Regulation of the Phosphoinositide Code by Phosphorylation of Membrane Readers
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Comparative Analysis of Membrane Binding Poses
3.2. Regulation Is Driven by PIP Specificity
3.3. Kinases Acting on PIP-Stops
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Overduin, M.; Cheever, M.L.; Kutateladze, T.G. Signaling with phosphoinositides: Better than binary. Mol. Interv. 2001, 1, 150–159. [Google Scholar]
- Sato, T.K.; Overduin, M.; Emr, S.D. Location, location, location: Membrane targeting directed by PX domains. Science 2001, 294, 1881–1885. [Google Scholar] [CrossRef]
- Chandra, M.; Chin, Y.K.; Mas, C.; Feathers, J.R.; Paul, B.; Datta, S.; Chen, K.E.; Jia, X.; Yang, Z.; Norwood, S.J.; et al. Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 2019, 10, 1528. [Google Scholar] [CrossRef]
- Samavarchi-Tehrani, P.; Abdouni, H.; Knight, J.D.; Astori, A.; Samson, R.; Lin, Z.Y.; Kim, D.K.; Knapp, J.J.; St-Germain, J.; Go, C.D.; et al. A SARS-CoV-2—Host proximity interactome. BioRxiv 2021. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic. Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef]
- Carlton, J.G.; Cullen, P.J. Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 2005, 15, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.; Radulovic, M.; Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef]
- Lenoir, M.; Ustunel, C.; Rajesh, S.; Kaur, J.; Moreau, D.; Gruenberg, J.; Overduin, M. Phosphorylation of conserved phosphoinositide binding pocket regulates sorting nexin membrane targeting. Nat. Commun. 2018, 9, 993. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic. Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview version 2: A Multiple Sequence Alignment and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Neves, M.A.; Totrov, M.; Abagyan, R. Docking and scoring with ICM: The benchmarking results and strategies for improvement. J. Comput. Aided Mol. Des. 2012, 26, 675–686. [Google Scholar] [CrossRef]
- Kufareva, I.; Lenoir, M.; Dancea, F.; Sridhar, P.; Raush, E.; Bissig, C.; Gruenberg, J.; Abagyan, R.; Overduin, M. Discovery of novel membrane binding structures and functions. Biochem. Cell Biol. 2014, 92, 555–563. [Google Scholar] [CrossRef]
- Hayashi, T.; Okabe, T.; Nasu-Nishimura, Y.; Sakaue, F.; Ohwada, S.; Matsuura, K.; Akiyama, T.; Nakamura, T. PX-RICS, a novel splicing variant of RICS, is a main isoform expressed during neural development. Genes Cells 2007, 12, 929–939. [Google Scholar] [CrossRef]
- Chiang, S.H.; Hwang, J.; Legendre, M.; Zhang, M.; Kimura, A.; Saltiel, A.R. TCGAP, a multidomain Rho GTPase-activating protein involved in insulin-stimulated glucose transport. EMBO J. 2003, 22, 2679–2691. [Google Scholar] [CrossRef]
- Holland, P.; Knaevelsrud, H.; Soreng, K.; Mathai, B.J.; Lystad, A.H.; Pankiv, S.; Bjorndal, G.T.; Schultz, S.W.; Lobert, V.H.; Chan, R.B.; et al. HS1BP3 negatively regulates autophagy by modulation of phosphatidic acid levels. Nat. Commun. 2016, 7, 13889. [Google Scholar] [CrossRef]
- Pyrpassopoulos, S.; Shuman, H.; Ostap, E.M. Adhesion force and attachment lifetime of the KIF16B-PX domain interaction with lipid membranes. Mol. Biol. Cell 2017, 28, 3315–3322. [Google Scholar] [CrossRef]
- Blatner, N.R.; Wilson, M.I.; Lei, C.; Hong, W.; Murray, D.; Williams, R.L.; Cho, W. The structural basis of novel endosome anchoring activity of KIF16B kinesin. EMBO J. 2007, 26, 3709–3719. [Google Scholar] [CrossRef]
- Lim, K.P.; Hong, W. Human Nischarin/imidazoline receptor antisera-selected protein is targeted to the endosomes by a combined action of a PX domain and a coiled-coil region. J. Biol. Chem 2004, 279, 54770–54782. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Lambeth, J.D. NOXO1, regulation of lipid binding, localization, and activation of Nox1 by the Phox homology (PX) domain. J. Biol. Chem 2004, 279, 4737–4742. [Google Scholar] [CrossRef] [PubMed]
- Ueyama, T.; Lekstrom, K.; Tsujibe, S.; Saito, N.; Leto, T.L. Subcellular localization and function of alternatively spliced Noxo1 isoforms. Free Radic Biol. Med. 2007, 42, 180–190. [Google Scholar] [CrossRef]
