Regulatory and Effector Cell Disequilibrium in Patients with Acute Cellular Rejection and Chronic Lung Allograft Dysfunction after Lung Transplantation: Comparison of Peripheral and Alveolar Distribution
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Lung Function Tests
2.3. Preparation and Storage of PBMCs
3. Bronchoalveolar Lavage Cell Processing and Collection
3.1. Lymphocyte Immunophenotyping by Flow Cytometry
3.2. Th17 Phenotyping by Flow Cytometry
4. Regulatory T Cell Detection by Flow Cytometry
5. Regulatory B Cell Detection by Flow Cytometry
Statistical Analysis
6. Results
6.1. Population
6.2. Peripheral Blood Mononuclear Cell Analysis
6.3. Broncho-Alveolar Lavage Cell Analysis
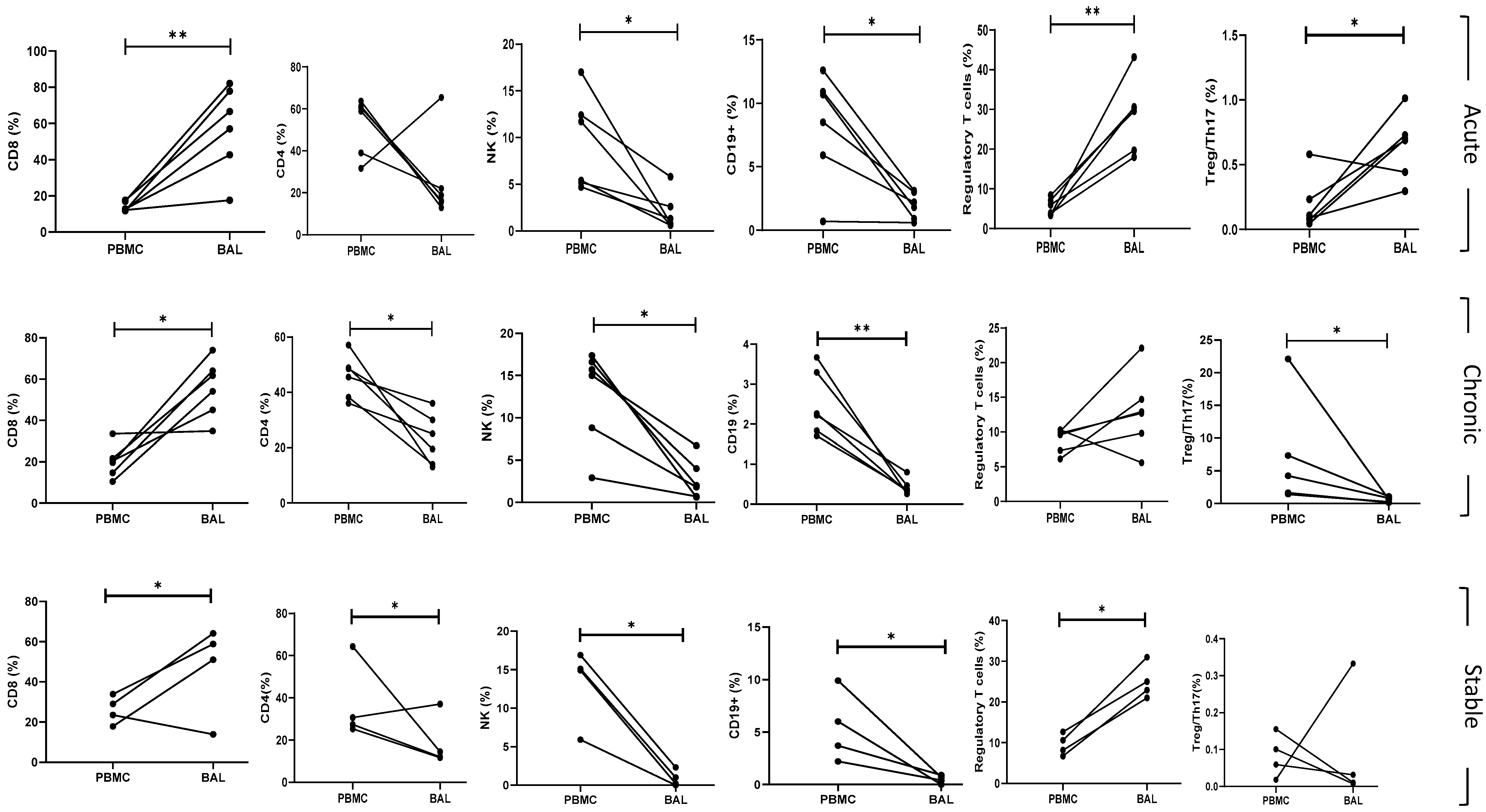
6.4. BAL and Peripheral Blood Comparison of Subpopulations
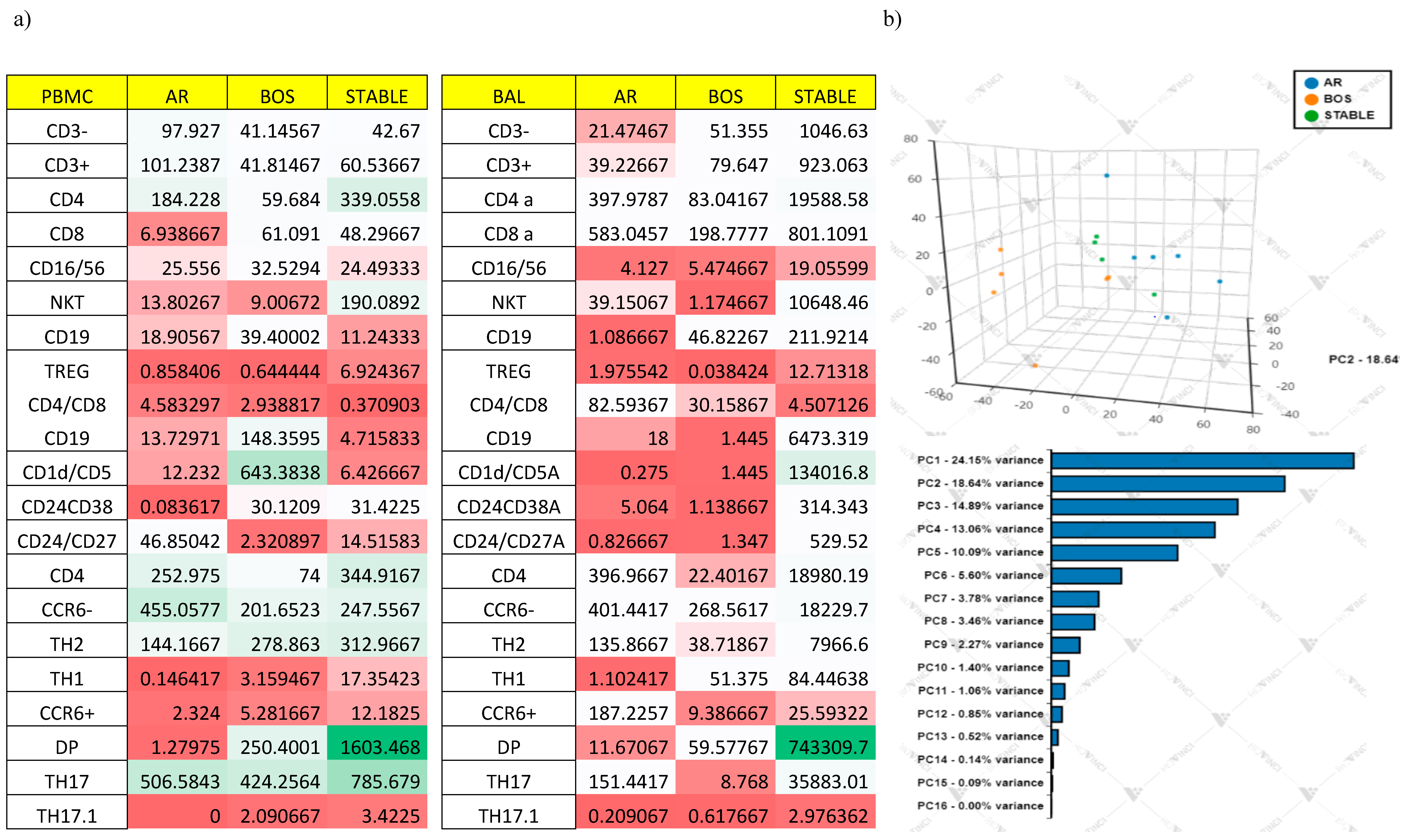
Multivariate Analysis
6.5. Correlation of Lung Function Parameters and Immune Cells
7. Discussion
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Chronic allograft dysfunction | CLAD |
| Lung transplant | LTx |
| Acute cellular rejection | AR |
| Chronic lung allograft dysfunction | CLAD |
| Bronchiolitis obliterans syndrome | BOS |
| Restrictive allograft syndrome | RAS |
| T helper | Th |
| Bronchoalveolar lavage | BAL |
| Peripheral blood mononuclear cells | PBMCs |
| Forced vital capacity | FVC |
| Forced expiratory volume in the first second | FEV1 |
| Diffusing capacity of the lung for carbon monoxide | DLCO |
References
- Boyton, R. The Role of Natural Killer T Cells in Lung Inflammation. J. Pathol. 2008, 214, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Bennett, D.; Fossi, A.; Bargagli, E.; Refini, R.M.; Pieroni, M.; Luzzi, L.; Ghiribelli, C.; Paladini, P.; Voltolini, L.; Rottoli, P. Mortality on the Waiting List for Lung Transplantation in Patients with Idiopathic Pulmonary Fibrosis: A Single-Centre Experience. Lung 2015, 193, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Vietri, L.; Bargagli, E.; Bennett, D.; Fossi, A.; Cameli, P.; Bergantini, L.; d’Alessandro, M.; Paladini, P.; Luzzi, L.; Gentili, F.; et al. Serum Amyloid A in Lung Transplantation. Sarcoidosis Vasc. Diffuse Lung Dis. Off. J. WASOG 2020, 37, 2–7. [Google Scholar] [CrossRef]
- Palleschi, A.; Gaudioso, G.; Edefonti, V.; Musso, V.; Terrasi, A.; Ambrogi, F.; Franzi, S.; Rosso, L.; Tarsia, P.; Morlacchi, L.C.; et al. Bronchoalveolar Lavage-MicroRNAs Are Potential Novel Biomarkers of Outcome After Lung Transplantation. Transplant. Direct 2020, 6. [Google Scholar] [CrossRef]
- Neujahr, D.C.; Cardona, A.C.; Ulukpo, O.; Rigby, M.; Pelaez, A.; Ramirez, A.; Gal, A.A.; Force, S.D.; Lawrence, E.C.; Kirk, A.D.; et al. Dynamics of Human Regulatory T Cells in Lung Lavages of Lung Transplant Recipients. Transplantation 2009, 88, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Levy, L.; Huszti, E.; Renaud-Picard, B.; Berra, G.; Kawashima, M.; Takahagi, A.; Fuchs, E.; Ghany, R.; Moshkelgosha, S.; Keshavjee, S.; et al. Risk Assessment of Chronic Lung Allograft Dysfunction Phenotypes: Validation and Proposed Refinement of the 2019 International Society for Heart and Lung Transplantation Classification System. J. Heart Lung Transplant. 2020, 39, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Sato, M. Bronchiolitis Obliterans Syndrome and Restrictive Allograft Syndrome after Lung Transplantation: Why Are There Two Distinct Forms of Chronic Lung Allograft Dysfunction? Ann. Transl. Med. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Bennett, D.; Lanzarone, N.; Fossi, A.; Perillo, F.; De Vita, E.; Luzzi, L.; Paladini, P.; Bargagli, E.; Sestini, P.; Rottoli, P. Pirfenidone in Chronic Lung Allograft Dysfunction: A Single Cohort Study. Panminerva Med. 2020, 62, 143–149. [Google Scholar] [CrossRef]
