Highly Species-Specific Foliar Metabolomes of Diverse Woody Species and Relationships with the Leaf Economics Spectrum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Species
2.2. Experimental Design and Plant Sampling
2.3. Chemical Analyses of Leaves
2.4. Data Processing and Statistical Analyses
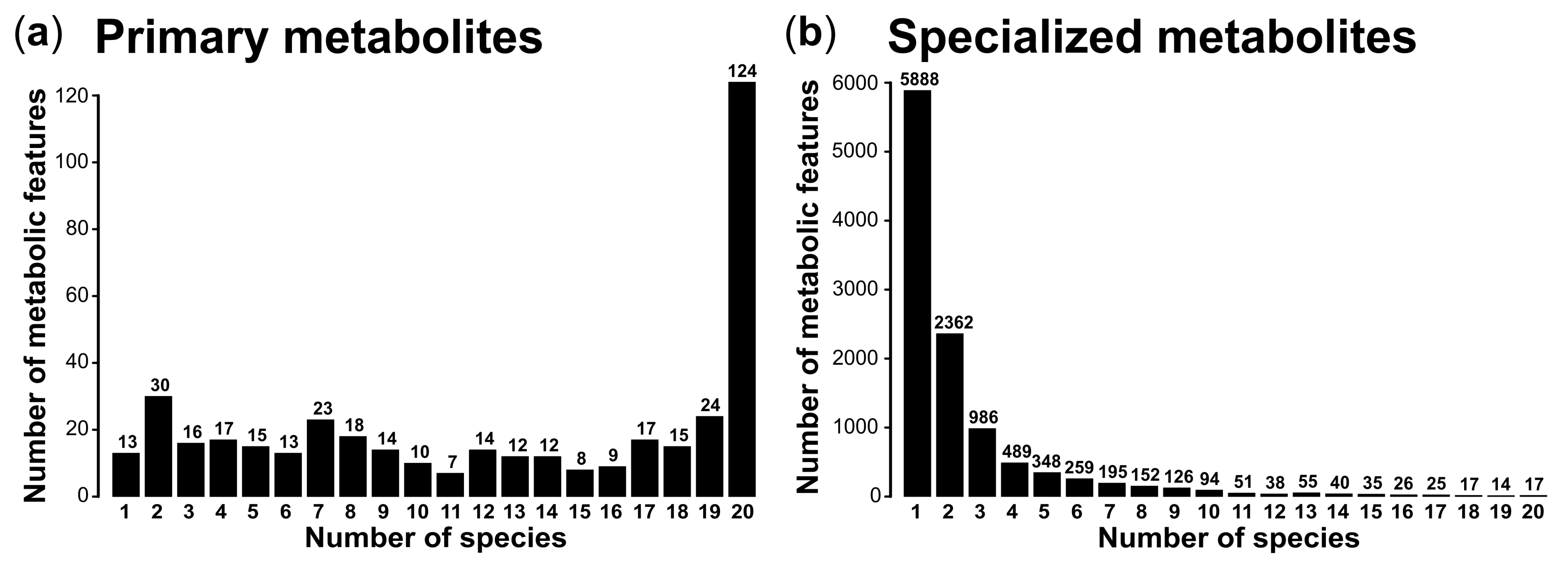
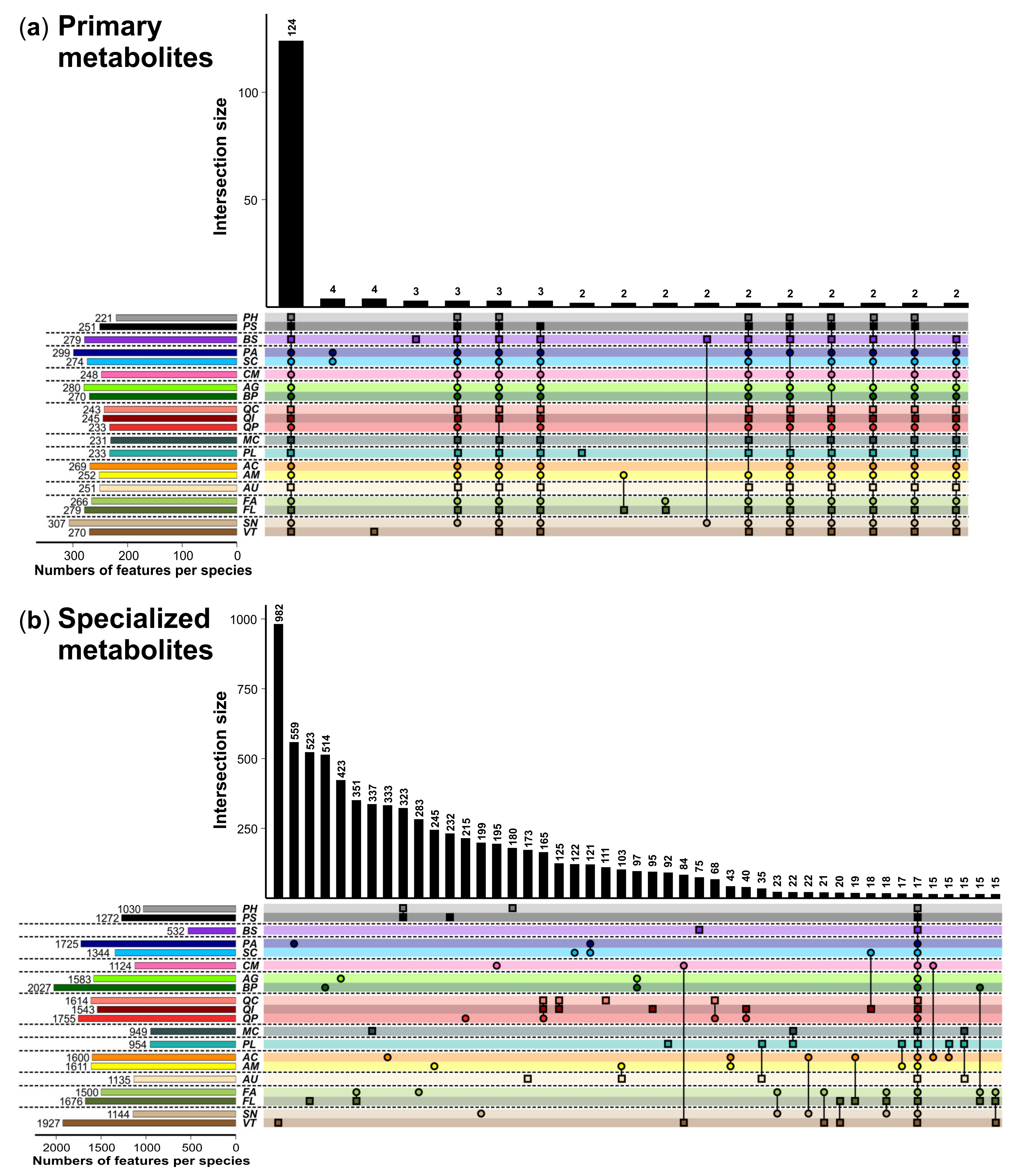
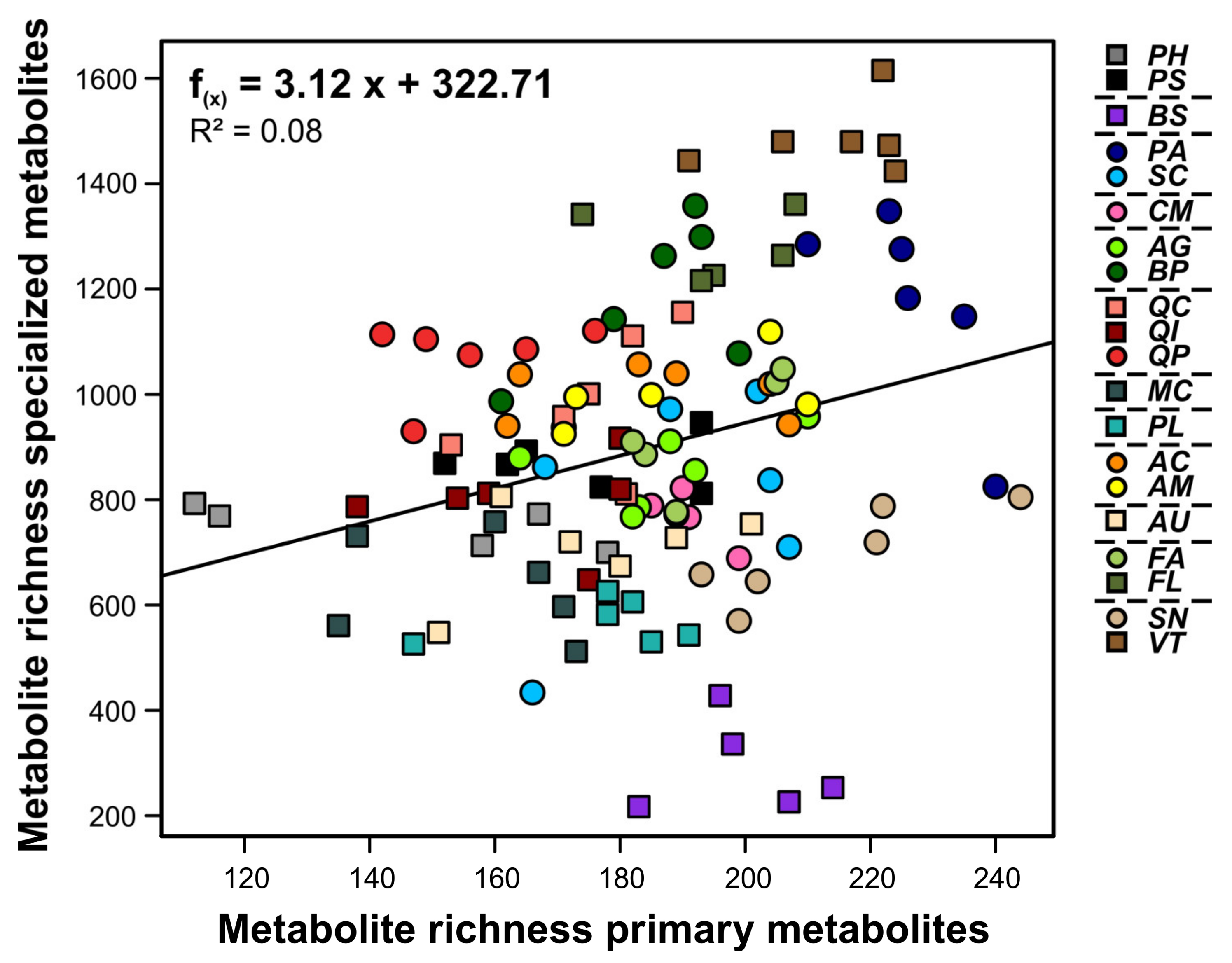
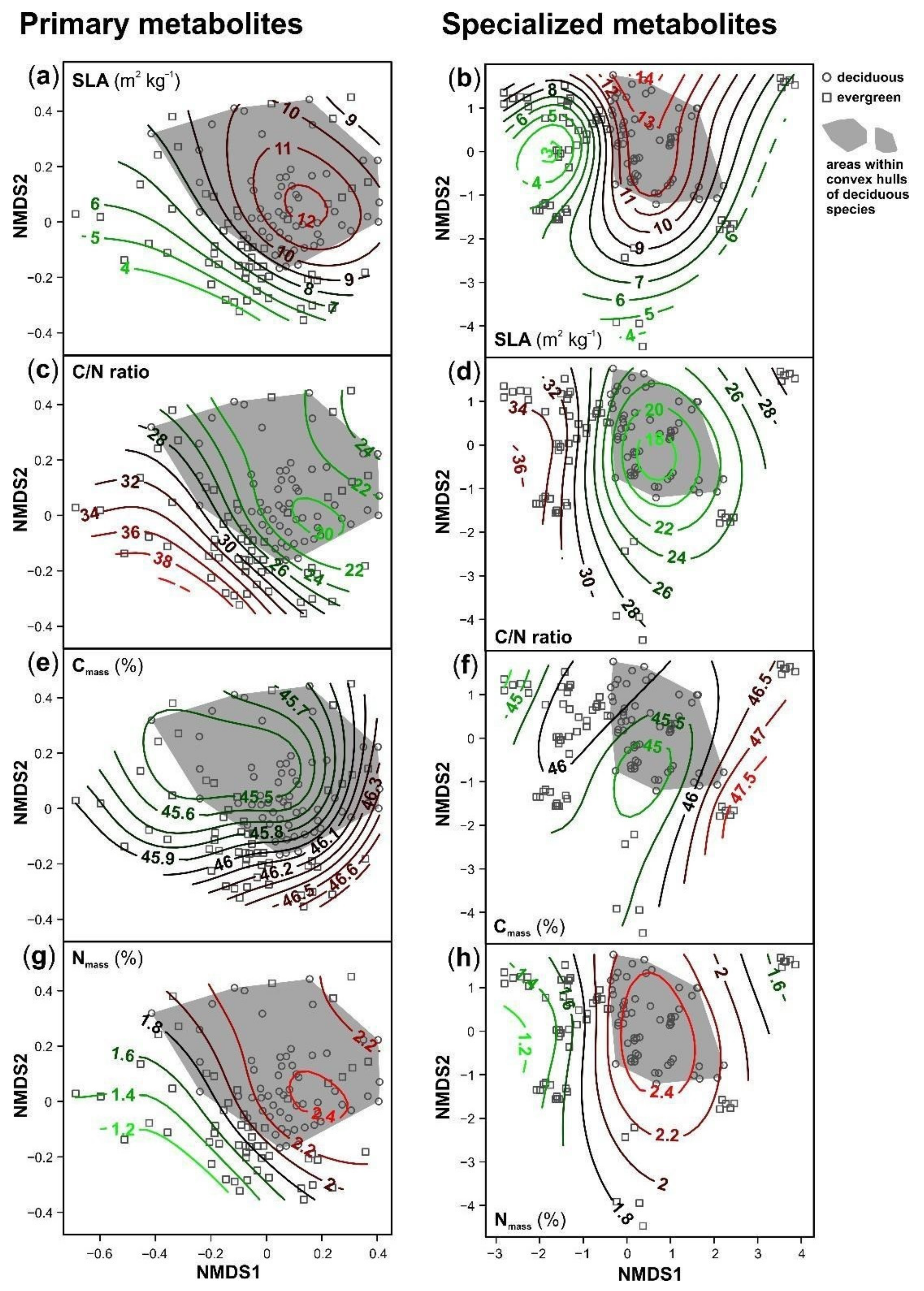
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Afendi, F.M.; Okada, T.; Yamazaki, M.; Hirai-Morita, A.; Nakamura, Y.; Nakamura, K.; Ikeda, S.; Takahashi, H.; Altaf-Ul-Amin, M.; Darusman, L.K.; et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012, 53, e1. [Google Scholar] [CrossRef]
- Pichersky, E.; Lewinsohn, E. Convergent evolution in plant specialized metabolism. Ann. Rev. Plant Biol. 2011, 62, 549–566. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, T. From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry 2007, 68, 2831–2846. [Google Scholar] [CrossRef] [PubMed]
- Kessler, A.; Kalske, A. Plant secondary metabolite diversity and species interactions. Ann. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- Rutz, A.; Dounoue-Kubo, M.; Ollivier, S.; Bisson, J.; Bagheri, M.; Saesong, T.; Ebrahimi, S.N.; Ingkaninan, K.; Wolfender, J.-L.; Allard, P.-M. Taxonomically informed scoring enhances confidence in natural products annotation. Front. Plant Sci. 2019, 10, 1329. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.A. Evolutionary diversification of primary metabolism and its contribution to plant chemical diversity. Front. Plant Sci. 2019, 10, 881. [Google Scholar] [CrossRef] [PubMed]
- Shirai, K.; Hanada, K. Contribution of functional divergence through copy number variations to the inter-species and intra-species diversity in specialized metabolites. Front. Plant Sci. 2019, 10, 1567. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, R.; Baier, M.C.; Persicke, M.; Müller, C. High specificity in plant leaf metabolic responses to arbuscular mycorrhiza. Nat. Commun. 2014, 5, 3886. [Google Scholar] [CrossRef] [PubMed]
- Dietz, S.; Herz, K.; Döll, S.; Haider, S.; Jandt, U.; Bruelheide, H.; Scheel, D. Semi-polar root exudates in natural grassland communities. Ecol. Evol. 2019, 9, 5526–5541. [Google Scholar] [CrossRef]
- Futuyma, D.J.; Agrawal, A.A. Macroevolution and the biological diversity of plants and herbivores. Proc. Natl. Acad. Sci. USA 2009, 106, 18054–18061. [Google Scholar] [CrossRef]
- Kissling, W.D.; Schleuning, M. Multispecies interactions across trophic levels at macroscales: Retrospective and future directions. Ecography 2015, 38, 346–357. [Google Scholar] [CrossRef]