- Davis, N.Y.; McPhail, L.C.; Horita, D.A. The NOXO1beta PX domain preferentially targets PtdIns(4,5)P2 and PtdIns(3,4,5)P3. J. Mol. Biol. 2012, 417, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Lambeth, J.D. Alternative mRNA splice forms of NOXO1: Differential tissue expression and regulation of Nox1 and Nox3. Gene 2005, 356, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xu, W.; Zhang, A.; Huang, G.; Liang, X.; Virbasius, J.V.; Czech, M.P.; Zhou, G.W. Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry 2001, 40, 8940–8944. [Google Scholar] [CrossRef]
- Stahelin, R.V.; Karathanassis, D.; Bruzik, K.S.; Waterfield, M.D.; Bravo, J.; Williams, R.L.; Cho, W. Structural and membrane binding analysis of the Phox homology domain of phosphoinositide 3-kinase-C2alpha. J. Biol. Chem. 2006, 281, 39396–39406. [Google Scholar] [CrossRef]
- Du, G.; Altshuller, Y.M.; Vitale, N.; Huang, P.; Chasserot-Golaz, S.; Morris, A.J.; Bader, M.F.; Frohman, M.A. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 2003, 162, 305–315. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, J.H.; Jang, I.H.; Kim, H.S.; Han, J.M.; Kazlauskas, A.; Yagisawa, H.; Suh, P.G.; Ryu, S.H. Phosphatidylinositol (3,4,5)-trisphosphate specifically interacts with the phox homology domain of phospholipase D1 and stimulates its activity. J. Cell Sci. 2005, 118, 4405–4413. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stahelin, R.V.; Ananthanarayanan, B.; Blatner, N.R.; Singh, S.; Bruzik, K.S.; Murray, D.; Cho, W. Mechanism of membrane binding of the phospholipase D1 PX domain. J. Biol. Chem. 2004, 279, 54918–54926. [Google Scholar] [CrossRef]
- Sciorra, V.A.; Rudge, S.A.; Prestwich, G.D.; Frohman, M.A.; Engebrecht, J.; Morris, A.J. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 1999, 18, 5911–5921. [Google Scholar] [CrossRef] [PubMed]
- Mahankali, M.; Henkels, K.M.; Gomez-Cambronero, J. A GEF-to-phospholipase molecular switch caused by phosphatidic acid, Rac and JAK tyrosine kinase that explains leukocyte cell migration. J. Cell Sci. 2013, 126, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Pastor, R.W.; Fenollar-Ferrer, C. PLD2-PI(4,5)P2 interactions in fluid phase membranes: Structural modeling and molecular dynamics simulations. PLoS ONE 2020, 15, e0236201. [Google Scholar] [CrossRef]
- Takeuchi, H.; Takeuchi, T.; Gao, J.; Cantley, L.C.; Hirata, M. Characterization of PXK as a protein involved in epidermal growth factor receptor trafficking. Mol. Cell Biol. 2010, 30, 1689–1702. [Google Scholar] [CrossRef]
- Kanai, F.; Liu, H.; Field, S.J.; Akbary, H.; Matsuo, T.; Brown, G.E.; Cantley, L.C.; Yaffe, M.B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001, 3, 675–678. [Google Scholar] [CrossRef]
- Ellson, C.D.; Gobert-Gosse, S.; Anderson, K.E.; Davidson, K.; Erdjument-Bromage, H.; Tempst, P.; Thuring, J.W.; Cooper, M.A.; Lim, Z.Y.; Holmes, A.B.; et al. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox). Nat. Cell Biol. 2001, 3, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.; Karathanassis, D.; Pacold, C.M.; Pacold, M.E.; Ellson, C.D.; Anderson, K.E.; Butler, P.J.; Lavenir, I.; Perisic, O.; Hawkins, P.T.; et al. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell 2001, 8, 829–839. [Google Scholar] [CrossRef]
- Ago, T.; Takeya, R.; Hiroaki, H.; Kuribayashi, F.; Ito, T.; Kohda, D.; Sumimoto, H. The PX domain as a novel phosphoinositide- binding module. Biochem. Biophys. Res. Commun. 2001, 287, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Okada, T.; Igarashi, N.; Fujita, T.; Jahangeer, S.; Nakamura, S. Identification and characterization of RPK118, a novel sphingosine kinase-1-binding protein. J. Biol. Chem. 2002, 277, 33319–33324. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, C.; Yuan, J.; Chen, X.; Xu, J.; Wei, Y.; Yang, J.; Lin, G.; Yu, L. RPK118, a PX domain-containing protein, interacts with peroxiredoxin-3 through pseudo-kinase domains. Mol. Cells 2005, 19, 39–45. [Google Scholar] [PubMed]
- Xu, J.; Liu, D.; Gill, G.; Songyang, Z. Regulation of cytokine-independent survival kinase (CISK) by the Phox homology domain and phosphoinositides. J. Cell Biol. 2001, 154, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Virbasius, J.V.; Song, X.; Pomerleau, D.P.; Zhan, Y.; Zhou, G.W.; Czech, M.P. Activation of the Akt-related cytokine-independent survival kinase requires interaction of its phox domain with endosomal phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA 2001, 98, 12908–12913. [Google Scholar] [CrossRef] [PubMed]