- Shilling, R.A.; Wilkes, D.S. Role of Th17 Cells and IL-17 in Lung Transplant Rejection. Semin. Immunopathol. 2011, 33, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Guo, B. IL-10 Modulates Th17 Pathogenicity during Autoimmune Diseases. J. Clin. Cell. Immunol. 2016, 7. [Google Scholar] [CrossRef]
- d’Alessandro, M.; Bergantini, L.; Cameli, P.; Fanetti, M.; Alderighi, L.; Armati, M.; Refini, R.M.; Alonzi, V.; Sestini, P.; Bargagli, E. Immunologic Responses to Antifibrotic Treatment in IPF Patients. Int. Immunopharmacol. 2021, 95, 107525. [Google Scholar] [CrossRef]
- Gregson, A.L.; Hoji, A.; Saggar, R.; Ross, D.J.; Kubak, B.M.; Jamieson, B.D.; Weigt, S.S.; Lynch, J.P.; Ardehali, A.; Belperio, J.A.; et al. Bronchoalveolar Immunologic Profile of Acute Human Lung Transplant Allograft Rejection. Transplantation 2008, 85, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Bergantini, L.; d’Alessandro, M.; Cameli, P.; Bono, C.; Perruzza, M.; Biagini, M.; Pini, L.; Bigliazzi, C.; Sestini, P.; Dotta, F.; et al. Regulatory T Cell Monitoring in Severe Eosinophilic Asthma Patients Treated with Mepolizumab. Scand. J. Immunol. 2021, e13031. [Google Scholar] [CrossRef]
- Bergantini, L.; Cameli, P.; d’Alessandro, M.; Vietri, L.; Perruzza, M.; Pieroni, M.; Lanzarone, N.; Refini, R.; Fossi, A.; Bargagli, E. Regulatory T Cells in Severe Persistent Asthma in the Era of Monoclonal Antibodies Target Therapies. Inflammation 2020, 43, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Downs-Canner, S.; Berkey, S.; Delgoffe, G.M.; Edwards, R.P.; Curiel, T.; Odunsi, K.; Bartlett, D.L.; Obermajer, N. Suppressive IL-17A+Foxp3+ and Ex-Th17 IL-17AnegFoxp3+ Treg Cells Are a Source of Tumour-Associated Treg Cells. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zhou, J.; Fan, C.; Zhao, N.; Liu, Y.; Wang, S.; Cui, X.; Huang, M.; Guan, H.; Li, Y.; et al. Increased Circulating Th17 but Decreased CD4+Foxp3+ Treg and CD19+CD1dhiCD5+ Breg Subsets in New-Onset Graves’ Disease. BioMed Res. Int. 2017, 2017. [Google Scholar] [CrossRef]
- Wang, W.; Yuan, X.; Chen, H.; Xie, G.; Ma, Y.; Zheng, Y.; Zhou, Y.; Shen, L. CD19+CD24hiCD38hiBregs Involved in Downregulate Helper T Cells and Upregulate Regulatory T Cells in Gastric Cancer. Oncotarget 2015, 6, 33486–33499. [Google Scholar] [CrossRef] [PubMed]
- Rosser, E.C.; Mauri, C. Regulatory B Cells: Origin, Phenotype, and Function. Immunity 2015, 42, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Ming, Y.; Yang, C. Regulatory B Cells: The Cutting Edge of Immune Tolerance in Kidney Transplantation. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Roden, A.C.; Aisner, D.L.; Allen, T.C.; Aubry, M.C.; Barrios, R.J.; Beasley, M.B.; Cagle, P.T.; Capelozzi, V.L.; Dacic, S.; Ge, Y.; et al. Diagnosis of Acute Cellular Rejection and Antibody-Mediated Rejection on Lung Transplant Biopsies: A Perspective From Members of the Pulmonary Pathology Society. Arch. Pathol. Lab. Med. 2017, 141, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.C.; Raghu, G.; Verleden, G.M.; Corris, P.A.; Aurora, P.; Wilson, K.C.; Brozek, J.; Glanville, A.R. ISHLT/ATS/ERS BOS Task Force Committee; ISHLT/ATS/ERS BOS Task Force Committee An International ISHLT/ATS/ERS Clinical Practice Guideline: Diagnosis and Management of Bronchiolitis Obliterans Syndrome. Eur. Respir. J. 2014, 44, 1479–1503. [Google Scholar] [CrossRef]
