The SOS Error-Prone DNA Polymerase V Mutasome and β-Sliding Clamp Acting in Concert on Undamaged DNA and during Translesion Synthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Oligonucleotides
2.2. Proteins
2.3. Pol V Mut Assembly
2.4. Activity Assays of Pol V Mut
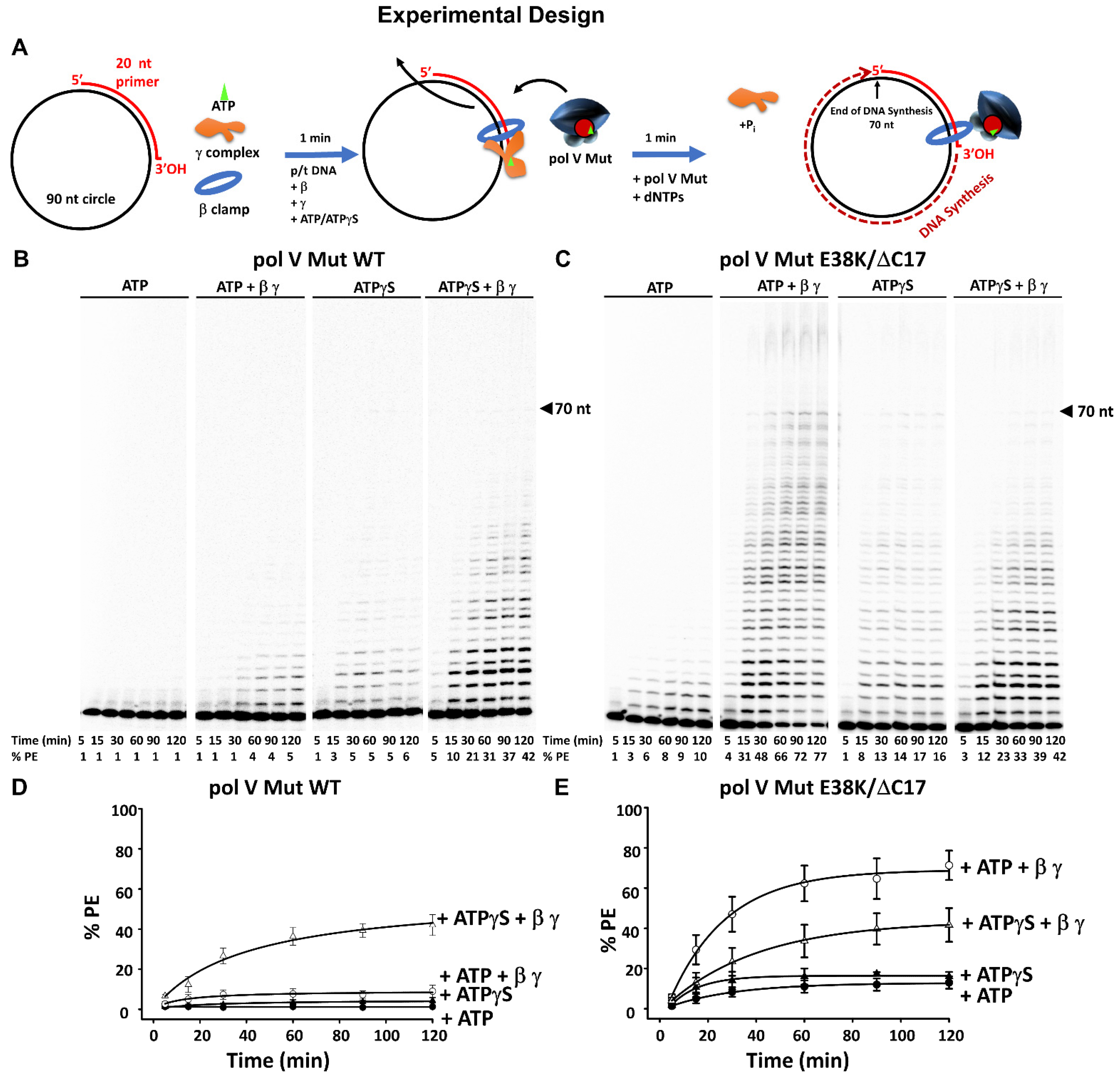
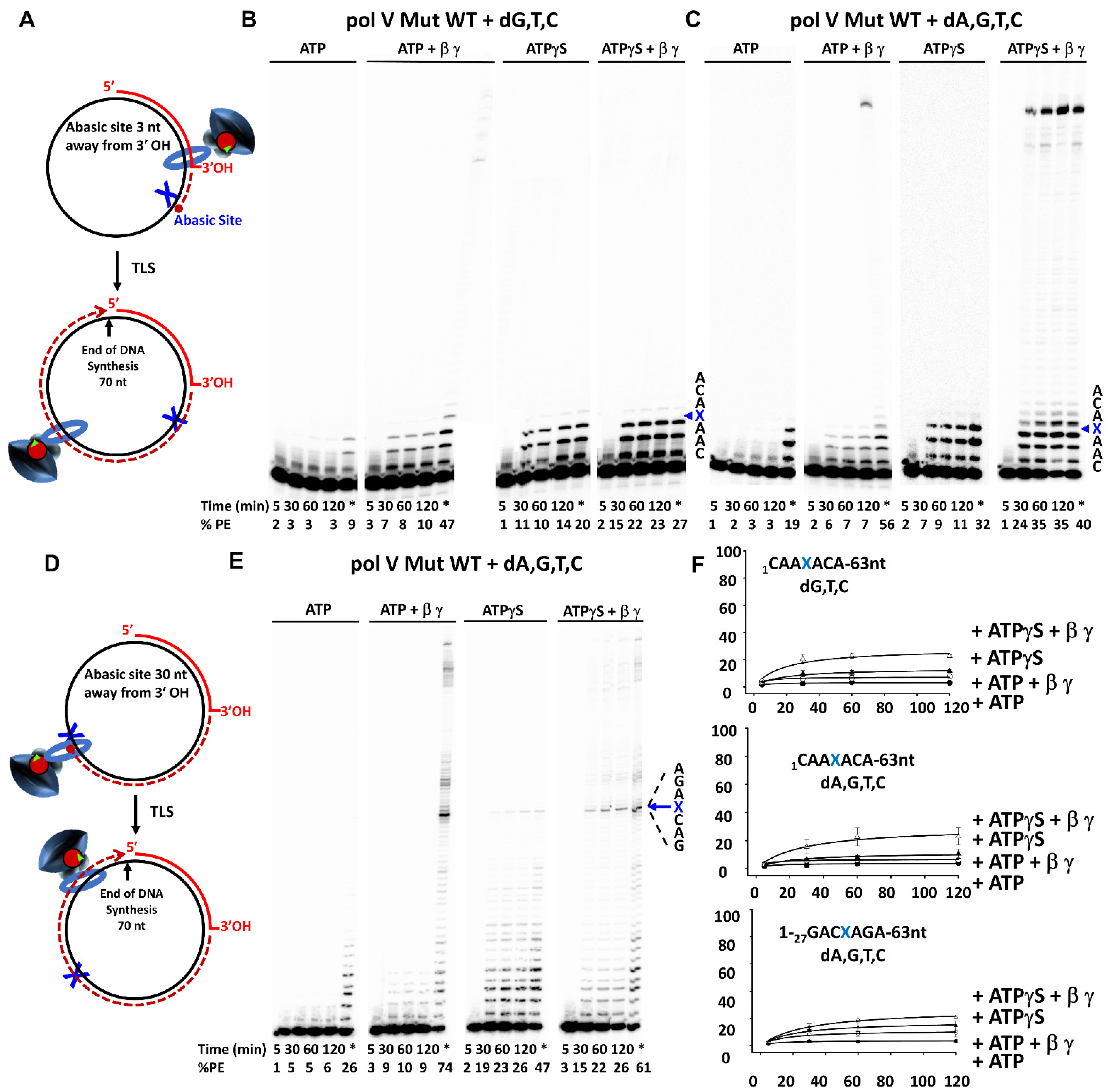
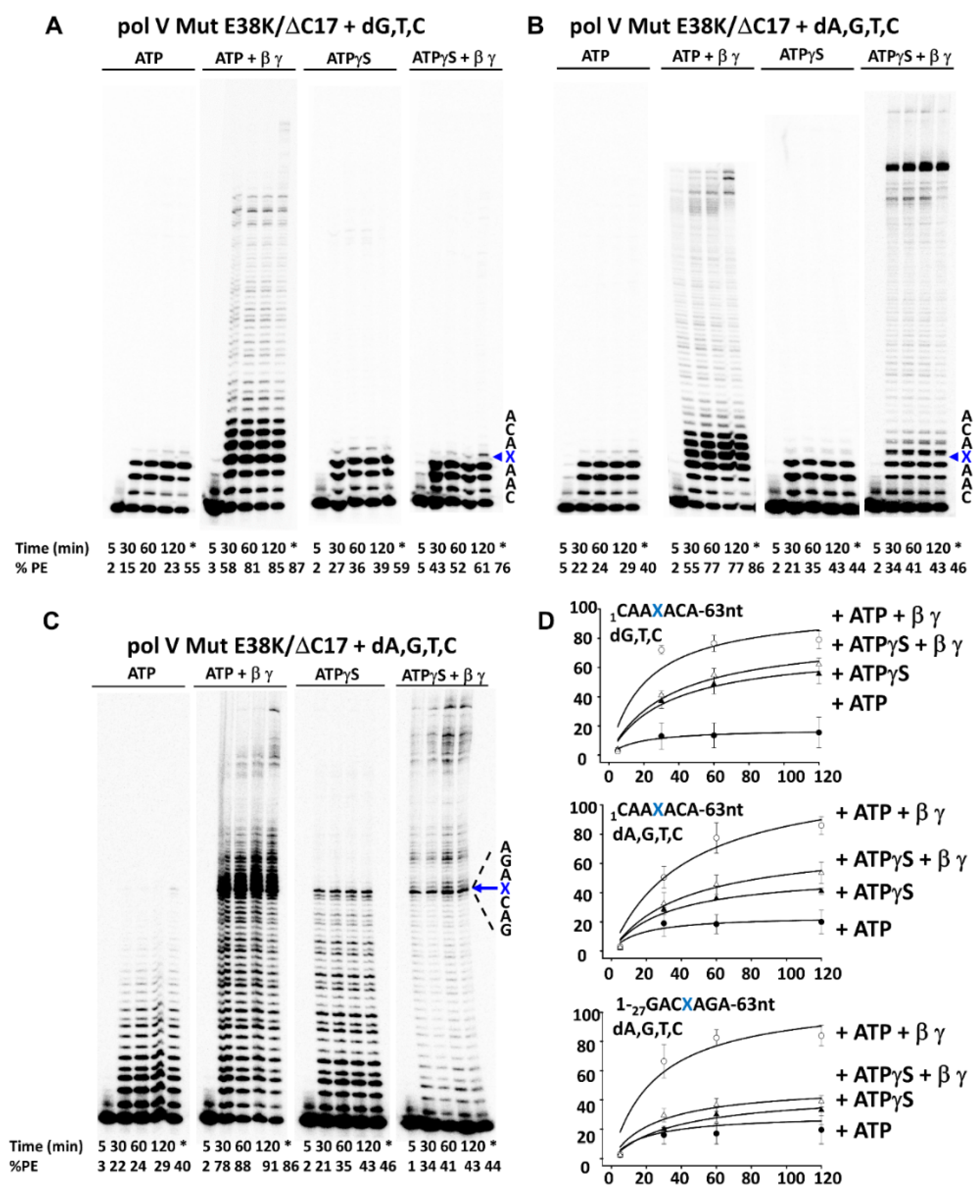
3. Results
3.1. Stimulation of Pol V Mut on Undamaged DNA
3.2. β-Clamp Is Required for Pol V Mut-Catalyzed TLS
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weigle, J.J. Induction of mutation in a bacterial virus. Proc. Natl. Acad. Sci. USA 1953, 39, 628–636. [Google Scholar] [CrossRef]
- Witkin, E.M. The radiation sensitivity of Escherichia coli B: A hypothesis relating filament formation and prophage induction. Proc. Natl. Acad. Sci. USA 1967, 57, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Little, J.W.; Kleid, D.G. Escherichia coli protein X is the recA gene product. J. Biol. Chem. 1977, 252, 6251–6252. [Google Scholar] [CrossRef]
- Little, J.W.; Edmiston, S.H.; Pacelli, L.Z.; Mount, D.W. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc. Natl. Acad. Sci. USA 1980, 77, 3225–3229. [Google Scholar] [CrossRef] [PubMed]
- Little, J.W.; Mount, D.W.; Yanisch-Perron, C.R. Purified LexA protein is a repressor of the recA and lexA genes. Proc. Natl. Acad. Sci. USA 1981, 78, 4199–4203. [Google Scholar] [CrossRef]
- Little, J.W.; Mount, D.W. The SOS regulatory system of Escherichia coli. Cell 1982, 29, 11–22. [Google Scholar] [CrossRef]
- Radman, M. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis. In Molecular and Environmental Aspects of Mutagenesis; Prakash, L., Sherman, F., Miller, M.W., Lawrence, C.W., Tabor, H.W., Eds.; Charles, C. Thomas: Springfield, IL, USA, 1974; pp. 128–142. [Google Scholar]
- Kato, T.; Shinoura, Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet. 1977, 156, 121–131. [Google Scholar] [CrossRef]
- Steinborn, G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol. Gen. Genet. 1978, 165, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Bagg, A.; Kenyon, C.J.; Walker, G.C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 1981, 78, 5749–5753. [Google Scholar] [CrossRef]