- Abram, C.L.; Seals, D.F.; Pass, I.; Salinsky, D.; Maurer, L.; Roth, T.M.; Courtneidge, S.A. The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J. Biol. Chem. 2003, 278, 16844–16851. [Google Scholar] [CrossRef]
- Buschman, M.D.; Bromann, P.A.; Cejudo-Martin, P.; Wen, F.; Pass, I.; Courtneidge, S.A. The novel adaptor protein Tks4 (SH3PXD2B) is required for functional podosome formation. Mol. Biol. Cell 2009, 20, 1302–1311. [Google Scholar] [CrossRef]
- Zhong, Q.; Lazar, C.S.; Tronchere, H.; Sato, T.; Meerloo, T.; Yeo, M.; Songyang, Z.; Emr, S.D.; Gill, G.N. Endosomal localization and function of sorting nexin 1. Proc. Natl. Acad. Sci. USA 2002, 99, 6767–6772. [Google Scholar] [CrossRef]
- Cozier, G.E.; Carlton, J.; McGregor, A.H.; Gleeson, P.A.; Teasdale, R.D.; Mellor, H.; Cullen, P.J. The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J. Biol. Chem. 2002, 277, 48730–48736. [Google Scholar] [CrossRef]
- Ceccato, L.; Chicanne, G.; Nahoum, V.; Pons, V.; Payrastre, B.; Gaits-Iacovoni, F.; Viaud, J. PLIF: A rapid, accurate method to detect and quantitatively assess protein-lipid interactions. Sci. Signal 2016, 9, rs2. [Google Scholar] [CrossRef] [PubMed]
- Catimel, B.; Schieber, C.; Condron, M.; Patsiouras, H.; Connolly, L.; Catimel, J.; Nice, E.C.; Burgess, A.W.; Holmes, A.B. The PI(3,5)P2 and PI(4,5)P2 interactomes. J. Proteome Res. 2008, 7, 5295–5313. [Google Scholar] [CrossRef] [PubMed]
- Carlton, J.; Bujny, M.; Peter, B.J.; Oorschot, V.M.; Rutherford, A.; Mellor, H.; Klumperman, J.; McMahon, H.T.; Cullen, P.J. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr. Biol. 2004, 14, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Carlton, J.G.; Bujny, M.V.; Peter, B.J.; Oorschot, V.M.; Rutherford, A.; Arkell, R.S.; Klumperman, J.; McMahon, H.T.; Cullen, P.J. Sorting nexin-2 is associated with tubular elements of the early endosome, but is not essential for retromer-mediated endosome-to-TGN transport. J. Cell Sci. 2005, 118, 4527–4539. [Google Scholar] [CrossRef]
- Xu, Y.; Hortsman, H.; Seet, L.; Wong, S.H.; Hong, W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 2001, 3, 658–666. [Google Scholar] [CrossRef]
- Traer, C.J.; Rutherford, A.C.; Palmer, K.J.; Wassmer, T.; Oakley, J.; Attar, N.; Carlton, J.G.; Kremerskothen, J.; Stephens, D.J.; Cullen, P.J. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol. 2007, 9, 1370–1380. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Z.Q.; Chen, C.X.; Magill, S.; Jiang, Y.; Liu, Y.J. Inhibitory regulation of EGF receptor degradation by sorting nexin 5. Biochem. Biophys. Res. Commun. 2006, 342, 537–546. [Google Scholar] [CrossRef]
- Koharudin, L.M.; Furey, W.; Liu, H.; Liu, Y.J.; Gronenborn, A.M. The phox domain of sorting nexin 5 lacks phosphatidylinositol 3-phosphate (PtdIns(3)P) specificity and preferentially binds to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2). J. Biol. Chem. 2009, 284, 23697–23707. [Google Scholar] [CrossRef]
- Catimel, B.; Yin, M.X.; Schieber, C.; Condron, M.; Patsiouras, H.; Catimel, J.; Robinson, D.E.; Wong, L.S.; Nice, E.C.; Holmes, A.B.; et al. PI(3,4,5)P3 Interactome. J. Proteome Res. 2009, 8, 3712–3726. [Google Scholar] [CrossRef]
- Merino-Trigo, A.; Kerr, M.C.; Houghton, F.; Lindberg, A.; Mitchell, C.; Teasdale, R.D.; Gleeson, P.A. Sorting nexin 5 is localized to a subdomain of the early endosomes and is recruited to the plasma membrane following EGF stimulation. J. Cell Sci. 2004, 117, 6413–6424. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, C.; Sun, Z.; Hong, Z.; Li, K.; Sun, D.; Yang, Y.; Tian, C.; Gong, W.; Liu, J.J. PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nat. Cell Biol. 2013, 15, 417–429. [Google Scholar] [CrossRef]
- van Weering, J.R.; Verkade, P.; Cullen, P.J. SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic 2012, 13, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Dyve, A.B.; Bergan, J.; Utskarpen, A.; Sandvig, K. Sorting nexin 8 regulates endosome-to-Golgi transport. Biochem. Biophys. Res. Commun. 2009, 390, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Lundmark, R.; Carlsson, S.R. Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J. Biol. Chem. 2003, 278, 46772–46781. [Google Scholar] [CrossRef] [PubMed]
