YY2 in Mouse Preimplantation Embryos and in Embryonic Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Oocyte and Embryo Collection and In Vitro Culture
2.2. Immunofluorescence and Confocal Microscopy
2.3. Immunological Reagents and Western Blot
2.4. Embryo Microinjection with siRNAs
2.5. Gene Expression in Preimplantation Embryos
2.6. Cell Culture, shRNA Knockdown, and RT-qPCR Analysis
3. Results
3.1. Expression of YY2 during Preimplantation Development
3.2. In Vitro Development of Yy2 Depleted Embryos
3.3. Yy2 Loss-of-Function Mouse Embryos Express Normal Levels of ERV Elements
3.4. Depletion of Yy2 Expression Alters Self-Renewal in Mouse ES Cells
3.5. Gene Expression in Yy2-Depleted ES Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rogers, M.B.; Hosler, A.B.; Gudas, L.J. Specific Expression of a Retinoic Acid-regulated, Zinc-finger Gene, Rex-1, in Preimplantation Embryos, Trophoblast and Spermatocytes. Development 1991, 113, 815–824. [Google Scholar] [CrossRef]
- Kim, J.; Faulk, C. Retroposition and Evolution of the DNA-binding Motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007, 35, 3442–3452. [Google Scholar] [CrossRef]
- Thomas, M.J.; Seto, E. Unlocking the Mechanisms of Transcription factor YY1: Are Chromatin Modifying Enzymes the Key? Gene 1999, 236, 197–208. [Google Scholar] [CrossRef]
- Atchison, M.L.; Basu, A.; Zaprazna, K.; Papasani, M. Mechanisms of Yin Yang 1 in Oncogenesis: The Importance of Indirect Effects. Crit. Rev. Oncog. 2011, 16, 143–161. [Google Scholar] [CrossRef]