- Elledge, S.J.; Walker, G.C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J. Mol. Biol. 1983, 164, 175–192. [Google Scholar] [CrossRef]
- Shinagawa, H.; Kato, T.; Ise, T.; Makino, K.; Nakata, A. Cloning and characterization of the umu operon responsible for inducible mutagenesis in Escherichia coli. Gene 1983, 23, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, S.E.; Woodgate, R.; Scheuermann, R.H.; Echols, H. UmuD mutagenesis protein of Escherichia coli: Overproduction, purification and cleavage by RecA. Proc. Natl. Acad. Sci. USA 1988, 85, 1811–1815. [Google Scholar] [CrossRef]
- Nohmi, T.; Battista, J.R.; Dodson, L.A.; Walker, G.C. RecA-mediated cleavage activates UmuD for mutagenesis: Mechanistic relationship between transcriptional derepression and posttranslational activation. Proc. Natl. Acad. Sci. USA 1988, 85, 1816–1820. [Google Scholar] [CrossRef]
- Shinagawa, H.; Iwasaki, H.; Kato, T.; Nakata, A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc. Natl. Acad. Sci. USA 1988, 85, 1806–1810. [Google Scholar] [CrossRef]
- Woodgate, R.; Rajagopalan, M.; Lu, C.; Echols, H. UmuC mutagenesis protein of Escherichia coli: Purification and interaction with UmuD and UmuD’. Proc. Natl. Acad. Sci. USA 1989, 86, 7301–7305. [Google Scholar] [CrossRef] [PubMed]
- Bruck, I.; Woodgate, R.; McEntee, K.; Goodman, M.F. Purification of a soluble UmuD’C complex from Escherichia coli: Cooperative binding of UmuD’C to single-stranded DNA. J. Biol. Chem. 1996, 271, 10767–10774. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Bruck, I.; Eritja, R.; Turner, J.; Frank, E.G.; Woodgate, R.; O’Donnell, M.; Goodman, M.F. Biochemical basis of SOS mutagenesis in Escherichia coli: Reconstitution of in vitro lesion bypass dependent on the UmuD’2C mutagenic complex and RecA protein. Proc. Natl. Acad. Sci. USA 1998, 95, 9755–9760. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Shen, X.; Frank, E.G.; O’Donnell, M.; Woodgate, R.; Goodman, M.F. UmuD’2C is an error-prone DNA polymerase, Escherichia coli, DNA pol V. Proc. Natl. Acad. Sci. USA 1999, 96, 8919–8924. [Google Scholar] [CrossRef] [PubMed]