- Pylypenko, O.; Lundmark, R.; Rasmuson, E.; Carlsson, S.R.; Rak, A. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007, 26, 4788–4800. [Google Scholar] [CrossRef] [PubMed]
- Yarar, D.; Waterman-Storer, C.M.; Schmid, S.L. SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev. Cell 2007, 13, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Yarar, D.; Surka, M.C.; Leonard, M.C.; Schmid, S.L. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic 2008, 9, 133–146. [Google Scholar] [CrossRef]
- Xu, J.; Xu, T.; Wu, B.; Ye, Y.; You, X.; Shu, X.; Pei, D.; Liu, J. Structure of sorting nexin 11 (SNX11) reveals a novel extended phox homology (PX) domain critical for inhibition of SNX10-induced vacuolation. J. Biol. Chem. 2013, 288, 16598–16605. [Google Scholar] [CrossRef]
- Xu, T.; Gan, Q.; Wu, B.; Yin, M.; Xu, J.; Shu, X.; Liu, J. Molecular Basis for PI(3,5)P2 Recognition by SNX11, a Protein Involved in Lysosomal Degradation and Endosome Homeostasis Regulation. J. Mol. Biol. 2020, 432, 4750–4761. [Google Scholar] [CrossRef]
- Pons, V.; Ustunel, C.; Rolland, C.; Torti, E.; Parton, R.G.; Gruenberg, J. SNX12 role in endosome membrane transport. PLoS ONE 2012, 7, e38949. [Google Scholar] [CrossRef]
- Mas, C.; Norwood, S.J.; Bugarcic, A.; Kinna, G.; Leneva, N.; Kovtun, O.; Ghai, R.; Ona Yanez, L.E.; Davis, J.L.; Teasdale, R.D.; et al. Structural basis for different phosphoinositide specificities of the PX domains of sorting nexins regulating G-protein signaling. J. Biol. Chem. 2014, 289, 28554–28568. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Ma, Y.C.; Ostrom, R.S.; Lavoie, C.; Gill, G.N.; Insel, P.A.; Huang, X.Y.; Farquhar, M.G. RGS-PX1, a GAP for GalphaS and sorting nexin in vesicular trafficking. Science 2001, 294, 1939–1942. [Google Scholar] [CrossRef]
- Danson, C.; Brown, E.; Hemmings, O.J.; McGough, I.J.; Yarwood, S.; Heesom, K.J.; Carlton, J.G.; Martin-Serrano, J.; May, M.T.; Verkade, P.; et al. SNX15 links clathrin endocytosis to the PtdIns3P early endosome independently of the APPL1 endosome. J. Cell Sci. 2013, 126, 4885–4899. [Google Scholar] [CrossRef]
- Barr, V.A.; Phillips, S.A.; Taylor, S.I.; Haft, C.R. Overexpression of a novel sorting nexin, SNX15, affects endosome morphology and protein trafficking. Traffic 2000, 1, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B.J.; Hong, W. Evidence for a role of SNX16 in regulating traffic between the early and later endosomal compartments. J. Biol. Chem. 2003, 278, 34617–34630. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, L.; Ye, Y.; Shan, Y.; Wan, C.; Wang, J.; Pei, D.; Shu, X.; Liu, J. SNX16 Regulates the Recycling of E-Cadherin through a Unique Mechanism of Coordinated Membrane and Cargo Binding. Structure 2017, 25, 1251–1263.E5. [Google Scholar] [CrossRef]
- Czubayko, M.; Knauth, P.; Schluter, T.; Florian, V.; Bohnensack, R. Sorting nexin 17, a non-self-assembling and a PtdIns(3)P high class affinity protein, interacts with the cerebral cavernous malformation related protein KRIT1. Biochem. Biophys. Res. Commun. 2006, 345, 1264–1272. [Google Scholar] [CrossRef] [PubMed]
- Knauth, P.; Schluter, T.; Czubayko, M.; Kirsch, C.; Florian, V.; Schreckenberger, S.; Hahn, H.; Bohnensack, R. Functions of sorting nexin 17 domains and recognition motif for P-selectin trafficking. J. Mol. Biol. 2005, 347, 813–825. [Google Scholar] [CrossRef]
- Haberg, K.; Lundmark, R.; Carlsson, S.R. SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J. Cell Sci. 2008, 121, 1495–1505. [Google Scholar] [CrossRef]
- Nakazawa, S.; Gotoh, N.; Matsumoto, H.; Murayama, C.; Suzuki, T.; Yamamoto, T. Expression of sorting nexin 18 (SNX18) is dynamically regulated in developing spinal motor neurons. J. Histochem. Cytochem. 2011, 59, 202–213. [Google Scholar] [CrossRef]
- Liebl, D.; Qi, X.; Zhe, Y.; Barnett, T.C.; Teasdale, R.D. SopB-Mediated Recruitment of SNX18 Facilitates Salmonella Typhimurium Internalization by the Host Cell. Front Cell Infect. Microbiol. 2017, 7, 257. [Google Scholar] [CrossRef] [PubMed]
