Extracellular Vesicles from Infected Cells Are Released Prior to Virion Release
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. EV Isolation and Ultracentrifugation
2.3. Virus Rescue Assay
2.4. EV and Virion Capture with Nanotrap Particles
2.5. Preparation of Whole Cell Extracts and Western Blot Analysis
2.6. RNA Isolation and RT-qPCR
2.7. Nanoparticle Tracking Analysis
2.8. Statistical Analysis
3. Results
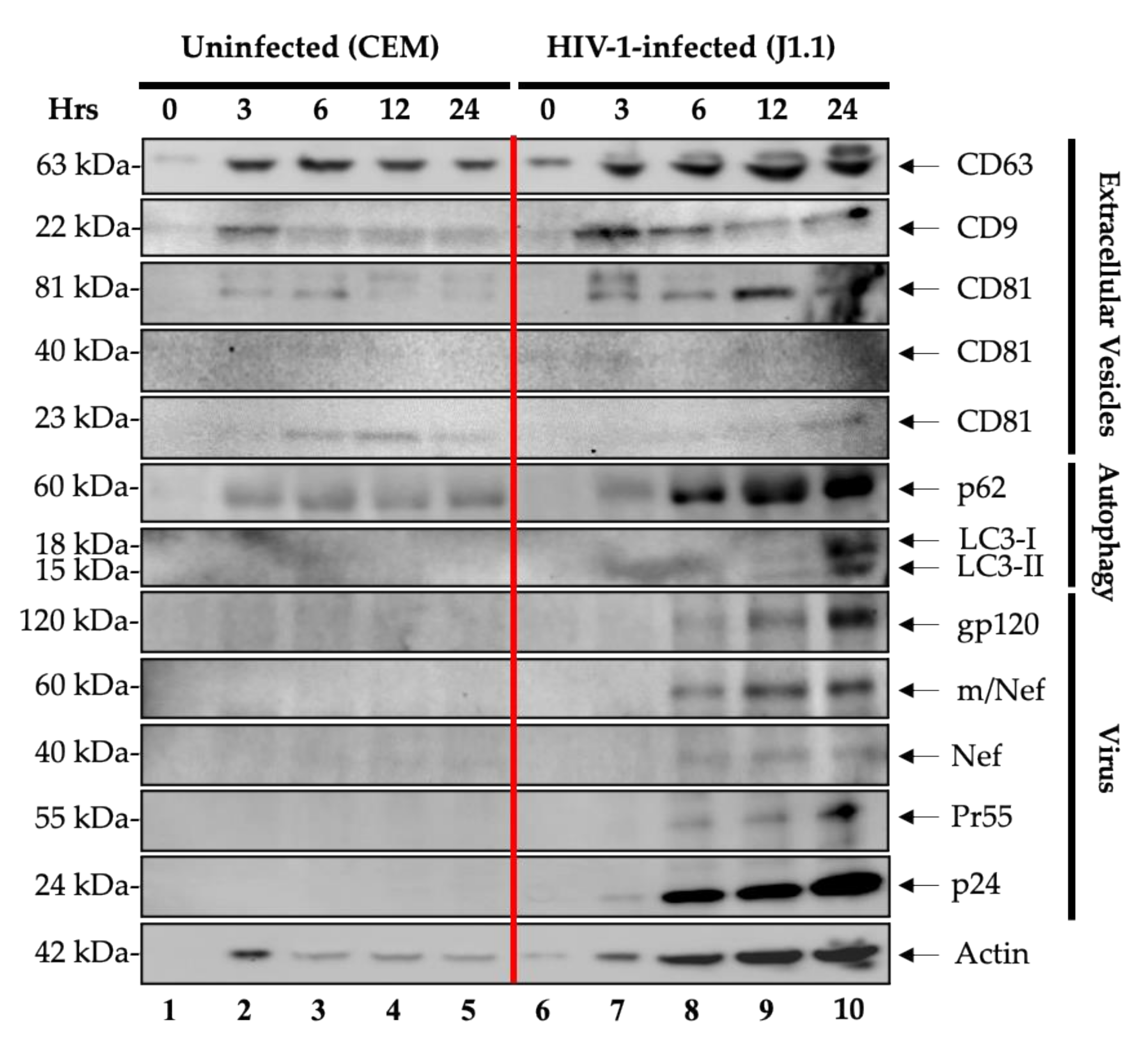
3.1. Enrichment and Characterization of the EVs and Virions Released over Time from HIV-1-Infected Cells
3.2. Increased Levels of Intracellular and Extracellular Viral RNA Post-Release
3.3. Functional HIV-1 Rescue from Released Cells
3.4. Evaluation of Viral and EV Release from Other Retrovirally Infected Cells (HTLV-1)
3.5. Detection of HTLV-1 RNA in Both Intracellular and Extracellular Environments
3.6. Virus Rescue Assay Using Various Susceptible Cells
3.7. Lack of Cell Death Following Release with Complete Media
3.8. HIV-1 and HTLV-1 EVs Promote Differential Expression of IL-8 and TNF-α in Myeloid and T Cell Lines
3.9. EV Contents from Infected Primary Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EV | extracellular vesicle |
| PHA | phytohemagglutinin |
| cART | combination antiretroviral therapy |
| TAR | transactivation response |
| ESCRT | endosomal sorting complex required for transport |
| HAND | HIV-1-associated neurocognitive disorder |
| RT-qPCR | real-time quantitative polymerase chain reaction |
| HAM/TSP | HTLV-1-associated myelopathy/tropical spastic paraparesis |
| NTA | nanoparticle tracking analysis |
| PBMC | peripheral blood mononuclear cell |
References
- AIDS by the Numbers. Available online: https://www.unaids.org/en/resources/documents/2019/aids-by-the-numbers (accessed on 19 October 2020).
- Kumar, A.; Abbas, W.; Herbein, G. HIV-1 Latency in Monocytes/Macrophages. Viruses 2014, 6, 1837–1860. [Google Scholar] [CrossRef]
- Archin, N.M.; Sung, J.M.; Garrido, C.; Soriano-Sarabia, N.; Margolis, D.M. Eradicating HIV-1 Infection: Seeking to Clear a Persistent Pathogen. Nat. Rev. Microbiol. 2014, 12, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.M.; Perelson, A.S.; Li, J.Z. Predictions of Time to HIV Viral Rebound Following ART Suspension That Incorporate Personal Biomarkers. PLoS Comput. Biol. 2019, 15, e1007229. [Google Scholar] [CrossRef] [PubMed]
- Frange, P.; Faye, A.; Avettand-Fenoël, V.; Bellaton, E.; Descamps, D.; Angin, M.; David, A.; Caillat-Zucman, S.; Peytavin, G.; Dollfus, C.; et al. HIV-1 Virological Remission Lasting More than 12 Years after Interruption of Early Antiretroviral Therapy in a Perinatally Infected Teenager Enrolled in the French ANRS EPF-CO10 Paediatric Cohort: A Case Report. Lancet HIV 2016, 3, e49–e54. [Google Scholar] [CrossRef]