- Reuven, N.B.; Arad, G.; Maor-Shoshani, A.; Livneh, Z. The mutagenic mrotein UmuC Is a DNA polymerase activated by UmuD’, RecA, and SSB and Is specialized for translesion replication. J. Biol. Chem. 1999, 274, 31763–31766. [Google Scholar] [CrossRef] [PubMed]
- Dutreix, M.; Moreau, P.L.; Bailone, A.; Galibert, F.; Battista, J.R.; Walker, G.C.; Devoret, R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J. Bacteriol. 1989, 171, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- Bailone, A.; Sommer, S.; Knezevic, J.; Dutreix, M.; Devoret, R. A RecA protein mutant deficient in its interaction with the UmuDC complex. Biochimie 1991, 73, 479–484. [Google Scholar] [CrossRef]
- Dutreix, M.; Burnett, B.; Bailone, A.; Radding, C.M.; Devoret, R. A partially deficient mutant, recA1730, that fails to form normal nucleoprotein filaments. Mol. Gen. Genet. 1992, 232, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Karata, K.; Woodgate, R.; Cox, M.M.; Goodman, M.F. The active form of DNA polymerase V is UmuD’(2)C-RecA-ATP. Nature 2009, 460, 359–363. [Google Scholar] [CrossRef]
- Erdem, A.L.; Jaszczur, M.; Bertram, J.G.; Woodgate, R.; Cox, M.M.; Goodman, M.F. DNA polymerase V activity is autoregulated by a novel intrinsic DNA-dependent ATPase. Elife 2014, 3, e02384. [Google Scholar] [CrossRef] [PubMed]
- Sweasy, J.B.; Witkin, E.M.; Sinha, N.; Roegner-Maniscalco, V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J. Bacteriol. 1990, 172, 3030–3036. [Google Scholar] [CrossRef] [PubMed]
- Isogawa, A.; Ong, J.L.; Potapov, V.; Fuchs, R.P.; Fujii, S. Pol V-mediated translesion synthesis elicits localized untargeted mutagenesis during post-replicative gap repair. Cell Rep. 2018, 24, 1290–1300. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Fuchs, R.P. A comprehensive view of translesion synthesis in Escherichia coli. MicroBiol. Mol. Biol. Rev. 2020, 84. [Google Scholar] [CrossRef] [PubMed]
- Frank, E.G.; Ennis, D.G.; Gonzalez, M.; Levine, A.S.; Woodgate, R. Regulation of SOS mutagenesis by proteolysis. Proc. Natl. Acad. Sci. USA 1996, 93, 10291–10296. [Google Scholar] [CrossRef]