- Gargallo-Garriga, A.; Sardans, J.; Granda, V.; Llusià, J.; Peguero, G.; Asensio, D.; Ogaya, R.; Urbina, I.; Van Langenhove, L.; Verryckt, L.T.; et al. Different “metabolomic niches” of the highly diverse tree species of the French Guiana rainforests. Sci. Rep. 2020, 10, 6937. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Oh, D.-G.; Lee, S.; Kim, G.R.; Lee, J.S.; Son, Y.K.; Bae, C.-H.; Yeo, J.; Lee, C.H. Chemotaxonomic metabolite profiling of 62 indigenous plant species and its correlation with bioactivities. Molecules 2015, 20, 19719–19734. [Google Scholar] [CrossRef] [PubMed]
- Peters, K.; Worrich, A.; Weinhold, A.; Alka, O.; Balcke, G.; Birkemeyer, C.; Bruelheide, H.; Calf, O.W.; Dietz, S.; Dührkop, K.; et al. Current challenges in plant eco-metabolomics. Int. J. Mol. Sci. 2018, 19, 1385. [Google Scholar] [CrossRef] [PubMed]
- Sardans, J.; Gargallo-Garriga, A.; Urban, O.; Klem, K.; Walker, T.W.N.; Holub, P.; Janssens, I.A.; Peñuelas, J. Ecometabolomics for a better understanding of plant responses and acclimation to abiotic factors linked to global change. Metabolites 2020, 10, 239. [Google Scholar] [CrossRef] [PubMed]
- Violle, C.; Navas, M.-L.; Vile, D.; Kazakou, E.; Fortunel, C.; Hummel, I.; Garnier, E. Let the concept of trait be functional! Oikos 2007, 116, 882–892. [Google Scholar] [CrossRef]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef]
- Wright, I.J.; Reich, P.B.; Cornelissen, J.H.C.; Falster, D.S.; Garnier, E.; Hikosaka, K.; Lamont, B.B.; Lee, W.; Oleksyn, J.; Osada, N.; et al. Assessing the generality of global leaf trait relationships. New Phytol. 2005, 166, 485–496. [Google Scholar] [CrossRef]