- Clairfeuille, T.; Norwood, S.J.; Qi, X.; Teasdale, R.D.; Collins, B.M. Structure and Membrane Binding Properties of the Endosomal Tetratricopeptide Repeat (TPR) Domain-containing Sorting Nexins SNX20 and SNX21. J. Biol. Chem. 2015, 290, 14504–14517. [Google Scholar] [CrossRef]
- Schaff, U.Y.; Shih, H.H.; Lorenz, M.; Sako, D.; Kriz, R.; Milarski, K.; Bates, B.; Tchernychev, B.; Shaw, G.D.; Simon, S.I. SLIC-1/sorting nexin 20: A novel sorting nexin that directs subcellular distribution of PSGL-1. Eur. J. Immunol. 2008, 38, 550–564. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhao, K.Q.; Newman, C.L.; Vinarov, D.A.; Markley, J.L. Solution structure of human sorting nexin 22. Protein Sci. 2007, 16, 807–814. [Google Scholar] [CrossRef][Green Version]
- Ghai, R.; Mobli, M.; Norwood, S.J.; Bugarcic, A.; Teasdale, R.D.; King, G.F.; Collins, B.M. Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases. Proc. Natl. Acad. Sci. USA 2011, 108, 7763–7768. [Google Scholar] [CrossRef] [PubMed]
- Lunn, M.L.; Nassirpour, R.; Arrabit, C.; Tan, J.; McLeod, I.; Arias, C.M.; Sawchenko, P.E.; Yates, J.R., 3rd; Slesinger, P.A. A unique sorting nexin regulates trafficking of potassium channels via a PDZ domain interaction. Nat. Neurosci. 2007, 10, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Rincon, E.; Saez de Guinoa, J.; Gharbi, S.I.; Sorzano, C.O.; Carrasco, Y.R.; Merida, I. Translocation dynamics of sorting nexin 27 in activated T cells. J. Cell Sci. 2011, 124, 776–788. [Google Scholar] [CrossRef]
- Vieira, N.; Deng, F.M.; Liang, F.X.; Liao, Y.; Chang, J.; Zhou, G.; Zheng, W.; Simon, J.P.; Ding, M.; Wu, X.R.; et al. SNX31: A novel sorting nexin associated with the uroplakin-degrading multivesicular bodies in terminally differentiated urothelial cells. PLoS ONE 2014, 9, e99644. [Google Scholar] [CrossRef]
- Almendinger, J.; Doukoumetzidis, K.; Kinchen, J.M.; Kaech, A.; Ravichandran, K.S.; Hengartner, M.O. A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PLoS ONE 2011, 6, e18325. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.P.; Chircop, M. SNX9, SNX18 and SNX33 are required for progression through and completion of mitosis. J. Cell Sci. 2012, 125, 4372–4382. [Google Scholar] [CrossRef]
- Yu, J.W.; Lemmon, M.A. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 2001, 276, 44179–44184. [Google Scholar] [CrossRef]
- Stahelin, R.V.; Karathanassis, D.; Murray, D.; Williams, R.L.; Cho, W. Structural and membrane binding analysis of the Phox homology domain of Bem1p: Basis of phosphatidylinositol 4-phosphate specificity. J. Biol. Chem. 2007, 282, 25737–25747. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Zhu, L.; Balogi, Z.; Stefan, C.; Pleiss, J.A.; Emr, S.D. Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J. Cell Biol. 2015, 210, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Varlakhanova, N.V.; Tornabene, B.A.; Ramachandran, R.; Zhang, P.; Ford, M.G.J. The cryo-EM structure of the SNX-BAR Mvp1 tetramer. Nat. Commun. 2020, 11, 1506. [Google Scholar] [CrossRef] [PubMed]
- Cheever, M.L.; Sato, T.K.; de Beer, T.; Kutateladze, T.G.; Emr, S.D.; Overduin, M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 2001, 3, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Burda, P.; Padilla, S.M.; Sarkar, S.; Emr, S.D. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 2002, 115, 3889–3900. [Google Scholar] [CrossRef]
- Lee, S.A.; Kovacs, J.; Stahelin, R.V.; Cheever, M.L.; Overduin, M.; Setty, T.G.; Burd, C.G.; Cho, W.; Kutateladze, T.G. Molecular mechanism of membrane docking by the Vam7p PX domain. J. Biol. Chem. 2006, 281. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.Y.; Huang, H.D.; Hung, J.H.; Huang, H.Y.; Yang, Y.S.; Wang, T.H. dbPTM: An information repository of protein post-translational modification. Nucleic Acids Res. 2006, 34, D622–D627. [Google Scholar] [CrossRef]
- Yu, K.; Zhang, Q.; Liu, Z.; Zhao, Q.; Zhang, X.; Wang, Y.; Wang, Z.X.; Jin, Y.; Li, X.; Liu, Z.X.; et al. qPhos: A database of protein phosphorylation dynamics in humans. Nucleic Acids Res. 2019, 47, D451–D458. [Google Scholar] [CrossRef]
- Giansanti, P.; Aye, T.T.; van den Toorn, H.; Peng, M.; van Breukelen, B.; Heck, A.J. An Augmented Multiple-Protease-Based Human Phosphopeptide Atlas. Cell Rep. 2015, 11, 1834–1843. [Google Scholar] [CrossRef]