- Garcia-Tuñon, I.; Guallar, D.; Alonso-Martin, S.; Benito, A.; Benítez-Lázaro, A.; Pérez-Palacios, R.; Muniesa, P.; Climent, M.; Sánchez, M.; Vidal, M.; et al. Association of Rex-1 to Target Genes Supports Its Interaction with Polycomb Function. Stem Cell Res. 2011, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jin, J.; Yao, T.; Gottschalk, A.J.; Swanson, S.K.; Wu, S.; Shi, Y.; Washburn, M.P.; Florens, L.; Conaway, R.C.; et al. YY1 Functions with INO80 to Activate Transcription. Nat. Struct. Mol. Biol. 2007, 14, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588. [Google Scholar] [CrossRef]
- Makhlouf, M.; Ouimette, J.-F.; Oldfield, A.; Navarro, P.; Neuillet, D.; Rougeulle, C. A Prominent and Conserved Role for YY1 in Xist Transcriptional Activation. Nat. Commun. 2014, 5, 4878. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, M.E.; Zhang, L.-F.; Xu, N.; Shi, Y.; Lee, J.T. Identification of a Ctcf Cofactor, Yy1, for the X Chromosome Binary Switch. Mol. Cell 2007, 25, 43–56. [Google Scholar] [CrossRef]
- Jeon, Y.; Lee, J.T. YY1 Tethers Xist RNA to the Inactive X Nucleation Center. Cell 2011, 146, 119–133. [Google Scholar] [CrossRef]
- Donohoe, M.E.; Zhang, X.; McGinnis, L.; Biggers, J.; Li, E.; Shi, Y. Targeted Disruption of Mouse Yin Yang 1 Transcription Factor Results in Peri-Implantation Lethality. Mol. Cell. Biol. 1999, 19, 7237–7244. [Google Scholar] [CrossRef]
- Ng, H.-H.; Surani, M.A. The Transcriptional and Signalling Networks of Pluripotency. Nat. Cell Biol. 2011, 13, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Climent, M.; Alonso-Martín, S.; Pérez-Palacios, R.; Guallar, D.; Benito, A.A.; Larraga, A.; Fernández-Juan, M.; Sanz, M.; De Diego, A.; Seisdedos, M.T.; et al. Functional Analysis of Rex1 during Preimplantation Development. Stem Cells Dev. 2013, 22, 459–472. [Google Scholar] [CrossRef]
- Kim, J.D.; Kim, H.; Ekram, M.B.; Yu, S.; Faulk, C.; Kim, J. Rex1/Zfp42 as an Epigenetic Regulator for Genomic Imprinting. Hum. Mol. Genet. 2011, 20, 1353–1362. [Google Scholar] [CrossRef]