- Sommer, S.; Boudsocq, F.; Devoret, R.; Bailone, A. Specific RecA amino acid changes affect RecA-UmuD’C interaction. Mol. MicroBiol. 1998, 28, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.; McDonald, J.P.; Caldas, V.E.; Patel, M.; Wood, E.A.; Punter, C.M.; Ghodke, H.; Cox, M.M.; Woodgate, R.; Goodman, M.F.; et al. Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet. 2015, 11, e1005482. [Google Scholar] [CrossRef] [PubMed]
- Jaszczur, M.M.; Vo, D.D.; Stanciauskas, R.; Bertram, J.G.; Sikand, A.; Cox, M.M.; Woodgate, R.; Mak, C.H.; Pinaud, F.; Goodman, M.F. Conformational regulation of Escherichia coli DNA polymerase V by RecA and ATP. PLoS Genet. 2019, 15, e1007956. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.J.; Erdem, A.L.; Sabat, G.; Karata, K.; Jaszczur, M.M.; Vo, D.D.; Olsen, T.M.; Woodgate, R.; Goodman, M.F.; Cox, M.M. A RecA protein surface required for activation of DNA polymerase V. PLoS Genet. 2015, 11, e1005066. [Google Scholar] [CrossRef] [PubMed]
- Jaszczur, M.; Bertram, J.G.; Robinson, A.; van Oijen, A.M.; Woodgate, R.; Cox, M.M.; Goodman, M.F. Mutations for worse or better: Low-fidelity DNA synthesis by SOS DNA polymerase V Is a tightly regulated double-edged sword. Biochemistry 2016, 55, 2309–2318. [Google Scholar] [CrossRef] [PubMed]
- Goodman, M.F.; McDonald, J.P.; Jaszczur, M.M.; Woodgate, R. Insights into the complex levels of regulation imposed on Escherichia coli DNA polymerase V. DNA Repair 2016, 44, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Karata, K.; Vaisman, A.; Goodman, M.F.; Woodgate, R. Simple and efficient purification of Escherichia coli DNA polymerase V: Cofactor requirements for optimal activity and processivity in vitro. DNA Repair 2012, 11, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Britt, R.L.; Haruta, N.; Lusetti, S.L.; Chitteni-Pattu, S.; Inman, R.B.; Cox, M.M. Disassembly of Escherichia coli RecA E38K/ΔC17 nucleoprotein filaments is required to complete DNA strand exchange. J. Biol. Chem. 2010, 285, 3211–3226. [Google Scholar] [CrossRef]
- Anderson, S.G.; Thompson, J.A.; Paschall, C.O.; O’Donnell, M.; Bloom, L.B. Temporal correlation of DNA binding, ATP hydrolysis, and clamp release in the clamp loading reaction catalyzed by the Escherichia coli ɣ-complex. Biochemistry 2009, 48, 8516–8527. [Google Scholar] [CrossRef][Green Version]