- Broos, C.E.; Koth, L.L.; Van Nimwegen, M.; Paulissen, S.M.; Van Hamburg, J.P.; Annema, J.T.; Heller-Baan, R.; Kleinjan, A.; Hoogsteden, H.C.; Wijsenbeek, M.S.; et al. Increased T-Helper 17.1 Cells in Sarcoidosis Mediastinal Lymph Nodes. Eur. Respir. J. 2018, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zheng, X.; Zhang, J.; Zhu, Y.; Zhu, X.; Liu, H.; Zeng, M.; Graner, M.W.; Zhou, B.; Chen, X. CD19+CD1d+CD5+ B Cell Frequencies Are Increased in Patients with Tuberculosis and Suppress Th17 Responses. Cell. Immunol. 2012, 274, 89–97. [Google Scholar] [CrossRef]
- Flores-Borja, F.; Bosma, A.; Ng, D.; Reddy, V.; Ehrenstein, M.R.; Isenberg, D.A.; Mauri, C. CD19+CD24hiCD38hi B Cells Maintain Regulatory T Cells While Limiting TH1 and TH17 Differentiation. Sci. Transl. Med. 2013, 5, 173ra23-173ra23. [Google Scholar] [CrossRef]
- Liu, Z.; Dang, E.; Li, B.; Qiao, H.; Jin, L.; Zhang, J.; Wang, G. Dysfunction of CD19+CD24hiCD27+ B Regulatory Cells in Patients with Bullous Pemphigoid. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Thompson-Snipes, L.; Klintmalm, G.; Demetris, A.J.; O’Leary, J.; Oh, S.; Joo, H. CD24hiCD38hi and CD24hiCD27+ Human Regulatory B Cells Display Common and Distinct Functional Characteristics. J. Immunol. 2019, 203, 2110–2120. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Dresel, J.; Eckstein, V.; Gellert, R.; Störch, H.; Venigalla, R.K.C.; Schwenger, V.; Max, R.; Blank, N.; Lorenz, H.-M.; et al. Impaired Suppressive Capacity of Activation-Induced Regulatory B Cells in Systemic Lupus Erythematosus. Arthritis Rheumatol. 2014, 66, 2849–2861. [Google Scholar] [CrossRef] [PubMed]
- Diller, M.L.; Kudchadkar, R.R.; Delman, K.A.; Lawson, D.H.; Ford, M.L. Balancing Inflammation: The Link between Th17 and Regulatory T Cells. Mediators Inflamm. 2016, 2016. [Google Scholar] [CrossRef]
- Bargagli, E.; Madioni, C.; Bianchi, N.; Refini, R.M.; Cappelli, R.; Rottoli, P. Serum Analysis of Coagulation Factors in IPF and NSIP. Inflammation 2014, 37, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.R. The Balance of Th17 versus Treg Cells in Autoimmunity. Int. J. Mol. Sci. 2018, 19, 730. [Google Scholar] [CrossRef]
- Knochelmann, H.M.; Dwyer, C.J.; Bailey, S.R.; Amaya, S.M.; Elston, D.M.; Mazza-McCrann, J.M.; Paulos, C.M. When Worlds Collide: Th17 and Treg Cells in Cancer and Autoimmunity. Cell. Mol. Immunol. 2018, 15, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Nakagiri, T.; Inoue, M.; Minami, M.; Shintani, Y.; Okumura, M. Immunology Mini-Review: The Basics of TH17 and Interleukin-6 in Transplantation. Transplant. Proc. 2012, 44, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Neujahr, D.C.; Larsen, C.P. Regulatory T Cells in Lung Transplantation—an Emerging Concept. Semin. Immunopathol. 2011, 33, 117–127. [Google Scholar] [CrossRef][Green Version]
- Increased Frequency of CD4+CD25highCD127low T Cells Early after Lung Transplant Is Associated with Improved Graft Survival—A Retrospective Study—Ius—2020—Transplant International—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1111/tri.13568 (accessed on 3 November 2020).