- Kumar, A.M.; Borodowsky, I.; Fernandez, B.; Gonzalez, L.; Kumar, M. Human Immunodeficiency Virus Type 1 RNA Levels in Different Regions of Human Brain: Quantification Using Real-Time Reverse Transcriptase-Polymerase Chain Reaction. J. Neurovirol. 2007, 13, 210–224. [Google Scholar] [CrossRef] [PubMed]
- Hatano, H.; Jain, V.; Hunt, P.W.; Lee, T.-H.; Sinclair, E.; Do, T.D.; Hoh, R.; Martin, J.N.; McCune, J.M.; Hecht, F.; et al. Cell-Based Measures of Viral Persistence Are Associated With Immune Activation and Programmed Cell Death Protein 1 (PD-1)–Expressing CD4+ T Cells. J. Infect. Dis. 2013, 208, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Zijlstra, A.; di Vizio, D. Size Matters in Nanoscale Communication. Nat. Cell Biol. 2018, 20, 228–230. [Google Scholar] [CrossRef]
- Kastelowitz, N.; Yin, H. Exosomes and Microvesicles: Identification and Targeting By Particle Size and Lipid Chemical Probes. Chembiochem Eur. J. Chem. Biol. 2014, 15, 923–928. [Google Scholar] [CrossRef]
- Kakizaki, M.; Yamamoto, Y.; Yabuta, S.; Kurosaki, N.; Kagawa, T.; Kotani, A. The Immunological Function of Extracellular Vesicles in Hepatitis B Virus-Infected Hepatocytes. PLoS ONE 2018, 13, e0205886. [Google Scholar] [CrossRef]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in Exosomes: Current Knowledge and the Way Forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.O.; DeMarino, C.; Pleet, M.L.; Cowen, M.; Branscome, H.; Al-Sharif, S.; Jones, J.; Dutartre, H.; Lepene, B.; Liotta, L.A.; et al. HTLV-1 Extracellular Vesicles Promote Cell-to-Cell Contact. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.A.; Sampey, G.C.; Lepene, B.; Akpamagbo, Y.; Barclay, R.A.; Iordanskiy, S.; Hakami, R.M.; Kashanchi, F. Presence of Viral RNA and Proteins in Exosomes from Cellular Clones Resistant to Rift Valley Fever Virus Infection. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Barclay, R.A.; Schwab, A.; de Marino, C.; Akpamagbo, Y.; Lepene, B.; Kassaye, S.; Iordanskiy, S.; Kashanchi, F. Exosomes from Uninfected Cells Activate Transcription of Latent HIV-1. J. Biol. Chem. 2017, 292, 11682–11701. [Google Scholar] [CrossRef]
- Pleet, M.L.; Mathiesen, A.; DeMarino, C.; Akpamagbo, Y.A.; Barclay, R.A.; Schwab, A.; Iordanskiy, S.; Sampey, G.C.; Lepene, B.; Nekhai, S.; et al. Ebola VP40 in Exosomes Can Cause Immune Cell Dysfunction. Front. Microbiol. 2016, 7, 1765. [Google Scholar] [CrossRef]
- Pinto, D.O.; Al-Sharif, S.; Mensah, G.; Cowen, M.; Khatkar, P.; Erickson, J.; Branscome, H.; Lattanze, T.; DeMarino, C.; Alem, F.; et al. Extracellular Vesicles from HTLV-1 Infected Cells Modulate Target Cells and Viral Spread. Retrovirology 2021, 18, 6. [Google Scholar] [CrossRef]