- Navarro, P.; Oldfield, A.; Legoupi, J.; Festuccia, N.; Dubois, A.; Attia, M.; Schoorlemmer, J.; Rougeulle, C.; Chambers, I.; Avner, P. Molecular Coupling of Tsix Regulation and Pluripotency. Nat. Cell Biol. 2010, 468, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Gontan, C.; Achame, E.M.; Demmers, J.; Barakat, T.S.; Rentmeester, E.; Van Ijcken, W.; Grootegoed, J.A.; Gribnau, J. RNF12 Initiates X-chromosome Inactivation by Targeting REX1 for Degradation. Nat. Cell Biol. 2012, 485, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Guallar, D.; Pérez-Palacios, R.; Climent, M.; Martínez-Abadía, I.; Larraga, A.; Fernández-Juan, M.; Vallejo, C.; Muniesa, P.; Schoorlemmer, J. Expression of Endogenous Retroviruses is Negatively Regulated by the Pluripotency Marker Rex1/Zfp42. Nucleic Acids Res. 2012, 40, 8993–9007. [Google Scholar] [CrossRef]
- Macfarlan, T.S.; Gifford, W.D.; Agarwal, S.; Driscoll, S.; Lettieri, K.; Wang, J.; Andrews, S.E.; Franco, L.; Rosenfeld, M.G.; Ren, B.; et al. Endogenous Retroviruses and Neighboring Genes are Coordinately Repressed by LSD1/KDM1A. Genes Dev. 2011, 25, 594–607. [Google Scholar] [CrossRef]
- Luo, C.; Lu, X.; Stubbs, L.; Kim, J. Rapid Evolution of a Recently Retroposed Transcription factor YY2 in Mammalian Genomes. Genomics 2006, 87, 348–355. [Google Scholar] [CrossRef][Green Version]
- Nguyen, N.; Zhang, X.; Olashaw, N.; Seto, E. Molecular Cloning and Functional Characterization of the Transcription Factor YY2. J. Biol. Chem. 2004, 279, 25927–25934. [Google Scholar] [CrossRef]
- Drews, D.; Klar, M.; Dame, C.; Bräuer, A.U. Developmental Expression Profile of the yy2 Gene in Mice. BMC Dev. Biol. 2009, 9, 45. [Google Scholar] [CrossRef]