- Piloni, D.; Morosini, M.; Magni, S.; Balderacchi, A.; Scudeller, L.; Cova, E.; Oggionni, T.; Stella, G.; Tinelli, C.; Antonacci, F.; et al. Analysis of Long Term CD4+CD25highCD127− T-Reg Cells Kinetics in Peripheral Blood of Lung Transplant Recipients. BMC Pulm. Med. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Bergantini, L.; Cameli, P.; d’Alessandro, M.; Vagaggini, C.; Refini, R.; Landi, C.; Pieroni, M.; Spalletti, M.; Sestini, P.; Bargagli, E. NK and NKT-like Cells in Granulomatous and Fibrotic Lung Diseases. Clin. Exp. Med. 2019, 19, 487–494. [Google Scholar] [CrossRef]
- d’Alessandro, M.; Carleo, A.; Cameli, P.; Bergantini, L.; Perrone, A.; Vietri, L.; Lanzarone, N.; Vagaggini, C.; Sestini, P.; Bargagli, E. BAL Biomarkers’ Panel for Differential Diagnosis of Interstitial Lung Diseases. Clin. Exp. Med. 2020, 20, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Lanzarone, N.; Gentili, F.; Alonzi, V.; Bergantini, L.; d’Alessandro, M.; Rottoli, P.; Refini, R.M.; Pieroni, M.; Vietri, L.; Bianchi, F.; et al. Bronchoalveolar Lavage and Serum KL-6 Concentrations in Chronic Hypersensitivity Pneumonitis: Correlations with Radiological and Immunological Features. Intern. Emerg. Med. 2020, 15, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- Paulissen, S.M.J.; van Hamburg, J.P.; Davelaar, N.; Vroman, H.; Hazes, J.M.W.; de Jong, P.H.P.; Lubberts, E. CCR6+ Th Cell Populations Distinguish ACPA Positive from ACPA Negative Rheumatoid Arthritis. Arthritis Res. Ther. 2015, 17, 344. [Google Scholar] [CrossRef]