- Stark, C.; Su, T.C.; Breitkreutz, A.; Lourenco, P.; Dahabieh, M.; Breitkreutz, B.J.; Tyers, M.; Sadowski, I. PhosphoGRID: A database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database 2010, 2010, bap026. [Google Scholar] [CrossRef]
- Lanz, M.C.; Yugandhar, K.; Gupta, S.; Sanford, E.J.; Faca, V.M.; Vega, S.; Joiner, A.M.N.; Fromme, J.C.; Yu, H.; Smolka, M.B. In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 2021, 22, e51121. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sopko, R.; Chung, V.; Foos, M.; Studer, R.A.; Landry, S.D.; Liu, D.; Rabinow, L.; Gnad, F.; Beltrao, P.; et al. iProteinDB: An Integrative Database of Drosophila Post-translational Modifications. G3 (Bethesda) 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Minguez, P.; Letunic, I.; Parca, L.; Garcia-Alonso, L.; Dopazo, J.; Huerta-Cepas, J.; Bork, P. PTMcode v2: A resource for functional associations of post-translational modifications within and between proteins. Nucleic Acids Res. 2015, 43, D494–D502. [Google Scholar] [CrossRef]
- Horn, H.; Schoof, E.M.; Kim, J.; Robin, X.; Miller, M.L.; Diella, F.; Palma, A.; Cesareni, G.; Jensen, L.J.; Linding, R. KinomeXplorer: An integrated platform for kinome biology studies. Nat. Methods 2014, 11, 603–604. [Google Scholar] [CrossRef]
- Hiroaki, H.; Ago, T.; Ito, T.; Sumimoto, H.; Kohda, D. Solution structure of the PX domain, a target of the SH3 domain. Nat. Struct Biol. 2001, 8, 526–530. [Google Scholar] [CrossRef]
- Jang, I.H.; Lee, S.; Park, J.B.; Kim, J.H.; Lee, C.S.; Hur, E.M.; Kim, I.S.; Kim, K.T.; Yagisawa, H.; Suh, P.G.; et al. The direct interaction of phospholipase C-gamma 1 with phospholipase D2 is important for epidermal growth factor signaling. J. Biol. Chem. 2003, 278, 18184–18190. [Google Scholar] [CrossRef]
- Elwell, C.A.; Czudnochowski, N.; von Dollen, J.; Johnson, J.R.; Nakagawa, R.; Mirrashidi, K.; Krogan, N.J.; Engel, J.N.; Rosenberg, O.S. Chlamydia interfere with an interaction between the mannose-6-phosphate receptor and sorting nexins to counteract host restriction. Elife 2017, 6. [Google Scholar] [CrossRef]
- Paul, B.; Kim, H.S.; Kerr, M.C.; Huston, W.M.; Teasdale, R.D.; Collins, B.M. Structural basis for the hijacking of endosomal sorting nexin proteins by Chlamydia trachomatis. Elife 2017, 6. [Google Scholar] [CrossRef]
- Zhou, C.Z.; De La Sierra-Gallay, I.L.; Quevillon-Cheruel, S.; Collinet, B.; Minard, P.; Blondeau, K.; Henckes, G.; Aufrère, R.; Leulliot, N.; Graille, M.; et al. Crystal Structure of the Yeast Phox Homology (PX) Domain Protein Grd19p Complexed to Phosphatidylinositol-3-phosphate. J. Biol. Chem. 2003, 278, 50371–50376. [Google Scholar] [CrossRef]
- Karathanassis, D.; Stahelin, R.V.; Bravo, J.; Perisic, O.; Pacold, C.M.; Cho, W.; Williams, R.L. Binding of the PX domain of p47phoxto phosphatidylinositol 3, 4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002, 21, 5057–5068. [Google Scholar] [CrossRef]
- Stampoulis, P.; Ueda, T.; Matsumoto, M.; Terasawa, H.; Miyano, K.; Sumimoto, H.; Shimada, I. Atypical membrane-embedded phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2)-binding site on p47(phox) Phox homology (PX) domain revealed by NMR. J. Biol. Chem. 2012, 287, 17848–17859. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Arcaro, A.; Zvelebil, M.J.; Wallasch, C.; Ullrich, A.; Waterfield, M.D.; Domin, J. Class II phosphoinositide 3-kinases are downstream targets of activated polypeptide growth factor receptors. Mol. Cell Biol. 2000, 20, 3817–3830. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Wu, R.F.; Xu, Y.C.; Ma, Z.; Nwariaku, F.E.; Sarosi, G.A., Jr.; Terada, L.S. Subcellular targeting of oxidants during endothelial cell migration. J. Cell Biol. 2005, 171, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Evensen, N.A.; Kuscu, C.; Nguyen, H.L.; Zarrabi, K.; Dufour, A.; Kadam, P.; Hu, Y.J.; Pulkoski-Gross, A.; Bahou, W.F.; Zucker, S.; et al. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J. Natl. Cancer Inst. 2013, 105, 1402–1416. [Google Scholar] [CrossRef]
- Haenig, C.; Atias, N.; Taylor, A.K.; Mazza, A.; Schaefer, M.H.; Russ, J.; Riechers, S.P.; Jain, S.; Coughlin, M.; Fontaine, J.F.; et al. Interactome Mapping Provides a Network of Neurodegenerative Disease Proteins and Uncovers Widespread Protein Aggregation in Affected Brains. Cell Rep. 2020, 32, 108050. [Google Scholar] [CrossRef]