- Klar, M.; Bode, J. Enhanceosome Formation over the Beta Interferon Promoter Underlies a Remote-Control Mechanism Mediated by YY1 and YY2. Mol. Cell. Biol. 2005, 25, 10159–10170. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shioda, T.; Coser, K.R.; Lynch, M.C.; Yang, C.; Schmidt, E.V. Genome-wide Analysis of YY2 versus YY1 Target Genes. Nucleic Acids Res. 2010, 38, 4011–4026. [Google Scholar] [CrossRef]
- Pérez-Palacios, R.; Macías-Redondo, S.; Climent, M.; Contreras-Moreira, B.; Muniesa, P.; Schoorlemmer, J. In vivo Chromatin Targets of the Transcription Factor Yin Yang 2 in Trophoblast Stem Cells. PLoS ONE 2016, 11, e0154268. [Google Scholar] [CrossRef] [PubMed]
- Tahmasebi, S.T.; Jafarnejad, S.M.; Tam, I.S.; Gonatopoulos-Pournatzis, T.; Matta-Camacho, E.; Tsukumo, Y.; Yanagiya, A.; Li, W.; Atlasi, Y.; Caron, M.; et al. Control of Embryonic Stem Cell Self-renewal and Differentiation via Coordinated Alternative Splicing and Translation of YY2. Proc. Natl. Acad. Sci. USA 2016, 113, 12360–12367. [Google Scholar] [CrossRef] [PubMed]
- Vella, P.; Barozzi, I.; Cuomo, A.; Bonaldi, T.; Pasini, D. Yin Yang 1 Extends the Myc-related Transcription Factors Network in Embryonic Stem Cells. Nucleic Acids Res. 2011, 40, 3403–3418. [Google Scholar] [CrossRef]
- Sigova, A.A.; Abraham, B.J.; Ji, X.; Molinie, B.; Hannett, N.M.; Guo, Y.E.; Jangi, M.R.; Giallourakis, C.C.; Sharp, P.A.; Young, R.A. Transcription Factor Trapping by RNA in Gene Regulatory Elements. Science 2015, 350, 978–981. [Google Scholar] [CrossRef] [PubMed]