- Lubberts, E. The IL-23-IL-17 Axis in Inflammatory Arthritis. Nat. Rev. Rheumatol. 2015, 11, 562. [Google Scholar] [CrossRef] [PubMed]
- Karlinsky, H. Alzheimer’s Disease in Down’s Syndrome: A Review. J. Am. Geriatr. Soc. 1986, 34, 728–734. [Google Scholar] [CrossRef] [PubMed]
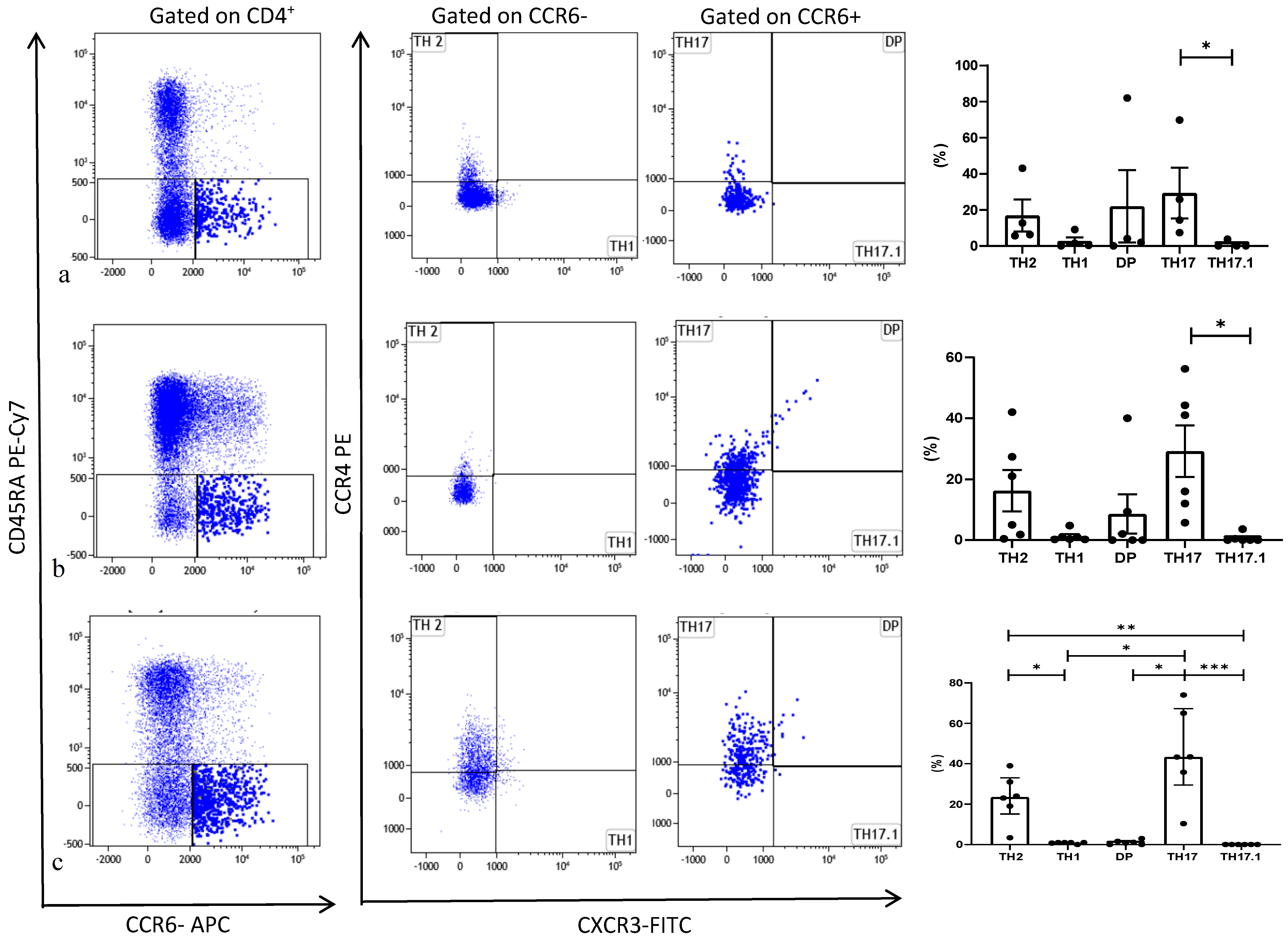
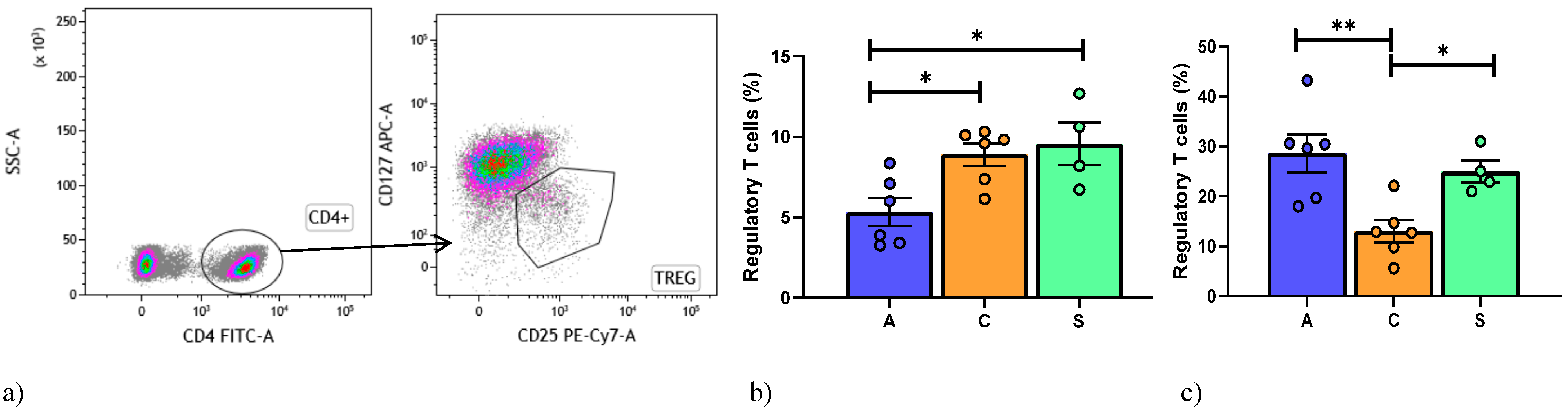
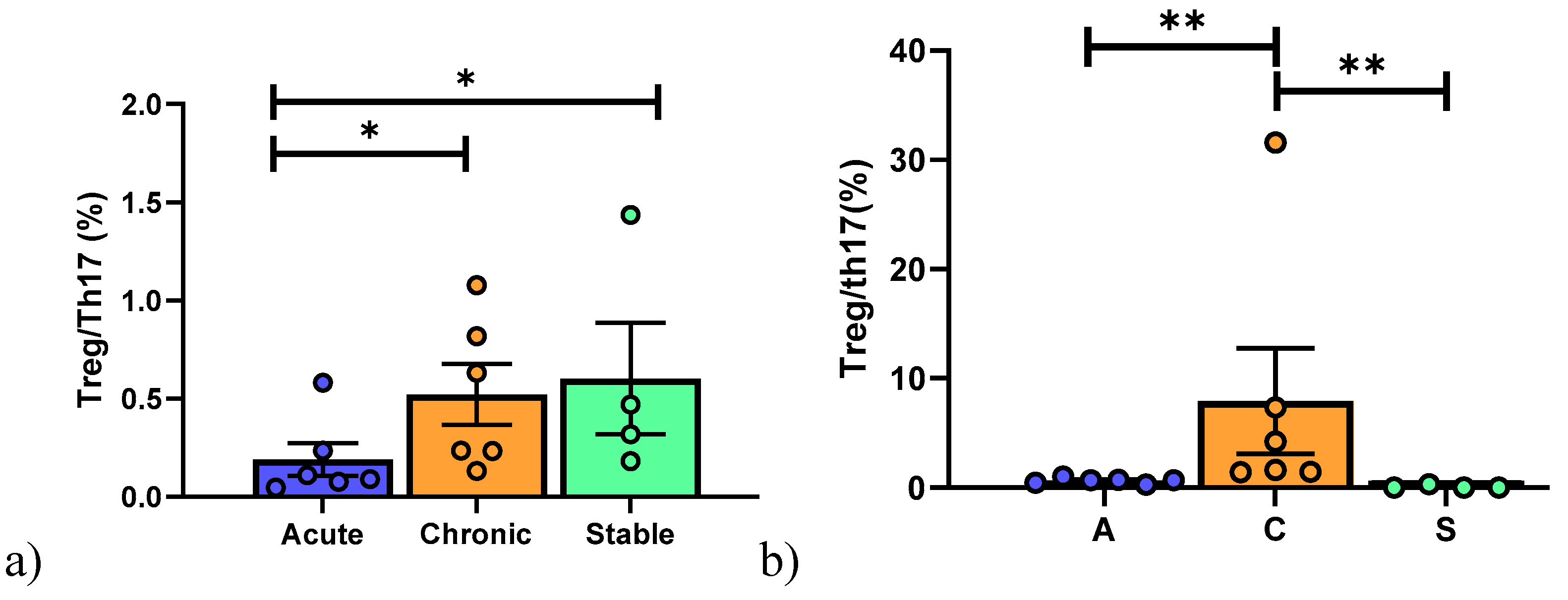
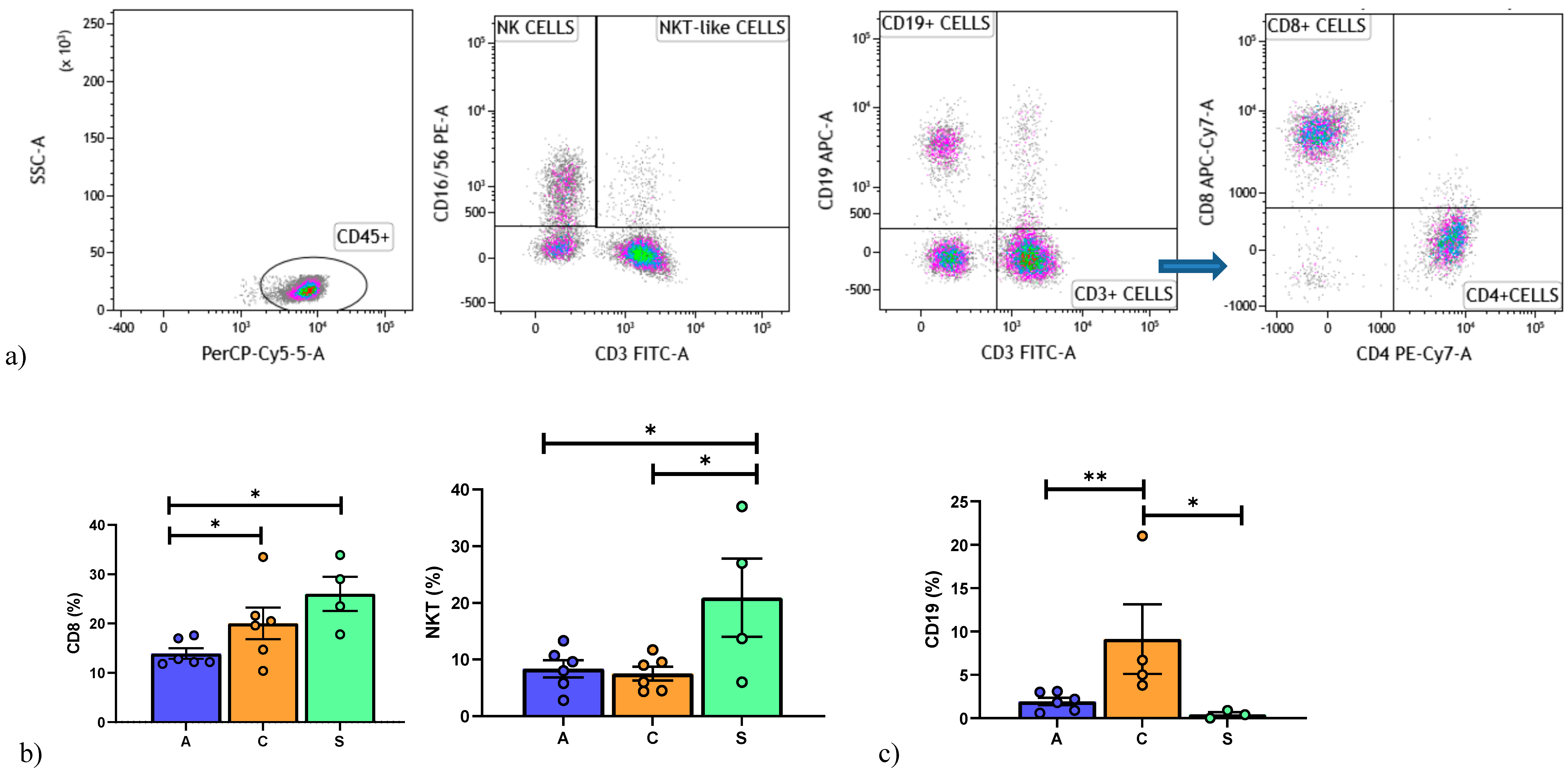

| Number of Patients/Sex | Age (yr) | Tx Indication | Type of TX | Months from Tx | Immunosuppression at Enrollment | Clinical Conditions |
|---|---|---|---|---|---|---|
| 1/M | 39 | CF | B | 17 | Pred + Tac + MMF | AR |
| 2/F | 63 | IPF | S | 7 | Pred + Tac + MMF | AR |
| 3/M | 62 | IPF | S | 2 | Pred + Tac + MMF | AR |
| 4/F | 38 | CF | B | 10 | Pred + Tac | AR |
| 5/M | 37 | CF | B | 2 | Pred + Tac | AR |
| 6/M | 65 | HP | S | 3 | Pred + Tac + MMF | AR |
| 7/M | 60 | CPFE | B | 29 | Pred + Tac | BOS |
| 8/M | 64 | COPD | S | 9 | Pred + Tac + MMF | BOS |
| 9/F | 33 | CF | B | 9 | Pred + Tac + MMF | BOS |
| 10/M | 68 | IPAF | S | 22 | Pred + Tac + MMF | BOS |
| 11/F | 37 | CF | B | 84 | Pred + Tac + MMF | BOS |
| 12/M | 52 | HP | S | 43 | Pred + Tac + MMF | BOS |
| 13/F | 44 | SARC | B | 5 | Pred + Tac | S |
| 14/F | 46 | CF | B | 84 | Pred + Tac + AZA | S |
| 15/F | 50 | PLCH | B | 7 | Pred + Tac + MMF | S |
| 16/M | 62 | IPF | B | 9 | Pred + Tac | S |
| PBMCs | AR | BOS | STABLE | p Value |
|---|---|---|---|---|
| Lymphocytes phenotyping: | ||||
| CD3+: | 76.8 (72–89) | 75.6 (72.6–79.9) | 77.9 (74.9–79.6) | ns |
| 59.6 (44–63.7) | 47 (40–48.8) | 28.9 (26.8–39) | ns |
