivity of the microtubule reassembly system. Difference between crude brain extract and purified microtubular proteins. J. Biochem. 1977, 82, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, A.C.; Zackroff, R.V.; Weisenberg, R.C. Magnesium stimulation of calcium binding to tubulin and calcium induced depolymerization of microtubules. FEBS Lett. 1976, 65, 144–147. [Google Scholar] [CrossRef]
- Weisenberg, R.C.; Deery, W.J. The mechanism of calcium-induced microtubule disassembly. Biochem. Biophys. Res. Commun. 1981, 102, 924–931. [Google Scholar] [CrossRef]
- Ziv, N.E.; Spira, M.E. Localized and Transient Elevations of Intracellular Ca2+ Induce the Dedifferentiation of Axonal Segments into Growth Cones. J. Neurosci. 1997, 17, 3568–3579. [Google Scholar] [CrossRef] [PubMed]
- Nwagbara, B.U.; Faris, A.E.; Bearce, E.A.; Erdogan, B.; Ebbert, P.T.; Evans, M.F.; Rutherford, E.L.; Enzenbacher, T.B.; Lowery, L.A. TACC3 is a microtubule plus end-tracking protein that promotes axon elongation and also regulates microtubule plus end dynamics in multiple embryonic cell types. Mol. Biol. Cell 2014, 25, 3350–3362. [Google Scholar] [CrossRef]
- Nawabi, H.; Belin, S.; Cartoni, R.; Williams, P.R.; Wang, C.; Latremolière, A.; Wang, X.; Zhu, J.; Taub, D.; Fu, X.; et al. Doublecortin-Like Kinases Promote Neuronal Survival and Induce Growth Cone Reformation via Distinct Mechanisms. Neuron 2015, 88, 704–719. [Google Scholar] [CrossRef] [PubMed]
- Song, A.-H.; Wang, D.; Chen, G.; Li, Y.; Luo, J.; Duan, S.; Poo, M.-M. A Selective Filter for Cytoplasmic Transport at the Axon Initial Segment. Cell 2009, 136, 1148–1160. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.L.; Svitkina, T.M. Axon Initial Segment Cytoskeleton: Architecture, Development, and Role in Neuron Polarity. Neural Plast. 2016, 2016, 6808293. [Google Scholar] [CrossRef] [PubMed]
- Arnold, D.B. Actin and Microtubule-Based Cytoskeletal Cues Direct Polarized Targeting of Proteins in Neurons. Sci. Signal. 2009, 2, pe49. [Google Scholar] [CrossRef]
- Lewis, T.L.; Mao, T.; Svoboda, K.; Arnold, D.B. Myosin-dependent targeting of transmembrane proteins to neuronal dendrites. Nat. Neurosci. 2009, 12, 568–576. [Google Scholar] [CrossRef]
- Eva, R.; Koseki, H.; Kanamarlapudi, V.; Fawcett, J.W. EFA6 regulates selective polarised transport and axon regeneration from the axon initial segment. J. Cell Sci. 2017, 130, 3663–3675. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez, G.; Chen, L. RETRACTED: EFA6 in Axon Regeneration, as a Microtubule Regulator and as a Guanine Nucleotide Exchange Factor. Cells 2021, 10, 1325. https://doi.org/10.3390/cells10061325
Gonzalez G, Chen L. RETRACTED: EFA6 in Axon Regeneration, as a Microtubule Regulator and as a Guanine Nucleotide Exchange Factor. Cells. 2021; 10(6):1325. https://doi.org/10.3390/cells10061325
Chicago/Turabian StyleGonzalez, Gilberto, and Lizhen Chen. 2021. "RETRACTED: EFA6 in Axon Regeneration, as a Microtubule Regulator and as a Guanine Nucleotide Exchange Factor" Cells 10, no. 6: 1325. https://doi.org/10.3390/cells10061325
APA StyleGonzalez, G., & Chen, L. (2021). RETRACTED: EFA6 in Axon Regeneration, as a Microtubule Regulator and as a Guanine Nucleotide Exchange Factor. Cells, 10(6), 1325. https://doi.org/10.3390/cells10061325