- Schoorlemmer, J.; Pérez-Palacios, R.; Climent, M.; Guallar, D.; Muniesa, P. Regulation of Mouse Retroelement MuERV-L/MERVL Expression by REX1 and Epigenetic Control of Stem Cell Potency. Front. Oncol. 2014, 4, 14. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, E.B.C.N.C.E.C. Regulatory Activities of Transposable Elements: From Conflicts to Benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef]
- Schlesinger, S.; Goff, S.P. Retroviral Transcriptional Regulation and Embryonic Stem Cells: War and Peace. Mol. Cell. Biol. 2014, 35, 770–777. [Google Scholar] [CrossRef]
- Cosby, R.L.; Chang, N.-C.; Feschotte, C. Host–transposon Interactions: Conflict, Cooperation, and Cooption. Genes Dev. 2019, 33, 1098–1116. [Google Scholar] [CrossRef]
- Macfarlan, T.S.; Gifford, W.D.; Driscoll, S.P.; Lettieri, K.; Rowe, H.M.; Bonanomi, D.; Firth, A.L.; Singer, O.; Trono, D.; Pfaff, S.L. Embryonic Stem Cell Potency Fluctuates with Endogenous Retrovirus Activity. Nat. Cell Biol. 2012, 487, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Beraldi, R.; Pittoggi, C.; Sciamanna, I.; Mattei, E.; Spadafora, C. Expression of LINE-1 Retroposons is Essential for Murine Preimplantation Development. Mol. Reprod. Dev. 2005, 73, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, X.; Wang, L.; Li, J.; Zhao, Y.; Bou, G.; Li, Y.; Jiao, G.; Shen, X.; Wei, R.; et al. A Novel Long Intergenic Noncoding RNA Indispensable for the Cleavage of Mouse Two-cell Embryos. EMBO Rep. 2016, 17, 1452–1470. [Google Scholar] [CrossRef]
- Jachowicz, J.W.; Bing, X.; Pontabry, J.; Bošković, A.; Rando, O.J.; Torres-Padilla, M.-E. LINE-1 Activation after Fertilization Regulates Global Chromatin Accessibility in the Early Mouse Embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Zalzman, M.; Falco, G.; Sharova, L.V.; Nishiyama, A.; Thomas, M.P.; Lee, S.-L.; Stagg, C.A.; Hoang, H.G.; Yang, H.-T.; Indig, F.E.; et al. Zscan4 Regulates Telomere Elongation and Genomic Stability in ES Cells. Nat. Cell Biol. 2010, 464, 858–863. [Google Scholar] [CrossRef]