- Narayanan, A.; Iordanskiy, S.; Das, R.; van Duyne, R.; Santos, S.; Jaworski, E.; Guendel, I.; Sampey, G.; Dalby, E.; Iglesias-Ussel, M.; et al. Exosomes Derived from HIV-1-Infected Cells Contain Trans-Activation Response Element RNA. J. Biol. Chem. 2013, 288, 20014–20033. [Google Scholar] [CrossRef]
- Sampey, G.C.; Saifuddin, M.; Schwab, A.; Barclay, R.; Punya, S.; Chung, M.-C.; Hakami, R.M.; Asad-Zadeh, M.; Lepene, B.; Klase, Z.A.; et al. Exosomes from HIV-1-Infected Cells Stimulate Production of Pro-Inflammatory Cytokines through Trans-Activating Response (TAR) RNA. J. Biol. Chem. 2016, 291, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Akpamagbo, Y.A.; de Marino, C.; Pleet, M.L.; Schwab, A.; Rodriguez, M.; Barclay, R.A.; Sampey, G.; Iordanskiy, S.; El-Hage, N.; Kashanchi, F. HIV-1 Transcription Inhibitors Increase the Synthesis of Viral Non-Coding RNA That Contribute to Latency. Curr. Pharm. Des. 2017, 23, 4133–4144. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Margolis, L.; Sadovsky, Y. The Biology of Extracellular Vesicles: The Known Unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef]
- Wang, J.; Reuschel, E.L.; Shackelford, J.M.; Jeang, L.; Shivers, D.K.; Diehl, J.A.; Yu, X.-F.; Finkel, T.H. HIV-1 Vif Promotes the G1- to S-Phase Cell-Cycle Transition. Blood 2011, 117, 1260–1269. [Google Scholar] [CrossRef]
- Re, F.; Braaten, D.; Franke, E.K.; Luban, J. Human Immunodeficiency Virus Type 1 Vpr Arrests the Cell Cycle in G2 by Inhibiting the Activation of P34cdc2-Cyclin B. J. Virol. 1995, 69, 6859–6864. [Google Scholar] [CrossRef]
- Coller, H.A.; Sang, L.; Roberts, J.M. A New Description of Cellular Quiescence. PLoS Biol. 2006, 4, e83. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, S.I.; Doherty, A.; Powell, M.D.; Obialo, C.; Huang, M.B.; Quarshie, A.; Mitchell, C.; Bashir, K.; Newman, G.W. Detection of HIV-1 and Human Proteins in Urinary Extracellular Vesicles from HIV+ Patients. Adv. Virol. 2018, 2018, 7863412. [Google Scholar] [CrossRef] [PubMed]
- Barclay, R.A.; Khatkar, P.; Mensah, G.; de Marino, C.; Chu, J.S.C.; Lepene, B.; Zhou, W.; Gillevet, P.; Torkzaban, B.; Khalili, K.; et al. An Omics Approach to Extracellular Vesicles from HIV-1 Infected Cells. Cells 2019, 8, 787. [Google Scholar] [CrossRef] [PubMed]
- Dreux, M.; Garaigorta, U.; Boyd, B.; Décembre, E.; Chung, J.; Whitten-Bauer, C.; Wieland, S.; Chisari, F.V. Short-Range Exosomal Transfer of Viral RNA from Infected Cells to Plasmacytoid Dendritic Cells Triggers Innate Immunity. Cell Host Microbe 2012, 12, 558–570. [Google Scholar] [CrossRef] [PubMed]
- DeMarino, C.; Pleet, M.L.; Cowen, M.; Barclay, R.A.; Akpamagbo, Y.; Erickson, J.; Ndembi, N.; Charurat, M.; Jumare, J.; Bwala, S.; et al. Antiretroviral Drugs Alter the Content of Extracellular Vesicles from HIV-1-Infected Cells. Sci. Rep. 2018, 8, 7653. [Google Scholar] [CrossRef]