| 12.5 (12.2–17.6) | 20 (15.9–21.3) | 26.2 (22–30.2) | 0.01 | |
| 8.8 (6.3–13.3) | 7.5 (4.8–9.4) | 20 (11.7–29.5) | ns | |
| 3.6 (3.2–5.1) | 2.2 (1.9–3) | 1.4 (1.2–1.7) | <0.0001 | |
| CD3−: | 17.9 (12.5–35.4) | 22.2 (18.45–25.1) | 19.4 (17.9–22.1) | ns |
| 9.6 (6.5–12.6) | 6.3 (4.2–11.3) | 4.8 (3.3–6.9) | ns |
| 8.35 (5.25–17) | 15.3 (10.3–16.3) | 15 (12.6–15.6) | ns | |
| Th cells subtypes: | ||||
| CD4+CCR6−: | 41.1 (25.8–67.3) | 46.2 (37.6–55.6) | 51.5 (42–57.8) | ns |
| 1 (0.7–1) | 0.7 (0.2–1) | 0.9 (0.2–3.3) | ns |
| 23.5 (20–39) | 13 (2.6–25) | 9.5 (6.1–20) | ns | |
| 0.03 (0.02–0.05) | 0.11 (0.06–0.17) | 0.04 (0.02–0.39) | ns | |
| CD4+CCR6+: | 4 (2.9–6.3) | 2.15 (1.25–4.9) | 3.25 (1.4–5.6) | ns |
| 43.3 (37.6–74) | 28.5 (13–43.4) | 20 (12.5–36.8) | ns |
| 0 (0–0) | 0 (0–0.3) | 0 (0–0.9) | ns | |
| 0.9 (1.8–3) | 1 (0–7.53) | 2.9 (1.4–23.5) | ns | |
| Regulatory B subtypes: | ||||
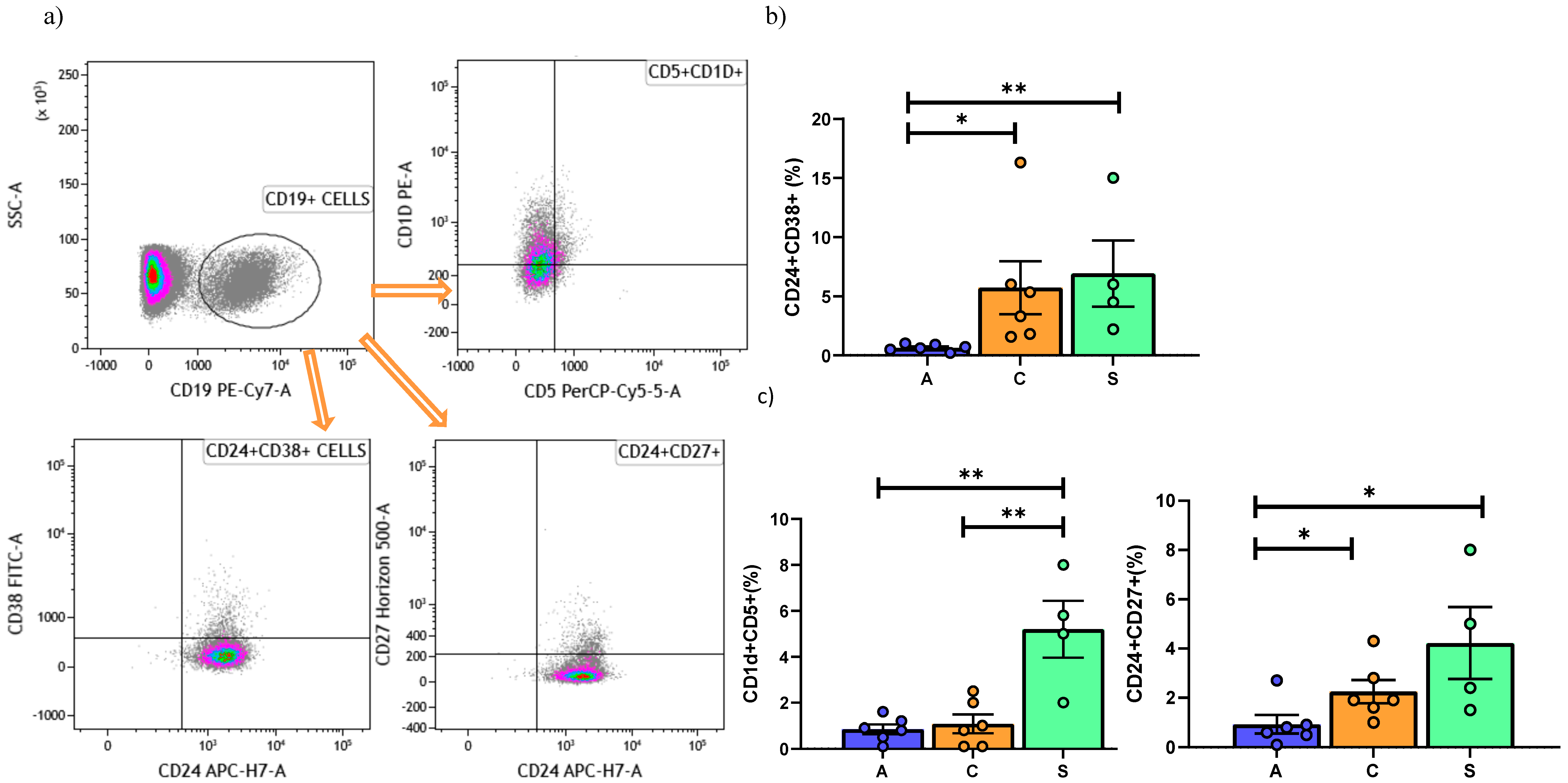
| 0.65 (0.5–1) | 4.3 (2.1–5.8) | 5.2 (3.9–8.2) | 0.0005 |
| 1.16 (0.6–17) | 1.8 (1.7–2.5) | 7.4 (5.2–9.7) | ns | |
| 2.45 (0.67–9.2) | 2.5 (7.6–3.1) | 2(1.9–3) | ns | |
| Regulatory T cells: | ||||
| 4.9 (3.5–8.3) | 9.4 (7.8–11.1) | 9.71 (7.9–10) | 0.0003 |
| 0.10(0.06–0.24) | 0.1(0.08–0.14) | 0.4 (0.28–0.71) | ns | |
| 4.8(3.7–5.9) | 16.4 (9.6–28.7) | 14.4 (6.5–20.9) | 0.004 | |
| BAL | AR | BOS | STABLE | p Value |
|---|---|---|---|---|
| Lymphocyte phenotyping: | ||||
| CD3+: | 85.9 (82.9–88.9) | 73.4 (69–79) | 88 (75.3–87.2) | ns |
| 17.5 (15.7–21.2) | 22.2 (15.4–28.7) | 13.2 (11.9–20) | ns |