- Nagy, A.; Gertsenstein, M.; Vintersten, K.; Behringer, R. Manipulating the Mouse Embryo: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2003. [Google Scholar]
- Palmieri, S.L.; Payne, J.; Stiles, C.D.; Biggers, J.D.; Mercola, M. Expression of Mouse PDGF-A and PDGF α-receptor Genes during Pre- and Post-implantation Development: Evidence for a Developmental Shift from an Autocrine to a Paracrine Mode of Action. Mech. Dev. 1992, 39, 181–191. [Google Scholar] [CrossRef]
- Torres-Padilla, M.-E.; Bannister, A.J.; Hurd, P.J.; Kouzarides, T.; Zernicka-Goetz, M. Dynamic Distribution of the Replacement Histone Variant H3.3 in the Mouse Oocyte and Preimplantation Embryos. Int. J. Dev. Biol. 2006, 50, 455–461. [Google Scholar] [CrossRef]
- Aubert, J.; Dunstan, H.; Chambers, I.; Smith, A.G. Functional Gene Screening in Embryonic Stem Cells Implicates Wnt Antagonism in Neural Differentiation. Nat. Biotechnol. 2002, 20, 1240–1245. [Google Scholar] [CrossRef]
- Ying, Q.-L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The Ground State of Embryonic Stem Cell Self-renewal. Nat. Cell Biol. 2008, 453, 519–523. [Google Scholar] [CrossRef]
- Velkey, J.M.; O’Shea, K.S. Oct4 RNA Interference Induces Trophectoderm Differentiation in Mouse Embryonic Stem Cells. Genesis 2003, 37, 18–24. [Google Scholar] [CrossRef]
- Santoyo, J.; Vaquerizas, J.M.; Dopazo, J.; Santoyo-Lopez, J. Highly Specific and Accurate Selection of siRNAs for High-throughput Functional Assays. Bioinformatics 2004, 21, 1376–1382. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wallingford, M.C.; Hiller, J.; Zhang, K.; Mager, J. YY1 Is Required for Posttranscriptional Stability of SOX2 and OCT4 Proteins. Cell Reprogram. 2017, 19, 263–269. [Google Scholar] [CrossRef]