- Sampey, G.C.; Meyering, S.S.; Asad-Zadeh, M.; Saifuddin, M.; Hakami, R.M.; Kashanchi, F. Exosomes and Their Role in CNS Viral Infections. J. Neurovirol. 2014, 20, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, E.; Narayanan, A.; van Duyne, R.; Shabbeer-Meyering, S.; Iordanskiy, S.; Saifuddin, M.; Das, R.; Afonso, P.V.; Sampey, G.C.; Chung, M.; et al. Human T-Lymphotropic Virus Type 1-Infected Cells Secrete Exosomes That Contain Tax Protein. J. Biol. Chem. 2014, 289, 22284–22305. [Google Scholar] [CrossRef] [PubMed]
- Pleet, M.L.; Erickson, J.; de Marino, C.; Barclay, R.A.; Cowen, M.; Lepene, B.; Liang, J.; Kuhn, J.H.; Prugar, L.; Stonier, S.W.; et al. Ebola Virus VP40 Modulates Cell Cycle and Biogenesis of Extracellular Vesicles. J. Infect. Dis. 2018, 218, S365–S387. [Google Scholar] [CrossRef] [PubMed]
- Shafagati, N.; Patanarut, A.; Luchini, A.; Lundberg, L.; Bailey, C.; Petricoin, E., III; Liotta, L.; Narayanan, A.; Lepene, B.; Kehn Hall, K. The Use of Nanotrap Particles for Biodefense and Emerging Infectious Disease Diagnostics. Pathog. Dis. 2014, 71, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Ojha, C.; Lapierre, J.; Rodriguez, M.; Dever, S.; Zadeh, M.; de Marino, C.; Pleet, M.; Kashanchi, F.; El-Hage, N. Interplay between Autophagy, Exosomes and HIV-1 Associated Neurological Disorders: New Insights for Diagnosis and Therapeutic Applications. Viruses 2017, 9, 176. [Google Scholar] [CrossRef]
- Yeung, M.L.; Bennasser, Y.; Watashi, K.; Le, S.-Y.; Houzet, L.; Jeang, K.-T. Pyrosequencing of Small Non-Coding RNAs in HIV-1 Infected Cells: Evidence for the Processing of a Viral-Cellular Double-Stranded RNA Hybrid. Nucleic Acids Res. 2009, 37, 6575–6586. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, D.L.; Plante, I.; Landry, P.; Barat, C.; Janelle, M.-E.; Flamand, L.; Tremblay, M.J.; Provost, P. Identification of Functional MicroRNAs Released through Asymmetrical Processing of HIV-1 TAR Element. Nucleic Acids Res. 2008, 36, 2353–2365. [Google Scholar] [CrossRef]
- DeMarino, C.; Cowen, M.; Pleet, M.L.; Pinto, D.O.; Khatkar, P.; Erickson, J.; Docken, S.S.; Russell, N.; Reichmuth, B.; Phan, T.; et al. Differences in Transcriptional Dynamics Between T-Cells and Macrophages as Determined by a Three-State Mathematical Model. Sci. Rep. 2020, 10, 2227. [Google Scholar] [CrossRef]
- Pinto, D.O.; Scott, T.A.; DeMarino, C.; Pleet, M.L.; Vo, T.T.; Saifuddin, M.; Kovalskyy, D.; Erickson, J.; Cowen, M.; Barclay, R.A.; et al. Effect of Transcription Inhibition and Generation of Suppressive Viral Non-Coding RNAs. Retrovirology 2019, 16, 13. [Google Scholar] [CrossRef]