| 61.8 (46.2–75) | 57.8 (47.2–63.4) | 54.9 (41.7–60.1) | ns | |
| 9.5 (3.3–14) | 2 (1.1–2) | 5.5 (4.3–7.1) | 0.04 | |
| 3.0 (2.1–3.1) | 1.28 (1–1.42) | 0.22 (0.2–0.8) | 0.0001 | |
| CD3−: | 11.3 (7.4–13.6) | 21.5 (15.2–25) | 14.5 (11.4–21) | ns |
| 2 (1.2–2.8) | 4.5 (3.8–6.2) | 0.5 (0.3–0.75) | 0.0023 |
| 1 (0.6–2.2) | 1.9 (0.97–3.5) | 0.6 (0.15–1.3) | ns | |
| Th cells subtypes: | ||||
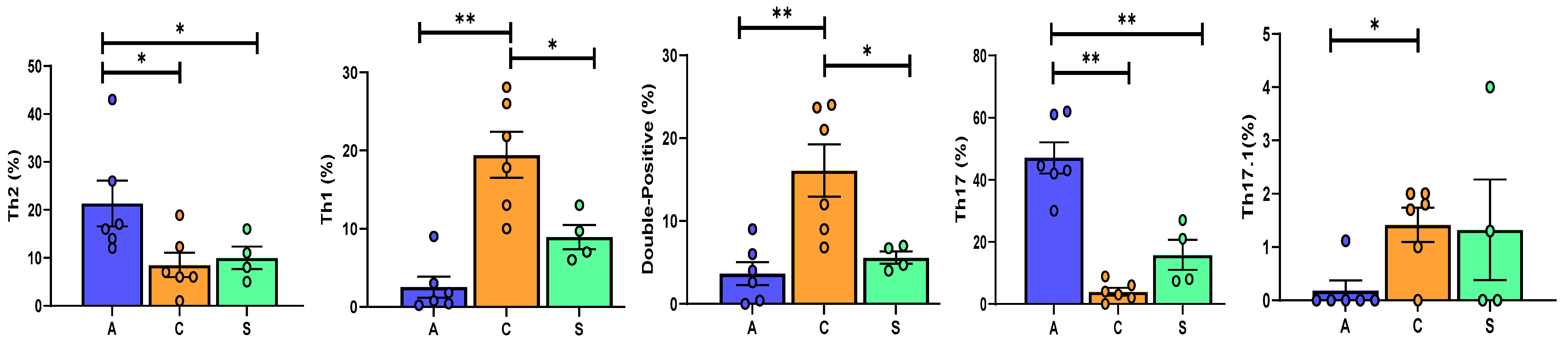
| CD4+CCR6− | 71.7 (65.2–89.2) | 81(70.5–93.5) | 40 (32.7–47.5) | 0.013 |
| 0.8 (0.4–1.5) | 19.8 (14.2–24.9) | 8.35(6.7–10.5) | <0.0001 |
| 16.5 (14.5–23.75) | 6.5(6–10.9) | 9.5 (7.2–12.2) | 0.04 | |
| 0.04 (0.01–0.11) | 2.26 (1.3–9.2) | 0.8 (0.6–1.6) | <0.0001 | |
| CD4+CCR6+ | 11.3 (7.4–19.25) | 6.9 (6–7.9) | 9.4 (8.5–10.7) | ns |
| 43.7 (42.2–56.8) | 3.5 (2.2–5.5) | 14.5 (7.8–22.5) | <0.0001 |
| 0 (0–0) | 1.75 (1.2–1.9) | 0.65 (0–1.9) | ns | |
| 1.6 (0.45–3.65) | 16.5 (9.7–23) | 5.7(4.5–6.7) | 0.0014 | |
| Regulatory B subtypes: | ||||
| 3.3 (2.2–5.1) | 1.85 (1.7–2.6) | 2 (1.2–4.9) | ns |
| 0.7 (0.52–0.87) | 1.9 (1.7–2.6) | 5 (2.8–8.3) | 0.0002 | |
| 0.85 (0.57–1.12) | 0.9 (0.52–1.4) | 5 (3.5–6.5) | 0.004 | |
| Regulatory T cells: | ||||
| 0.31 (0.23–0.38) | 0.35 (0.32–0.44) | 23.9 (22.4–26.5) | 0.01 |
| 0.005 (0.003–0.008) | 0.12 (0.06–0.18) | 1.9 (1.13–2.9) | <0.0001 | |
| 18.5 (15.6–41.9) | 0.02 (0.01–0.03) | 2.8 (2.8–3.4) | <0.0001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergantini, L.; d’Alessandro, M.; De Vita, E.; Perillo, F.; Fossi, A.; Luzzi, L.; Paladini, P.; Perrone, A.; Rottoli, P.; Sestini, P.; et