- Schlesinger, S.; Lee, A.H.; Wang, G.Z.; Green, L.; Goff, S.P. Proviral Silencing in Embryonic Cells Is Regulated by Yin Yang 1. Cell Rep. 2013, 4, 50–58. [Google Scholar] [CrossRef]
- Enver, T.; Pera, M.; Peterson, C.; Andrews, P.W. Stem Cell States, Fates, and the Rules of Attraction. Cell Stem Cell 2009, 4, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Toyooka, Y.; Shimosato, D.; Murakami, K.; Takahashi, K.; Niwa, H. Identification and Characterization of Subpopulations in Undifferentiated ES Cell Culture. Development 2008, 135, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Lopes, S.M.C.D.S.; Tang, F.; Surani, M.A. Dynamic Equilibrium and Heterogeneity of Mouse Pluripotent Stem Cells with Distinct Functional and Epigenetic States. Cell Stem Cell 2008, 3, 391–401. [Google Scholar] [CrossRef]
- Wray, J.; Kalkan, T.; Gomez-Lopez, S.; Eckardt, D.; Cook, A.D.; Kemler, R.; Smith, A. Inhibition of Glycogen Synthase Kinase-3 Alleviates Tcf3 Repression of the Pluripotency Network and Increases Embryonic Stem Cell Resistance to Differentiation. Nat. Cell Biol. 2011, 13, 838–845. [Google Scholar] [CrossRef]
- Osorno, R.; Chambers, I. Transcription Factor Heterogeneity and Epiblast Pluripotency. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 2230–2237. [Google Scholar] [CrossRef]
- Lee, S.-L.; Falco, G.; Stanghellini, I.; Bassey, U.C.; Hamatani, T.; Ko, M.S.H. Zscan4: A Novel Gene Expressed Exclusively in Late 2-cell Embryos. Biol. Reprod. 2007, 77, 79. [Google Scholar] [CrossRef]