- Coffin, J.M. The Discovery of HTLV-1, the First Pathogenic Human Retrovirus. Proc. Natl. Acad. Sci. USA 2015, 112, 15525–15529. [Google Scholar] [CrossRef]
- Kehn, K.; Deng, L.; Wu, K.; Maddukuri, A.; Baylor, S.; Rufner, R.; Pumfery, A.; Bottazzi, M.E.; Kashanchi, F. The Role of Cyclin D2 and P21/Waf1 in Human T-Cell Leukemia Virus Type 1 Infected Cells. Retrovirology 2004, 17, 1–7. [Google Scholar]
- Kehn, K.; Fuente, C.; Strouss, K.; Berro, R.; Jiang, H.; Brady, J.; Mahieux, R.; Pumfery, A.; Bottazzi, M.E.; Kashanchi, F. The HTLV-I Tax Oncoprotein Targets the Retinoblastoma Protein for Proteasomal Degradation. Oncogene 2005, 24, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Thali, M. The Roles of Tetraspanins in HIV-1 Replication. In HIV Interactions with Host Cell Proteins; Spearman, P., Freed, E.O., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2009; Volume 339, pp. 85–102. ISBN 978-3-642-02174-9. [Google Scholar]
- Suárez, H.; Rocha-Perugini, V.; Álvarez, S.; Yáñez-Mó, M. Tetraspanins, Another Piece in the HIV-1 Replication Puzzle. Front. Immunol. 2018, 9, 1811. [Google Scholar] [CrossRef] [PubMed]
- Böker, K.O.; Lemus-Diaz, N.; Rinaldi-Ferreira, R.; Schiller, L.; Schneider, S.; Gruber, J. The Impact of the CD9 Tetraspanin on Lentivirus Infectivity and Exosome Secretion. Mol. Ther. 2018, 26, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Grigorov, B.; Attuil-Audenis, V.; Perugi, F.; Nedelec, M.; Watson, S.; Pique, C.; Darlix, J.-L.; Conjeaud, H.; Muriaux, D. A Role for CD81 on the Late Steps of HIV-1 Replication in a Chronically Infected T Cell Line. Retrovirology 2009, 6, 28. [Google Scholar] [CrossRef]
- Perez-Hernandez, D. The Intracellular Interactome of Tetraspanin-Enriched Microdomains Reveals Their Function as Sorting Machineries toward Exosomes. J. Biol. Chem. 2013, 288, 13. [Google Scholar] [CrossRef]
- Leidal, A.M.; Huang, H.H.; Marsh, T.; Solvik, T.; Zhang, D.; Ye, J.; Kai, F.; Goldsmith, J.; Liu, J.Y.; Huang, Y.-H.; et al. The LC3-Conjugation Machinery Specifies the Loading of RNA-Binding Proteins into Extracellular Vesicles. Nat. Cell Biol. 2020, 22, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Dubois, N.; Khoo, K.K.; Ghossein, S.; Seissler, T.; Wolff, P.; McKinstry, W.J.; Mak, J.; Paillart, J.-C.; Marquet, R.; Bernacchi, S. The C-Terminal P6 Domain of the HIV-1 Pr55 Gag Precursor Is Required for Specific Binding to the Genomic RNA. RNA Biol. 2018, 15, 923–936. [Google Scholar] [CrossRef]
- Campbell, J.H.; Hearps, A.C.; Martin, G.E.; Williams, K.C.; Crowe, S.M. The Importance of Monocytes and Macrophages in HIV Pathogenesis, Treatment, and Cure. AIDS 2014, 28, 2175–2187. [Google Scholar] [CrossRef]