- Douma, L.G.; Yu, K.K.; England, J.K.; Levitus, M.; Bloom, L.B. Mechanism of opening a sliding clamp. Nucleic Acids Res. 2017, 45, 10178–10189. [Google Scholar] [CrossRef] [PubMed]
- Bertram, J.G.; Bloom, L.B.; Hingorani, M.M.; Beechem, J.M.; O’Donnell, M.; Goodman, M.F. Molecular mechanism and energetics of clamp assembly in Escherichia coli. The role of ATP hydrolysis when ɣ complex loads β on DNA. J. Biol. Chem. 2000, 275, 28413–28420. [Google Scholar] [CrossRef]
- Kelman, Z.; O’Donnell, M. DNA polymerase III holoenzyme: Structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 1995, 64, 171–200. [Google Scholar] [CrossRef]
- Bloom, L.B.; Goodman, M.F. A sliding clamp monkey wrench. Nat. Struct Biol. 2001, 8, 829–831. [Google Scholar] [CrossRef]
- Lopez de Saro, F.J.; Georgescu, R.E.; Goodman, M.F.; O’Donnell, M. Competitive processivity-clamp usage by DNA polymerases during DNA replication and repair. EMBO J. 2003, 22, 6408–6418. [Google Scholar] [CrossRef] [PubMed]
- Dalrymple, B.P.; Kongsuwan, K.; Wijffels, G.; Dixon, N.E.; Jennings, P.A. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 2001, 98, 11627–11632. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.; Bertram, J.G.; O’Donnell, M.; Woodgate, R.; Goodman, M.F. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature 2001, 409, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Onrust, R.; Stukenberg, P.T.; O’Donnell, M. Analysis of the ATPase subassembly which initiates processive DNA synthesis by DNA polymerase III holoenzyme. J. Biol. Chem. 1991, 266, 21681–21686. [Google Scholar] [CrossRef]
- Fujii, S.; Fuchs, R.P. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004, 23, 4342–4352. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Fuchs, R.P. Biochemical basis for the essential genetic requirements of RecA and the β-clamp in Pol V activation. Proc. Natl. Acad. Sci. USA 2009, 106, 14825–14830. [Google Scholar] [CrossRef]
- Schlacher, K.; Cox, M.M.; Woodgate, R.; Goodman, M.F. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 2006, 442, 883–887. [Google Scholar] [CrossRef]