- Parks, W.T.; Frank, D.B.; Huff, C.; Renfrew Haft, C.; Martin, J.; Meng, X.; de Caestecker, M.P.; McNally, J.G.; Reddi, A.; Taylor, S.I.; et al. Sorting nexin 6, a novel SNX, interacts with the transforming growth factor-beta family of receptor serine-threonine kinases. J. Biol. Chem. 2001, 276, 19332–19339. [Google Scholar] [CrossRef]
- Behrmann, I.; Smyczek, T.; Heinrich, P.C.; Schmitz-Van de Leur, H.; Komyod, W.; Giese, B.; Muller-Newen, G.; Haan, S.; Haan, C. Janus kinase (Jak) subcellular localization revisited: The exclusive membrane localization of endogenous Janus kinase 1 by cytokine receptor interaction uncovers the Jak.receptor complex to be equivalent to a receptor tyrosine kinase. J. Biol. Chem. 2004, 279, 35486–35493. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, Q.; Song, Y.; Wang, X.; Guo, Q.; Zhang, J.; Li, J.; Han, Y.; Miao, Z.; Li, F. PAK1 regulates RUFY3-mediated gastric cancer cell migration and invasion. Cell Death Dis. 2015, 6, e1682. [Google Scholar] [CrossRef]
- Semenova, G.; Chernoff, J. Targeting PAK1. Biochem. Soc. Trans. 2017, 45, 79–88. [Google Scholar] [CrossRef]
- Wong, W.; Scott, J.D. AKAP signalling complexes: Focal points in space and time. Nat. Rev. Mol. Cell Biol. 2004, 5, 959–970. [Google Scholar] [CrossRef]
- Zhang, H.; Kong, Q.; Wang, J.; Jiang, Y.; Hua, H. Complex roles of cAMP-PKA-CREB signaling in cancer. Exp. Hematol. Oncol. 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Parker, P.J.; Brown, S.J.; Calleja, V.; Chakravarty, P.; Cobbaut, M.; Linch, M.; Marshall, J.J.T.; Martini, S.; McDonald, N.Q.; Soliman, T.; et al. Equivocal, explicit and emergent actions of PKC isoforms in cancer. Nat. Rev. Cancer 2021, 21, 51–63. [Google Scholar] [CrossRef]
- Breitkopf, S.B.; Yuan, M.; Helenius, K.P.; Lyssiotis, C.A.; Asara, J.M. Triomics Analysis of Imatinib-Treated Myeloma Cells Connects Kinase Inhibition to RNA Processing and Decreased Lipid Biosynthesis. Anal. Chem. 2015, 87, 10995–11006. [Google Scholar] [CrossRef]
- Stuart, S.A.; Houel, S.; Lee, T.; Wang, N.; Old, W.M.; Ahn, N.G. A Phosphoproteomic Comparison of B-RAFV600E and MKK1/2 Inhibitors in Melanoma Cells. Mol. Cell Proteom. 2015, 14, 1599–1615. [Google Scholar] [CrossRef]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef]
- Chen, E.J.; Sowalsky, A.G.; Gao, S.; Cai, C.; Voznesensky, O.; Schaefer, R.; Loda, M.; True, L.D.; Ye, H.; Troncoso, P.; et al. Abiraterone treatment in castration-resistant prostate cancer selects for progesterone responsive mutant androgen receptors. Clin. Cancer Res. 2015, 21, 1273–1280. [Google Scholar] [CrossRef]
- Ago, T.; Kuribayashi, F.; Hiroaki, H.; Takeya, R.; Ito, T.; Kohda, D.; Sumimoto, H. Phosphorylation of p47phox directs phox homology domain from SH3 domain toward phosphoinositides, leading to phagocyte NADPH oxidase activation. Proc. Natl. Acad. Sci. USA 2003, 100, 4474–4479. [Google Scholar] [CrossRef]
- McMillan, E.A.; Ryu, M.J.; Diep, C.H.; Mendiratta, S.; Clemenceau, J.R.; Vaden, R.M.; Kim, J.H.; Motoyaji, T.; Covington, K.R.; Peyton, M.; et al. Chemistry-First Approach for Nomination of Personalized Treatment in Lung Cancer. Cell 2018, 173, 864–878.E29. [Google Scholar] [CrossRef]
- Martin, D.; Abba, M.C.; Molinolo, A.A.; Vitale-Cross, L.; Wang, Z.; Zaida, M.; Delic, N.C.; Samuels, Y.; Lyons, J.G.; Gutkind, J.S. The head and neck cancer cell oncogenome: A platform for the development of precision molecular therapies. Oncotarget 2014, 5, 8906–8923. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, J.; Deng, Y.; Kelly, J.A.; Kim, K.; Bang, S.Y.; Lee, H.S.; Li, Q.Z.; Wakeland, E.K.; Qiu, R.; et al. A missense variant in NCF1 is associated with susceptibility to multiple autoimmune diseases. Nat. Genet. 2017, 49, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Z.; Cejudo-Martin, P.; de Brouwer, A.; van der Zwaag, B.; Ruiz-Lozano, P.; Scimia, M.C.; Lindsey, J.D.; Weinreb, R.; Albrecht, B.; Megarbane, A.; et al. Disruption of the podosome adaptor protein TKS4 (SH3PXD2B) causes the skeletal dysplasia, eye, and cardiac abnormalities of Frank-Ter Haar Syndrome. Am. J. Hum. Genet. 2010, 86, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gao, Z.; Li, F.; Li, X.; Sun, Y.; Wang, M.; Li, D.; Wang, R.; Li, F.; Fang, R.; et al. Whole Exome Sequencing Identifies Frequent Somatic Mutations in Cell-Cell Adhesion Genes in Chinese Patients with Lung Squamous Cell Carcinoma. Sci. Rep. 2015, 5, 14237. [Google Scholar] [CrossRef]