- Chen, Q.; Lu, X.; Guo, X.; Guo, Q.; Li, D. Metabolomics characterization of two Apocynaceae plants, Catharanthus roseus and Vinca minor, using GC-MS and LC-MS methods in combination. Molecules 2017, 22, 997. [Google Scholar] [CrossRef]
- Gago, J.; de Menezes Daloso, D.; Figueroa, C.M.; Flexas, J.; Fernie, A.R.; Nikoloski, Z. Relationships of leaf net photosynthesis, stomatal conductance, and mesophyll conductance to primary metabolism: A multispecies meta-analysis approach. Plant Physiol. 2016, 171, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Herms, D.A.; Mattson, W.J. The dilemma of plants: To grow or defend. Q. Rev. Biol. 1992, 67, 283–335. [Google Scholar] [CrossRef]
- Sardans, J.; Llusia, J.; Owen, S.M.; Niinemets, Ü.; Peñuelas, J. Screening study of leaf terpene concentration of 75 Borneo rainforest plant species: Relationships with leaf elemental concentrations and morphology. Rec. Nat. Prod. 2015, 9, 19–40. [Google Scholar]
- Mason, C.M.; Bowsher, A.W.; Crowell, B.L.; Celoy, R.M.; Tsai, C.-J.; Donovan, L.A. Macroevolution of leaf defenses and secondary metabolites across the genus Helianthus. New Phytol. 2016, 209, 1720–1733. [Google Scholar] [CrossRef]
- De Oliveira, A.C.P.; Nunes, A.; Rodrigues, R.G.; Branquinho, C. The response of plant functional traits to aridity in a tropical dry forest. Sci. Total Environ. 2020, 747, 141177. [Google Scholar] [CrossRef]