- Nolte-‘t Hoen, E.; Cremer, T.; Gallo, R.C.; Margolis, L.B. Extracellular Vesicles and Viruses: Are They Close Relatives? Proc. Natl. Acad. Sci. USA 2016, 113, 9155. [Google Scholar] [CrossRef]
- Rezaie, J.; Aslan, C.; Ahmadi, M.; Zolbanin, N.M.; Kashanchi, F.; Jafari, R. The Versatile Role of Exosomes in Human Retroviral Infections: From Immunopathogenesis to Clinical Application. Cell Biosci. 2021, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Jaworski, E.; van Duyne, R.; Iordanskiy, S.; Guendel, I.; Das, R.; Currer, R.; Sampey, G.; Chung, M.; Kehn-Hall, K.; et al. Exosomes Derived from HTLV-1 Infected Cells Contain the Viral Protein Tax. Retrovirology 2014, 11, O46. [Google Scholar] [CrossRef]
- Mazurov, D.; Heidecker, G.; Derse, D. The Inner Loop of Tetraspanins CD82 and CD81 Mediates Interactions with Human T Cell Lymphotrophic Virus Type 1 Gag Protein. J. Biol. Chem. 2006, 282, 3896–3903. [Google Scholar] [CrossRef]
- Ren, T.; Takahashi, Y.; Liu, X.; Loughran, T.P.; Sun, S.-C.; Wang, H.-G.; Cheng, H. HTLV-1 Tax Deregulates Autophagy by Recruiting Autophagic Molecules into Lipid Raft Microdomains. Oncogene 2015, 34, 334–345. [Google Scholar] [CrossRef]
- Al Sharif, S.; Pinto, D.O.; Mensah, G.A.; Dehbandi, F.; Khatkar, P.; Kim, Y.; Branscome, H.; Kashanchi, F. Extracellular Vesicles in HTLV-1 Communication: The Story of an Invisible Messenger. Viruses 2020, 12, 1422. [Google Scholar] [CrossRef] [PubMed]
- Azran, I.; Schavinsky-Khrapunsky, Y.; Aboud, M. Role of Tax Protein in Human T-Cell Leukemia Virus Type-I Leukemogenicity. Retrovirology 2004, 1, 20. [Google Scholar] [CrossRef] [PubMed]
- Gross, C.; Thoma-Kress, A.K. Molecular Mechanisms of HTLV-1 Cell-to-Cell Transmission. Viruses 2016, 8, 74. [Google Scholar] [CrossRef]
- Nicot, C. HTLV-I Tax-Mediated Inactivation of Cell Cycle Checkpoints and DNA Repair Pathways Contribute to Cellular Transformation: “A Random Mutagenesis Model”. J. Cancer Sci. 2015, 2. [Google Scholar] [CrossRef]
- Tsubata, C.; Higuchi, M.; Takahashi, M.; Oie, M.; Tanaka, Y.; Gejyo, F.; Fujii, M. PDZ Domain-Binding Motif of Human T-Cell Leukemia Virus Type 1 Tax Oncoprotein Is Essential for the Interleukin 2 Independent Growth Induction of a T-Cell Line. Retrovirology 2005, 2, 46. [Google Scholar] [CrossRef][Green Version]
- Kulkarni, G.V.; McCulloch, C.A. Serum Deprivation Induces Apoptotic Cell Death in a Subset of Balb/c 3T3 Fibroblasts. J. Cell Sci. 1994, 107, 1169. [Google Scholar]
- Higuchi, A.; Shimmura, S.; Takeuchi, T.; Suematsu, M.; Tsubota, K. Elucidation of Apoptosis Induced by Serum Deprivation in Cultured Conjunctival Epithelial Cells. Br. J. Ophthalmol. 2006, 90, 760–764. [Google Scholar] [CrossRef]