- Bonner, C.A.; Randall, S.K.; Rayssiguier, C.; Radman, M.; Eritja, R.; Kaplan, B.E.; McEntee, K.; Goodman, M.F. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J. Biol. Chem. 1988, 263, 18946–18952. [Google Scholar] [CrossRef]
- Iwasaki, H.; Nakata, A.; Walker, G.C.; Shinagawa, H. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J. Bacteriol. 1990, 172, 6268–6273. [Google Scholar] [CrossRef]
- Wagner, J.; Gruz, P.; Kim, S.R.; Yamada, M.; Matsui, K.; Fuchs, R.P.P.; Nohmi, T. The dinB gene encodes an novel Escherichia coli DNA polymerase (DNA pol IV) involved in mutagenesis. Mol. Cell 1999, 4, 281–286. [Google Scholar] [CrossRef]
- Yeiser, B.; Pepper, E.D.; Goodman, M.F.; Finkel, S.E. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. USA 2002, 99, 8737–8741. [Google Scholar] [CrossRef] [PubMed]
- Corzett, C.H.; Goodman, M.F.; Finkel, S.E. Competitive fitness during feast and famine: How SOS DNA polymerases influence physiology and evolution in Escherichia coli. Genetics 2013, 194, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Woodgate, R.; Ennis, D.G. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol. Gen. Genet. 1991, 229, 10–16. [Google Scholar] [CrossRef]
- Witkin, E.M.; McCall, J.O.; Volkert, M.R.; Wermundsen, I.E. Constitutive expression of SOS functions and modulation of mutagenesis resulting from resolution of genetic instability at or near the recA locus of Escherichia coli. Mol. Gen. Genet. 1982, 185, 43–50. [Google Scholar] [CrossRef]
- Ennis, D.G.; Levine, A.S.; Koch, W.H.; Woodgate, R. Analysis of recA mutants with altered SOS functions. Mutat. Res. 1995, 336, 39–48. [Google Scholar] [CrossRef]
- Woodgate, R.; Bridges, B.A.; Kelly, C. Non-mutability by ultraviolet light in uvrD recB derivatives of Escherichia coli WP2 uvrA is due to inhibition of RecA protein activation. Mutat. Res. 1989, 226, 141–144. [Google Scholar] [CrossRef]
- Bridges, B.A.; Woodgate, R. The two-step model of bacterial UV mutagenesis. Mutat. Res. 1985, 150, 133–139. [Google Scholar] [CrossRef]
- Woodgate, R. Evolution of the two-step model for UV-mutagenesis. Mutat. Res. 2001, 485, 83–92. [Google Scholar] [CrossRef]
- Echols, H.; Goodman, M.F. Mutation induced by DNA damage: A many protein affair. Mutat. Res. 1990, 236, 301–311. [Google Scholar] [CrossRef]