- Doblas-Miranda, E.; Alonso, R.; Arnan, X.; Bermejo, V.; Brotons, L.; de las Heras, J.; Estiarte, M.; Hódar, J.A.; Llorens, P.; Lloret, F.; et al. A review of the combination among global change factors in forests, shrublands and pastures of the Mediterranean Region: Beyond drought effects. Glob. Planet. Chang. 2017, 148, 42–54. [Google Scholar] [CrossRef]
- Garzón, M.B.; Alía, R.; Robson, T.M.; Zavala, M.A. Intra-specific variability and plasticity influence potential tree species distributions under climate change. Glob. Ecol. Biogeogr. 2011, 20, 766–778. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Kováts, E. Gaschromatographische Charakterisierung organischer Verbindungen. 1. Retentionsindices aliphatischer Halogenide, Alkohole, Aldehyde und Ketone. Helv. Chim. Act. 1958, 41, 1915–1932. [Google Scholar] [CrossRef]
- Schweiger, R.; Padilla-Arizmendi, F.; Nogueira-López, G.; Rostás, M.; Lawry, R.; Brown, C.; Hampton, J.; Steyaert, J.M.; Müller, C.; Mendoza-Mendoza, A. Insights into metabolic changes caused by the Trichoderma virens-maize root interaction. Mol. Plant Microbe Interact. 2020, MPMI04200081R. [Google Scholar] [CrossRef] [PubMed]
- Ottensmann, M.; Stoffel, M.A.; Nichols, H.J.; Hoffman, J.I. GCalignR: An R package for aligning gas-chromatography data for ecological and evolutionary studies. PLoS ONE 2018, 13, e0198311. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 15 January 2021).