- Carloni, M.; Meschini, R.; Ovidi, L.; Palitti, F. PHA-Induced Cell Proliferation Rescues Human Peripheral Blood Lymphocytes from X-Ray-Induced Apoptosis. Mutagenesis 2001, 16, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, H.-M.; Hieronymus, T.; Grünke, M.; Manger, B.; Kalden, J.R. Differential Role for IL-2 and IL-15 in the Inhibition of Apoptosis in Short Term Activated Human Lymphocytes. Scand. J. Immunol. 1997, 45, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Kedzierska, K.; Crowe, S.M. Cytokines and HIV-1: Interactions and Clinical Implications. Antivir. Chem. Chemother. 2001, 12, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Breen, E.C. Pro- and Anti-Inflammatory Cytokines in Human Immunodeficiency Virus Infection and Acquired Immunodeficiency Syndrome. Pharmacol. Ther. 2002, 95, 295–304. [Google Scholar] [CrossRef]
- Mori, N.; Murakami, S.; Oda, S.; Prager, D.; Eto, S. Production of Interleukin 8 in Adult T-Cell Leukemia Cells: Possible Transactivation of the Interleukin 8 Gene by Human T-Cell Leukemia Virus Type I tax. Cancer Res. 1995, 55, 3592. [Google Scholar]
- Zargari, R.; Mahdifar, M.; Mohammadi, A.; Vahidi, Z.; Hassanshahi, G.; Rafatpanah, H. The Role of Chemokines in the Pathogenesis of HTLV-1. Front. Microbiol. 2020, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.O.; DeMarino, C.; Vo, T.T.; Cowen, M.; Kim, Y.; Pleet, M.L.; Barclay, R.A.; Noren-Hooten, N.; Evans, M.K.; Heredia, A.; et al. Low-Level Ionizing Radiation Induces Selective Killing of HIV-1-Infected Cells with Reversal of Cytokine Induction Using MTOR Inhibitors. Viruses 2020, 12, 885. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Mensah, G.A.; Al Sharif, S.; Pinto, D.O.; Branscome, H.; Yelamanchili, S.V.; Cowen, M.; Erickson, J.; Khatkar, P.; Mahieux, R.; et al. Extracellular Vesicles from Infected Cells Are Released Prior to Virion Release. Cells 2021, 10, 781. https://doi.org/10.3390/cells10040781
Kim Y, Mensah GA, Al Sharif S, Pinto DO, Branscome H, Yelamanchili SV, Cowen M, Erickson J, Khatkar P, Mahieux R, et al. Extracellular Vesicles from Infected Cells Are Released Prior to Virion Release. Cells. 2021; 10(4):781. https://doi.org/10.3390/cells10040781
Chicago/Turabian StyleKim, Yuriy, Gifty A. Mensah, Sarah Al Sharif, Daniel O. Pinto, Heather Branscome, Sowmya V. Yelamanchili, Maria Cowen, James Erickson, Pooja Khatkar, Renaud Mahieux, and et al. 2021. "Extracellular Vesicles from Infected Cells Are Released Prior to Virion Release" Cells 10, no. 4: 781. https://doi.org/10.3390/cells10040781
APA StyleKim, Y., Mensah, G. A., Al Sharif, S., Pinto, D. O., Branscome, H., Yelamanchili, S. V., Cowen, M., Erickson, J., Khatkar, P., Mahieux, R., & Kashanchi, F. (2021). Extracellular Vesicles from Infected Cells Are Released Prior to Virion Release. Cells, 10(4), 781. https://doi.org/10.3390/cells10040781