- Rajagopalan, M.; Lu, C.; Woodgate, R.; O’Donnell, M.; Goodman, M.F.; Echols, H. Activity of the purified mutagenesis proteins UmuC, UmuD’, and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc. Natl. Acad. Sci. USA 1992, 89, 10777–10781. [Google Scholar] [CrossRef] [PubMed]


| Name | Sequence |
|---|---|
| 20 mer primer | 5’—TTT GGA TGA AGG TGA TTT CT—3’ |
| Regular template | 5’—ATG ACA AGA CAA GAC AAG ACA AGA CAA GAC AAG ACA AGA CAA GAC AAG AAA TCA CCA TCA TCC AAA TCC ACT AAA CCA TAT CCA TCC TCG—3’ |
| Template V1—Abasic site 3 nt away from primer terminus | 5’—ATG ACA AGA CAA GAC XAG ACA AGA CAA GAC AAG ACA AGA CAA GAC AAG AAA TCA CCT TCA TCC AAA TCC ACT AAA CCA TAT CCA TCC TC—3’ |
| Template V2—Abasic site 30 nt away from primer terminus | 5’—ATG ACA AGA CAA GAC AAG ACA AGA CAA GAC AAG ACA AGA CAA XAC AAG AAA TCA CCT TCA TCC AAA TCC ACT AAA CCA TAT CCA TCC TCG—3’ |
| 20 mer linker for circularization | 5’—GTC TTG TCA TCG AGG ATG GA—3’ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sikand, A.; Jaszczur, M.; Bloom, L.B.; Woodgate, R.; Cox, M.M.; Goodman, M.F. The SOS Error-Prone DNA Polymerase V Mutasome and β-Sliding Clamp Acting in Concert on Undamaged DNA and during Translesion Synthesis. Cells 2021, 10, 1083. https://doi.org/10.3390/cells10051083
Sikand A, Jaszczur M, Bloom LB, Woodgate R, Cox MM, Goodman MF. The SOS Error-Prone DNA Polymerase V Mutasome and β-Sliding Clamp Acting in Concert on Undamaged DNA and during Translesion Synthesis. Cells. 2021; 10(5):1083. https://doi.org/10.3390/cells10051083
Chicago/Turabian StyleSikand, Adhirath, Malgorzata Jaszczur, Linda B. Bloom, Roger Woodgate, Michael M. Cox, and Myron F. Goodman. 2021. "The SOS Error-Prone DNA Polymerase V Mutasome and β-Sliding Clamp Acting in Concert on Undamaged DNA and during Translesion Synthesis" Cells 10, no. 5: 1083. https://doi.org/10.3390/cells10051083
APA StyleSikand, A., Jaszczur, M., Bloom, L. B., Woodgate, R., Cox, M. M., & Goodman, M. F. (2021). The SOS Error-Prone DNA Polymerase V Mutasome and β-Sliding Clamp Acting in Concert on Undamaged DNA and during Translesion Synthesis. Cells, 10(5), 1083. https://doi.org/10.3390/cells10051083