- Moore, B.D.; Andrew, R.L.; Külheim, C.; Foley, W.J. Explaining intraspecific diversity in plant secondary metabolites in an ecological context. New Phytol. 2014, 201, 733–750. [Google Scholar] [CrossRef]
- Wen, W.; Alseekh, S.; Fernie, A.R. Conservation and diversification of flavonoid metabolism in the plant kingdom. Curr. Opin. Plant Biol. 2020, 55, 100–108. [Google Scholar] [CrossRef]
- Boachon, B.; Buell, C.R.; Crisovan, E.; Dudareva, N.; Garcia, N.; Godden, G.; Henry, L.; Kamileen, M.O.; Kates, H.R.; Kilgore, M.B.; et al. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol. Plant 2018, 11, 1084–1096. [Google Scholar] [CrossRef]
- Weng, J.-K.; Philippe, R.N.; Noel, J.P. The rise of chemodiversity in plants. Science 2012, 336, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef]
- Austen, N.; Walker, H.J.; Lake, J.A.; Phoenix, G.K.; Cameron, D.D. The regulation of plant secondary metabolism in response to abiotic stress: Interactions between heat shock and elevated CO2. Front. Plant Sci. 2019, 10, 1463. [Google Scholar] [CrossRef] [PubMed]
- Schwachtje, J.; Fischer, A.; Erban, A.; Kopka, J. Primed primary metabolism in systemic leaves: A functional systems analysis. Sci. Rep. 2018, 8, 216. [Google Scholar] [CrossRef] [PubMed]
- Hilker, M. New synthesis: Parallels between biodiversity and chemodiversity. J. Chem. Ecol. 2014, 40, 225–226. [Google Scholar] [CrossRef]
- Weinhold, A.; Döll, S.; Liu, M.; Schedl, A.; Xu, X.; Neumann, S.; van Dam, N.M. Plot diversity differentially affects the chemical composition of leaves, roots and root exudates in four subtropical tree species. bioRxiv 2020, preprint. [Google Scholar] [CrossRef]
- Carrington, Y.; Guo, J.; Le, C.H.; Fillo, A.; Kwon, J.; Tran, L.T.; Ehlting, J. Evolution of a secondary metabolic pathway from primary metabolism: Shikimate and quinate biosynthesis in plants. Plant J. 2018, 95, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.B.; Walters, M.B.; Ellsworth, D.S. From tropics to tundra: Global convergence in plant functioning. Proc. Natl. Acad. Sci. USA 1997, 94, 13730–13734. [Google Scholar] [CrossRef] [PubMed]
- Donovan, L.A.; Maherali, H.; Caruso, C.M.; Huber, H.; de Kroon, H. The evolution of the worldwide leaf economics spectrum. Trends Ecol. Evol. 2011, 26, 88–95. [Google Scholar] [CrossRef]
- Pan, Y.; Cieraad, E.; Armstrong, J.; Armstrong, W.; Clarkson, B.R.; Colmer, T.D.; Pedersen, O.; Visser, E.J.W.; Voesenek, L.A.C.J.; van Bodegom, P.M. Global patterns of the leaf economics spectrum in wetlands. Nat. Commun. 2020, 11, 4519. [Google Scholar] [CrossRef]