- Lucas, M.; Gershlick, D.C.; Vidaurrazaga, A.; Rojas, A.L.; Bonifacino, J.S.; Hierro, A. Structural Mechanism for Cargo Recognition by the Retromer Complex. Cell 2016, 167, 1623–1635.e14. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Trieber, C.; Overduin, M. Structural biology of endogenous membrane protein assemblies in native nanodiscs. Curr. Opin. Struct Biol. 2021, 69, 70–77. [Google Scholar] [CrossRef]

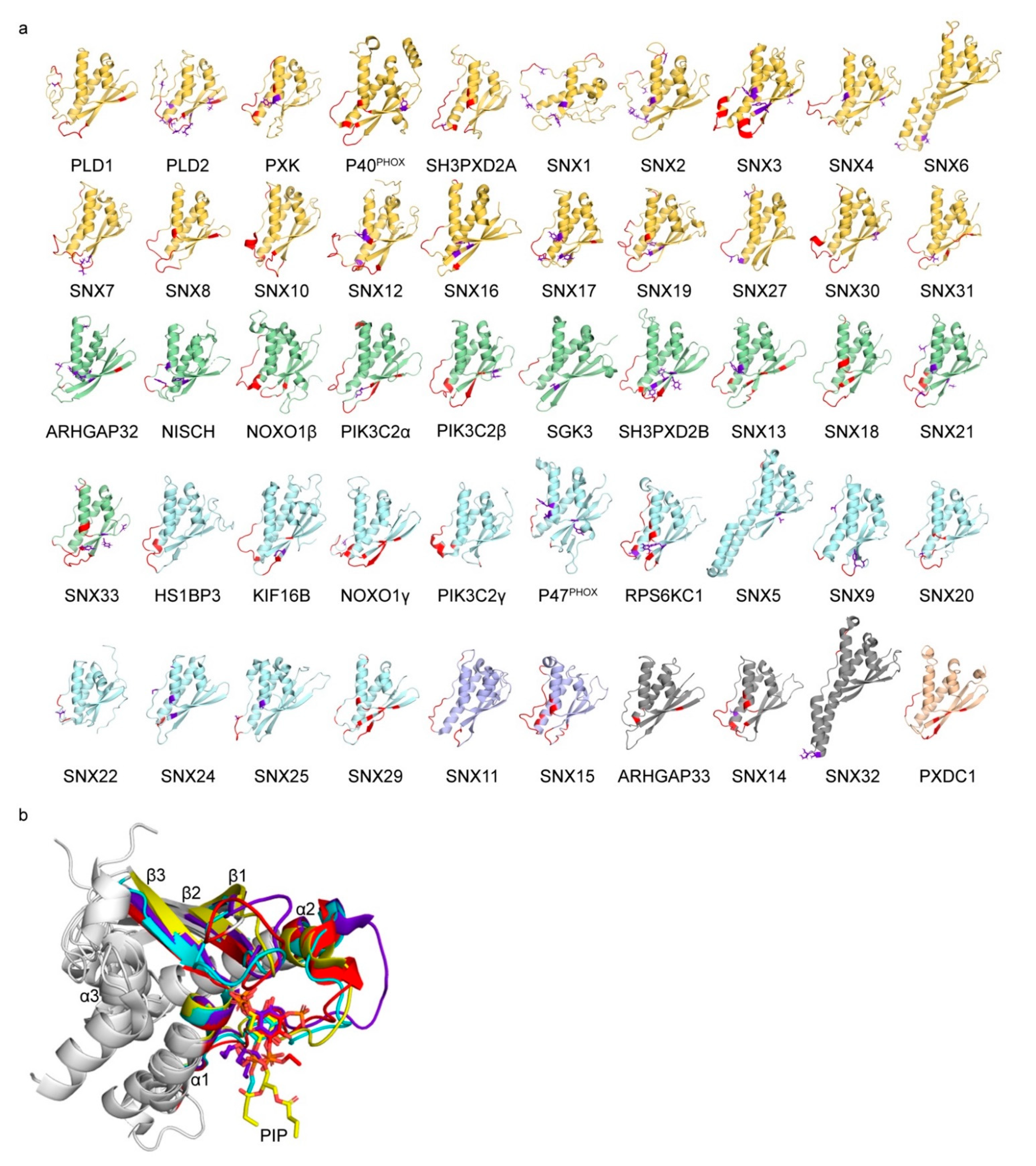
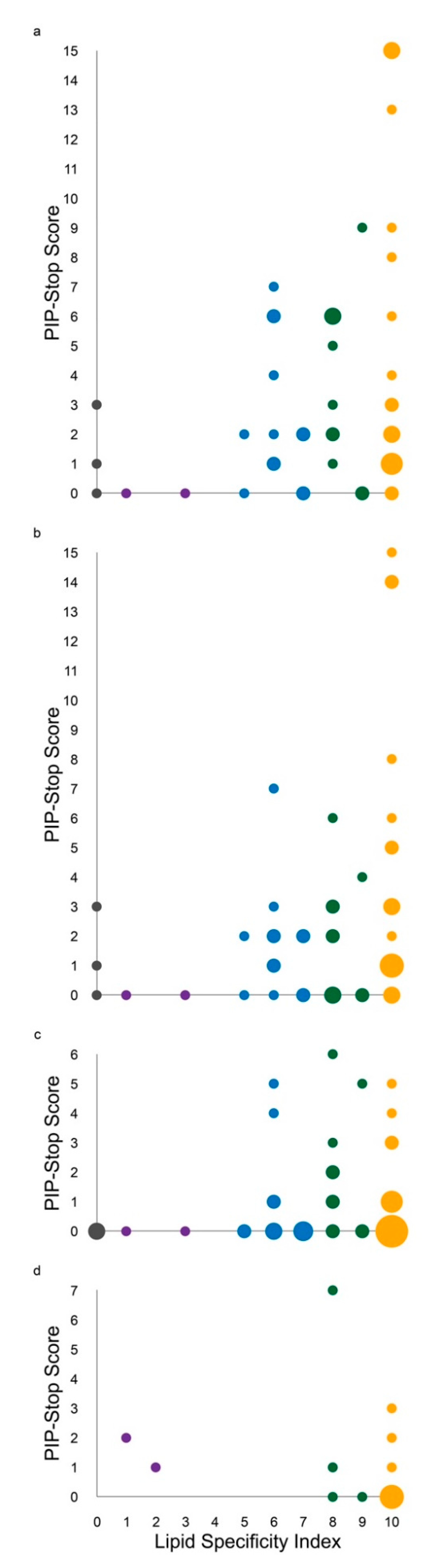
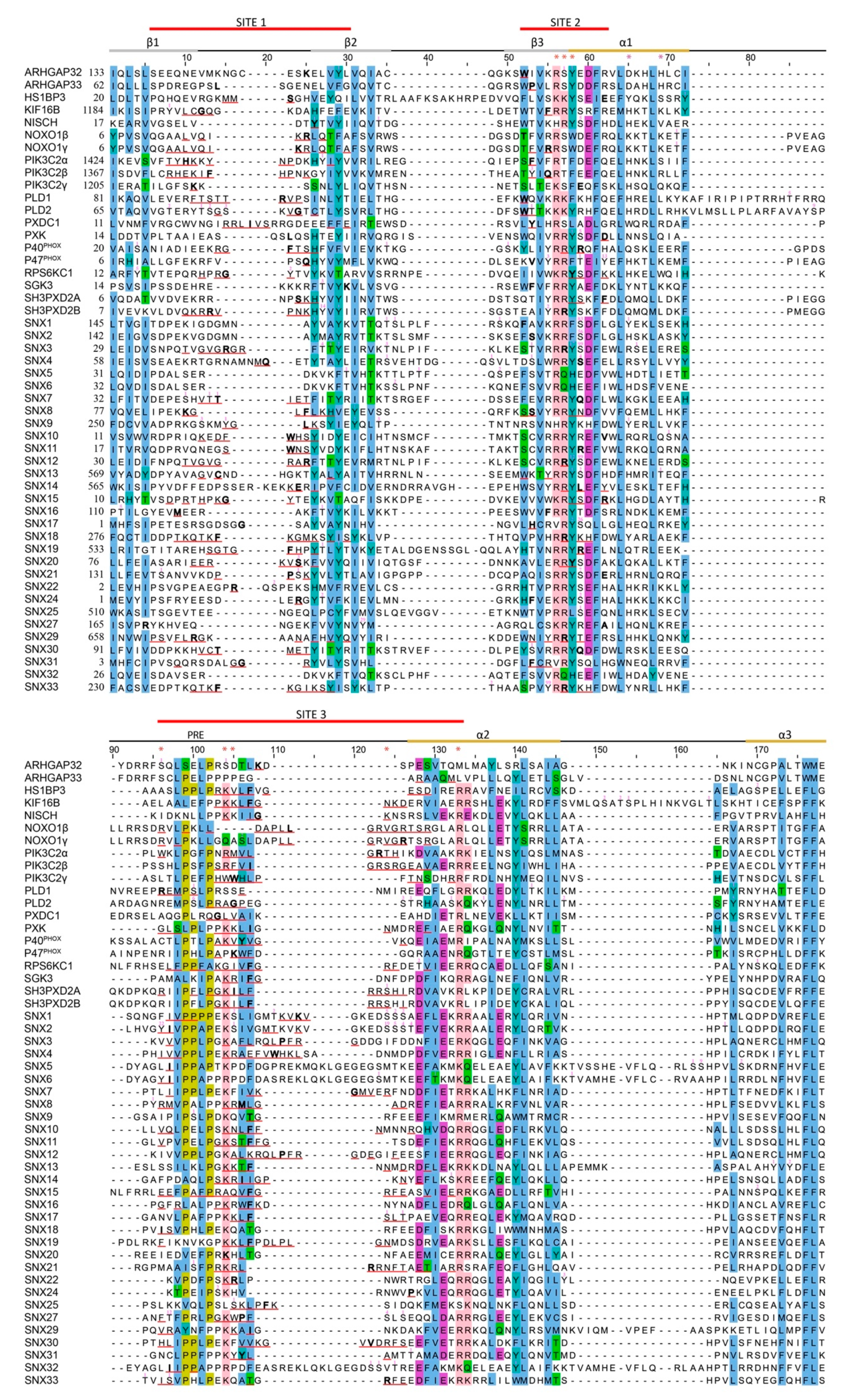

| H.s. Protein | LSI | PIP Ligands | PSS | MAI | PDB | ||||
| PLD1 | 10 | 345 | 1 | S | IT | ||||
| PLD2 | 10 | 45 | 16 | W | IT | ||||
| PXK | 10 | 3 | 4 | W | IT | ||||
| p40phox | 10 | 3 | 1 | S | 1h6h | ||||
| SH3PXD2A | 10 | 3 | 1 | S | IT | ||||
| SNX1 | 10 | 34 | 15 | S | 2i4k | ||||
| SNX2 | 10 | 34 | 17 | S | IT | ||||
| SNX3 | 10 | 3 | 13 | S | 5f0j | ||||
| SNX4 | 10 | 3 | 3 | W | IT | ||||
| SNX6 | 10 | 4 | 2 | W | IT | ||||
| SNX7 | 10 | 3 | 3 | W | IT | ||||
| SNX8 | 10 | 3 | 0 | S | IT | ||||
| SNX10 | 10 | 3 | 0 | W | 4on3 | ||||
| SNX12 | 10 | 3 | 8 | S | 2csk | ||||
| SNX16 | 10 | 3 | 2 | S | 5gw0 | ||||
| SNX17 | 10 | 3 | 9 | S | IT | ||||
| SNX19 | 10 | 3 | 2 | S | IT | ||||
| SNX27 | 10 | 3 | 6 | S | 4has | ||||
| SNX30 | 10 | 3 | 1 | W | IT | ||||
| SNX31 | 10 | 3 | 1 | S | IT | ||||
| NOXO1β | 9 | 45,345 | 0 | W | 2l73 | ||||
| SNX18 | 9 | 34,45 | 0 | S | IT | ||||
| SNX33 | 9 | 34,45 | 9 | S | IT | ||||
| ARHGAP32 | 8 | 3,4,5 | 5 | W | IT | ||||
| NISCH | 8 | 3,34 | 6 | W | 3p0c | ||||
| PIK3C2α | 8 | 34,35,45 | 1 | S | 2ar5 | ||||
| PIK3C2β | 8 | 34,45,345 | 2 | S | IT | ||||
| SGK3 | 8 | 3,34 | 2 | S | 1xte | ||||
| SH3PXD2B | 8 | 3,34 | 6 | S | IT | ||||
| SNX13 | 8 | 3,34 | 3 | W | IT | ||||
| SNX21 | 8 | 3,45 | 6 | S | IT | ||||
| NOXO1γ | 7 | 4,5,35 | 2 | W | IT | ||||
| PIK3C2γ | 7 | 34,35,45,345 | 0 | W | 2wwe | ||||
| SNX25 | 7 | 34,35,45,345 | 2 | S | 5woe | ||||
| SNX29 | 7 | 3,34,45 | 0 | S | IT | ||||
| KIF16B | 6 | 3,34,45,345 | 1 | S | 2v14 | ||||
| p47phox | 6 | 3,34,45,345 | 6 | S | 1kq6 | ||||
| RPS6KC1 | 6 | 3,34,45,345 | 6 | S | IT | ||||
| SNX5 | 6 | 3,34,35,45 | 1 | W | 3hpc | ||||
| SNX9 | 6 | 3,34,45,345 | 4 | W | 2raj | ||||
| SNX22 | 6 | 3,34,45,345 | 2 | S | 2ett | ||||
| SNX24 | 6 | 3,34,35,45 | 7 | S | 4az9 | ||||
| HS1BP3 | 5 | 3,34,35,45,345 | 0 | S | IT | ||||
| SNX20 | 5 | 3,5,35,45 | 2 | S | IT | ||||
| SNX15 | 3 | 3,4,34,35,45,345 | 0 | S | IT | ||||
| SNX11 | 1 | 3,4,5,34,35,45,345 | 0 | S | 4ikb | ||||
| ARHGAP33 | 0 | 0 | 0 | N | IT | ||||
| SNX14 | 0 | 0 | 1 | N | IT | ||||
| SNX32 | 0 | 0 | 3 | N | 6e8r | ||||
| PXDC1 | nd | nd | 0 | n.d. | IT | ||||
| S.c. Protein | LSI | PIP Ligands | PSS | PDB | |||||
| Bem1 | 10 | 4 | 0 | 2v6v,2czo | |||||
| Bem3 | 10 | 3 | 2 | 6fsf | |||||
| Grd19 | 10 | 3 | 3 | 1ocs,1ocu | |||||
| Mdm1 | 10 | 3 | 0 | ||||||
| Mvp1 | 10 | 3 | 0 | 6p0x | |||||
| Spo14 | 10 | 3 | 0 | ||||||
| Vam7 | 10 | 3 | 1 | 1kmd | |||||
| Ykr078w | 10 | 3 | 0 | ||||||
| Ypt35 | 10 | 3 | 0 | ||||||
| Vps5 | 9 | 3,5 | 0 | ||||||
| Atg20 | 8 | 3,4,5 | 1 | ||||||
| Snx4 | 8 | 3,45 | 0 | ||||||
| Snx41 | 8 | 3,4,5 | 7 | ||||||
| Vps17 | 2 | 3,4,5,34,35,45 | 1 | ||||||
| Ypr097w | 1 | 3,4,5,34,35,45,345 | 2 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kervin, T.A.; Overduin, M. Regulation of the Phosphoinositide Code by Phosphorylation of Membrane Readers. Cells 2021, 10, 1205. https://doi.org/10.3390/cells10051205
Kervin TA, Overduin M. Regulation of the Phosphoinositide Code by Phosphorylation of Membrane Readers. Cells. 2021; 10(5):1205. https://doi.org/10.3390/cells10051205
Chicago/Turabian StyleKervin, Troy A., and Michael Overduin. 2021. "Regulation of the Phosphoinositide Code by Phosphorylation of Membrane Readers" Cells 10, no. 5: 1205. https://doi.org/10.3390/cells10051205
APA StyleKervin, T. A., & Overduin, M. (2021). Regulation of the Phosphoinositide Code by Phosphorylation of Membrane Readers. Cells, 10(5), 1205. https://doi.org/10.3390/cells10051205