al. Regulatory and Effector Cell Disequilibrium in Patients with Acute Cellular Rejection and Chronic Lung Allograft Dysfunction after Lung Transplantation: Comparison of Peripheral and Alveolar Distribution. Cells 2021, 10, 780. https://doi.org/10.3390/cells10040780
Bergantini L, d’Alessandro M, De Vita E, Perillo F, Fossi A, Luzzi L, Paladini P, Perrone A, Rottoli P, Sestini P, et al. Regulatory and Effector Cell Disequilibrium in Patients with Acute Cellular Rejection and Chronic Lung Allograft Dysfunction after Lung Transplantation: Comparison of Peripheral and Alveolar Distribution. Cells. 2021; 10(4):780. https://doi.org/10.3390/cells10040780
Chicago/Turabian StyleBergantini, Laura, Miriana d’Alessandro, Elda De Vita, Felice Perillo, Antonella Fossi, Luca Luzzi, Piero Paladini, Anna Perrone, Paola Rottoli, Piersante Sestini, and et al. 2021. "Regulatory and Effector Cell Disequilibrium in Patients with Acute Cellular Rejection and Chronic Lung Allograft Dysfunction after Lung Transplantation: Comparison of Peripheral and Alveolar Distribution" Cells 10, no. 4: 780. https://doi.org/10.3390/cells10040780
APA StyleBergantini, L., d’Alessandro, M., De Vita, E., Perillo, F., Fossi, A., Luzzi, L., Paladini, P., Perrone, A., Rottoli, P., Sestini, P., Bargagli, E., & Bennett, D. (2021). Regulatory and Effector Cell Disequilibrium in Patients with Acute Cellular Rejection and Chronic Lung Allograft Dysfunction after Lung Transplantation: Comparison of Peripheral and Alveolar Distribution. Cells, 10(4), 780. https://doi.org/10.3390/cells10040780