- Damesin, C.; Rambal, S.; Joffre, R. Co-occurrence of trees with different leaf habit: A functional approach on Mediterranean oaks. Acta Oecol. 1998, 19, 195–204. [Google Scholar] [CrossRef]
- Mediavilla, S.; Escudero, A. Relative growth rate of leaf biomass and leaf nitrogen content in several mediterranean woody species. Plant Ecol. 2003, 168, 321–332. [Google Scholar] [CrossRef]
- Takashima, T.; Hikosaka, K.; Hirose, T. Photosynthesis or persistence: Nitrogen allocation in leaves of evergreen and deciduous Quercus species. Plant Cell Environ. 2004, 27, 1047–1054. [Google Scholar] [CrossRef]
- Palacio, S.; Camarero, J.J.; Maestro, M.; Alla, A.Q.; Lahoz, E.; Montserrat-Martí, G. Are storage and tree growth related? Seasonal nutrient and carbohydrate dynamics in evergreen and deciduous Mediterranean oaks. Trees 2018, 32, 777–790. [Google Scholar] [CrossRef]
- Miyazawa, S.-I.; Satomi, S.; Terashima, I. Slow leaf development of evergreen broad-leaved tree species in Japanese warm temperate forests. Ann. Bot. 1998, 82, 859–869. [Google Scholar] [CrossRef]
- Bryant, J.P.; Chapin, F.S., III; Klein, D.R. Carbon/nutrient balance of boreal plants in relation to vertebrate herbivory. Oikos 1983, 40, 357–368. [Google Scholar] [CrossRef]
- Coley, P.D.; Bryant, J.P.; Chapin, F.S., III. Resource availability and plant antiherbivore defense. Science 1985, 230, 895–899. [Google Scholar] [CrossRef]
- Moreira, X.; Pearse, I.S. Leaf habit does not determine the investment in both physical and chemical defences and pair-wise correlations between these defensive traits. Plant Biol. 2017, 19, 354–359. [Google Scholar] [CrossRef]
- Pringle, E.G.; Adams, R.I.; Broadbent, E.; Busby, P.E.; Donatti, C.I.; Kurten, E.L.; Renton, K.; Dirzo, R. Distinct leaf-trait syndromes of evergreen and deciduous trees in a seasonally dry tropical forest. Biotropica 2011, 43, 299–308. [Google Scholar] [CrossRef]
- Hamilton, J.G.; Zangerl, A.R.; DeLucia, E.H.; Berenbaum, M.R. The carbon-nutrient balance hypothesis: Its rise and fall. Ecol. Lett. 2001, 4, 86–95. [Google Scholar] [CrossRef]