- Carter, M.G.; Stagg, C.A.; Falco, G.; Yoshikawa, T.; Bassey, U.C.; Aiba, K.; Sharova, L.V.; Shaik, N.; Ko, M.S. An in situ Hybridization-based Screen for Heterogeneously Expressed Genes in Mouse ES Cells. Gene Expr. Patterns 2008, 8, 181–198. [Google Scholar] [CrossRef] [PubMed]
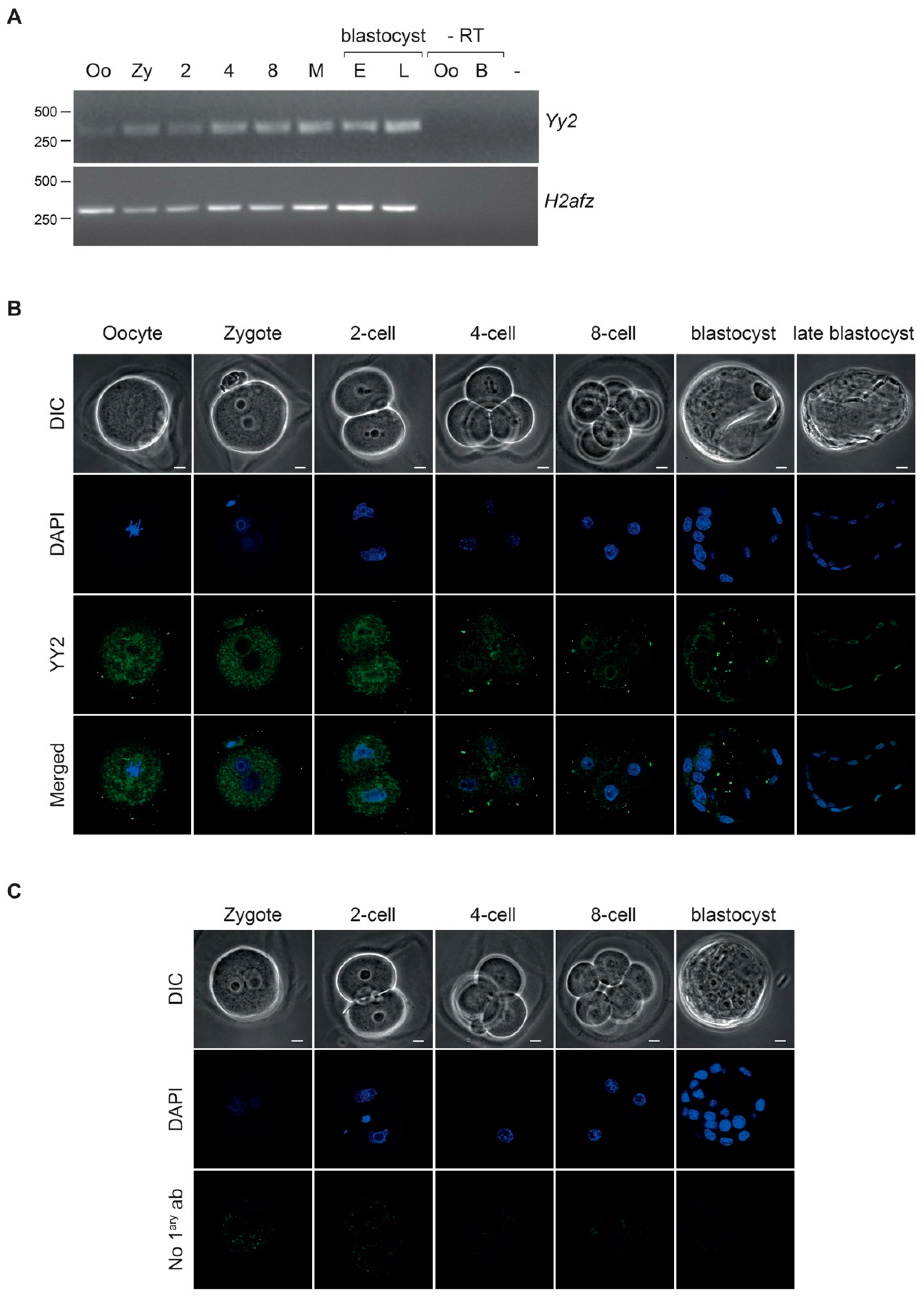
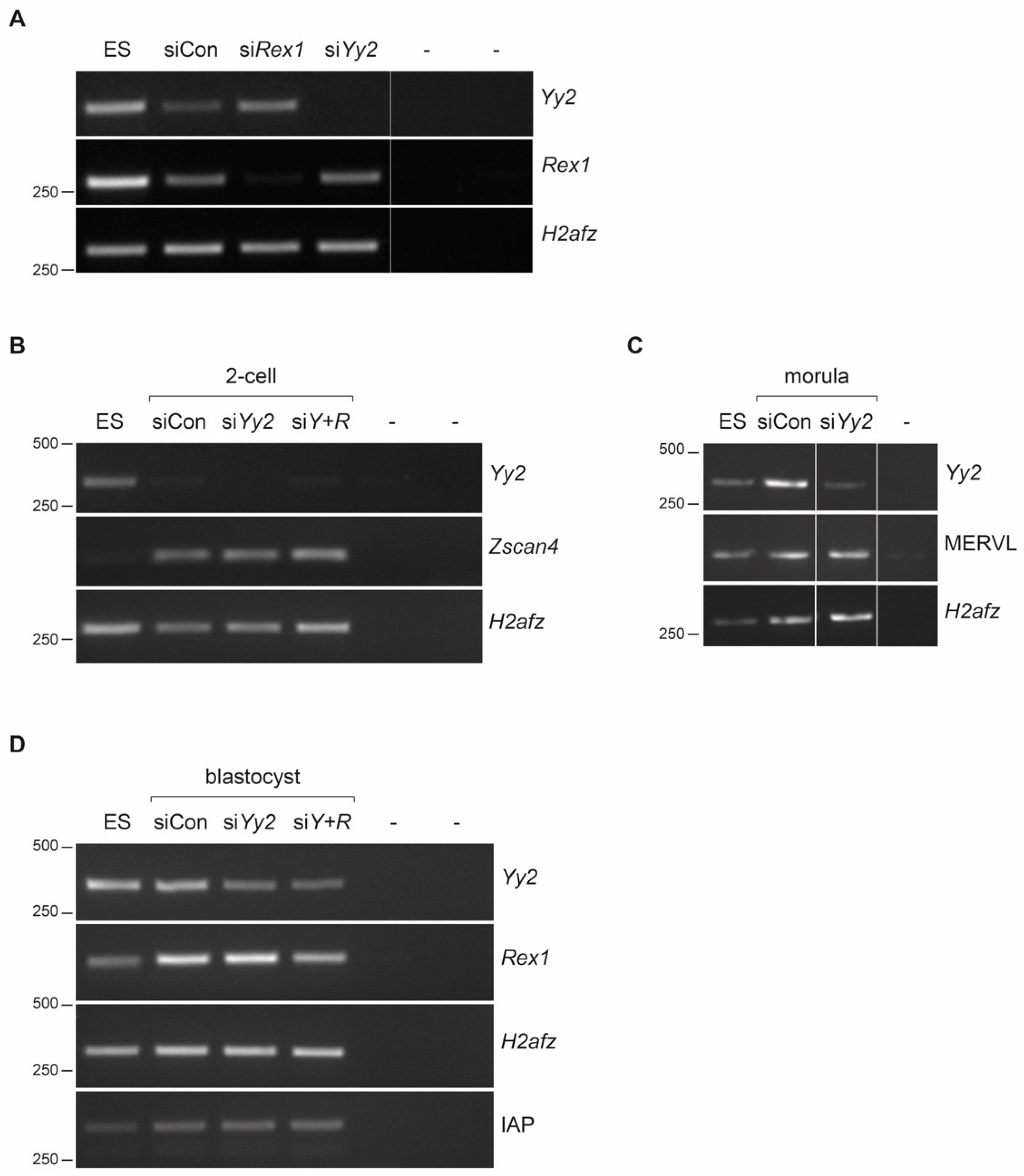
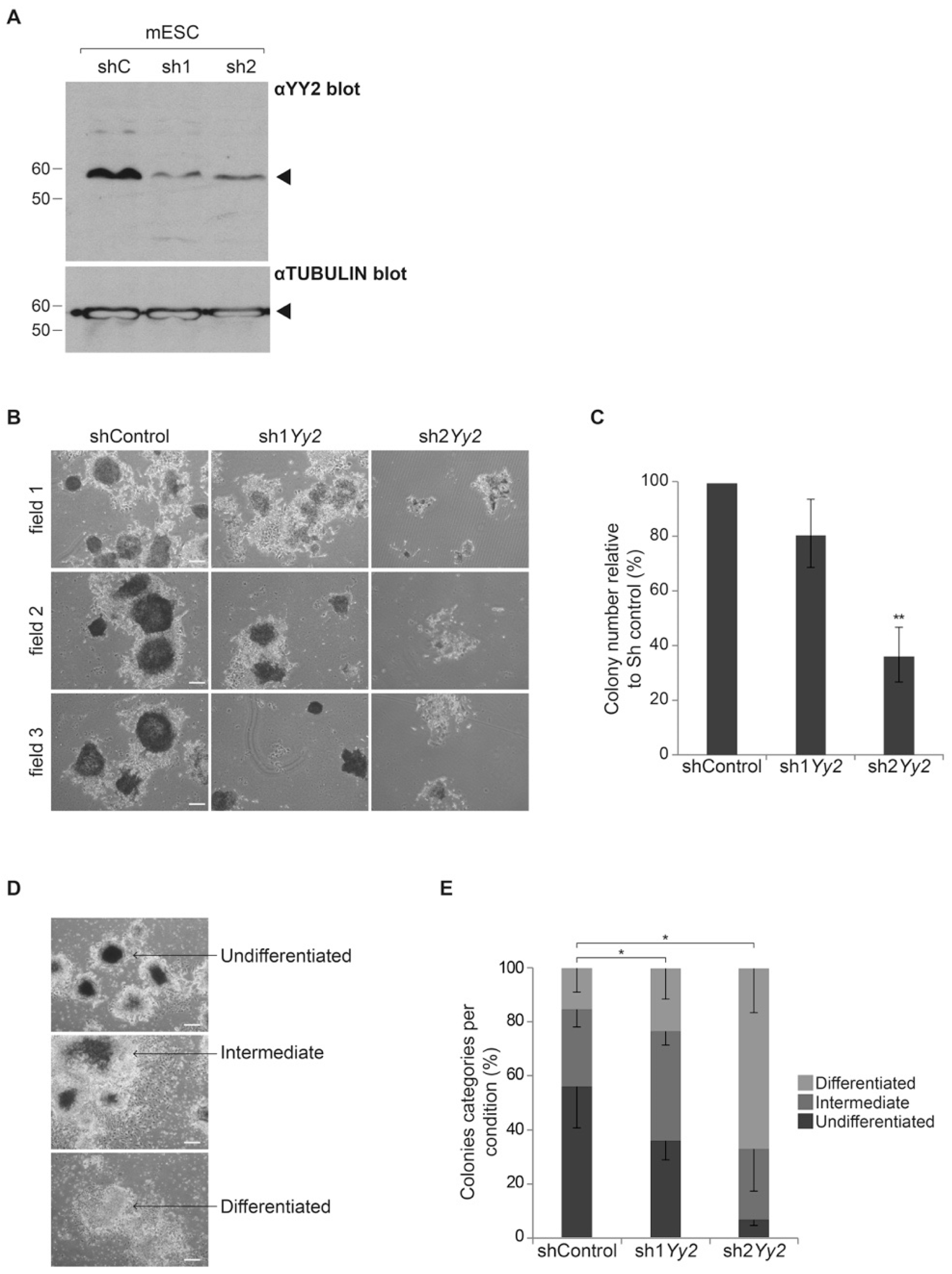
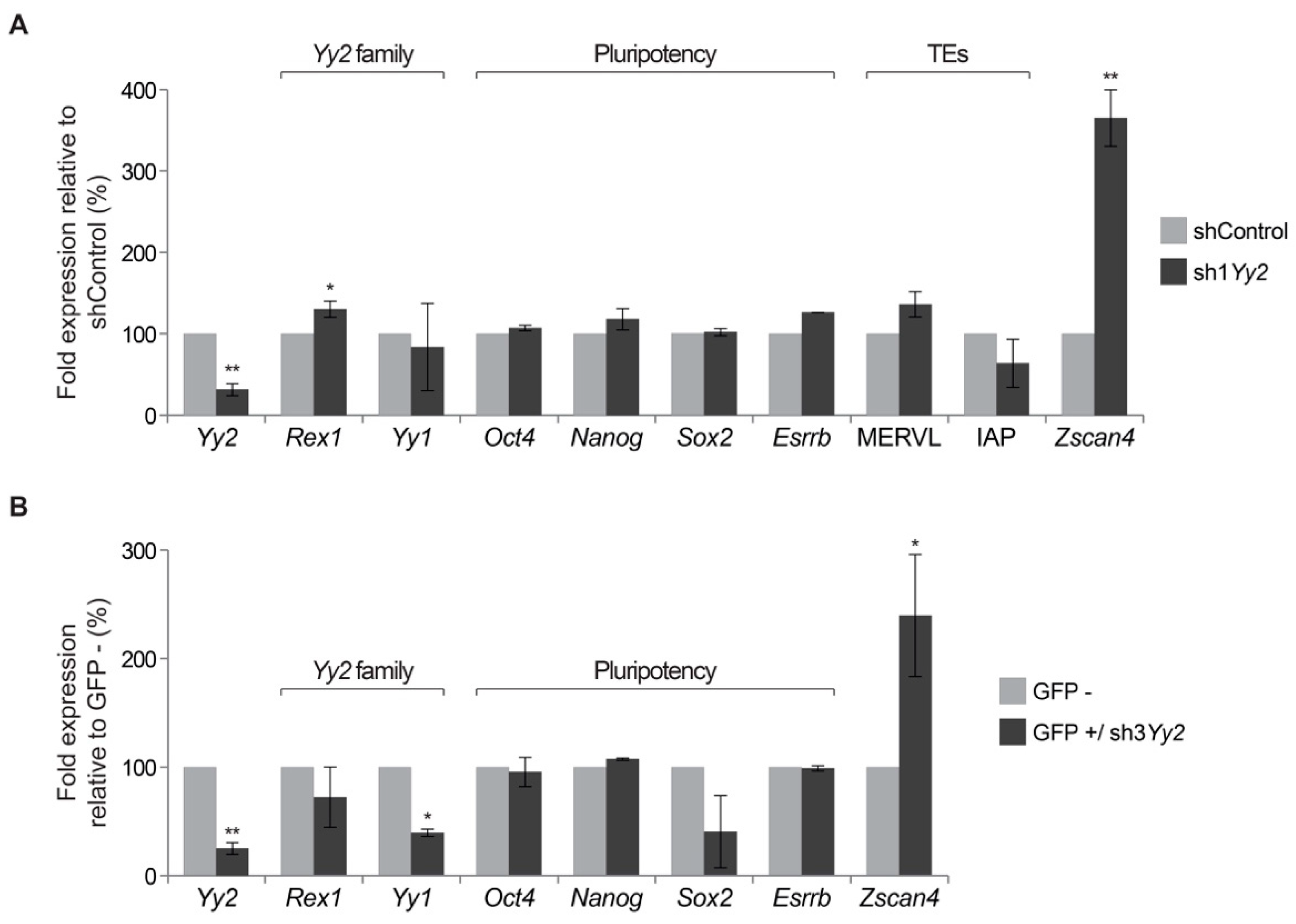
| 2-Cell | 5–8 Cells | ≥Morula | Blastocyst | |
|---|---|---|---|---|
| Day 2 | Day 3 | Day 4 | Day 5 | |
| siControl | 104/104 | 81/92 | 72/80 | 21/26 |
| (100%) | (88.0%) | (90.0%) | (80.8%) | |
| siYy2 | 113/114 | 83/101 | 80/86 | 58/76 |
| (99.1%) | (82.2%) | (93.0%) | (76.3%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Palacios, R.; Climent, M.; Santiago-Arcos, J.; Macías-Redondo, S.; Klar, M.; Muniesa, P.; Schoorlemmer, J. YY2 in Mouse Preimplantation Embryos and in Embryonic Stem Cells. Cells 2021, 10, 1123. https://doi.org/10.3390/cells10051123
Pérez-Palacios R, Climent M, Santiago-Arcos J, Macías-Redondo S, Klar M, Muniesa P, Schoorlemmer J. YY2 in Mouse Preimplantation Embryos and in Embryonic Stem Cells. Cells. 2021; 10(5):1123. https://doi.org/10.3390/cells10051123
Chicago/Turabian StylePérez-Palacios, Raquel, María Climent, Javier Santiago-Arcos, Sofía Macías-Redondo, Martin Klar, Pedro Muniesa, and Jon Schoorlemmer. 2021. "YY2 in Mouse Preimplantation Embryos and in Embryonic Stem Cells" Cells 10, no. 5: 1123. https://doi.org/10.3390/cells10051123
APA StylePérez-Palacios, R., Climent, M., Santiago-Arcos, J., Macías-Redondo, S., Klar, M., Muniesa, P., & Schoorlemmer, J. (2021). YY2 in Mouse Preimplantation Embryos and in Embryonic Stem Cells. Cells, 10(5), 1123. https://doi.org/10.3390/cells10051123