- Villar, R.; Ruiz Robleto, J.; de Jong, Y.; Poorter, H. Differences in construction costs and chemical composition between deciduous and evergreen woody species are small as compared to differences among families. Plant Cell Environ. 2006, 29, 1629–1643. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Zhao, H.; Yu, S.; Yu, J.; Cai, K.; Sun, W.; Liu, X.; Li, X.; Yu, M.; Ali, S.; et al. Comparative transcriptome and metabolomic profiling reveal the complex mechanisms underlying the developmental dynamics of tobacco leaves. Genomics 2020, 112, 4009–4022. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Dörken, V.M.; Arnold, A.I.-M.; Adamczyk, B. Environmental conditions and species identity drive metabolite levels in green leaves and leaf litter of 14 temperate woody species. Forests 2018, 9, 775. [Google Scholar] [CrossRef]
- Ficken, C.D.; Wright, J.P. Nitrogen uptake and biomass resprouting show contrasting relationships with resource acquisitive and conservative plant traits. J. Veg. Sci. 2019, 30, 65–74. [Google Scholar] [CrossRef]
- De la Riva, E.G.; Villar, R.; Pérez-Ramos, I.M.; Quero, J.L.; Matías, L.; Poorter, L.; Marañón, T. Relationships between leaf mass per area and nutrient concentrations in 98 Mediterranean woody species are determined by phylogeny, habitat and leaf habit. Trees 2018, 32, 497–510. [Google Scholar] [CrossRef]
- Carmona, D.; Lajeunesse, M.J.; Johnson, M.T.J. Plant traits that predict resistance to herbivores. Funct. Ecol. 2011, 25, 358–367. [Google Scholar] [CrossRef]
- Haug, K.; Salek, R.M.; Conesa, P.; Hastings, J.; de Matos, P.; Rijnbeek, M.; Mahendraker, T.; Williams, M.; Neumann, S.; Rocca-Serra, P.; et al. MetaboLights—An open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013, 41, D781–D786. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O′Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed]

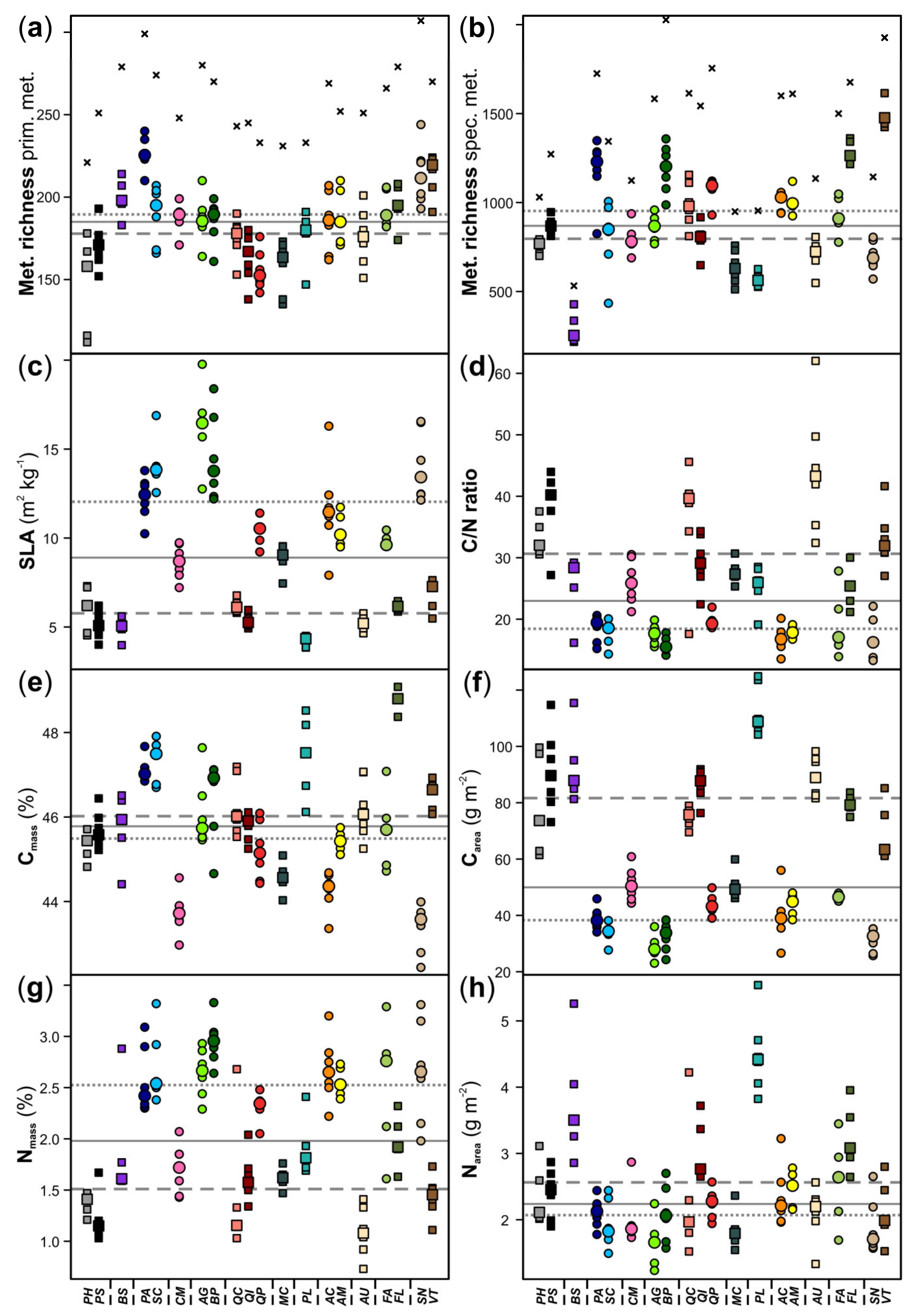
| Species | Abbr. | Class | Family | Leaf Habit 1 |
|---|---|---|---|---|
| Pinus halepensis | PH | Gymnospermae | Pinaceae | e |
| Pinus sylvestris | PS | e | ||
| Buxus sempervirens | BS | Angiospermae | Buxaceae | e |
| Populus alba | PA | Salicaceae | d | |
| Salix cinerea | SC | d | ||
| Crataegus monogyna | CM | Rosaceae | d | |
| Alnus glutinosa | AG | Betulaceae | d | |
| Betula pubescens | BP | d | ||
| Quercus coccifera | QC | Fagaceae | e | |
| Quercus ilex | QI | e | ||
| Quercus petraea | QP | d | ||
| Myrtus communis | MC | Myrtaceae | e | |
| Pistacia lentiscus | PL | Anacardiaceae | e | |
| Acer campestre | AC | Sapindaceae | d | |
| Acer monspessulanum | AM | d | ||
| Arbutus unedo | AU | Ericaceae | e | |
| Fraxinus angustifolia | FA | Oleaceae | d | |
| Phillyrea latifolia | FL | e | ||
| Sambucus nigra | SN | Adoxaceae | d | |
| Viburnum tinus | VT | e |
| NMDS1 | NMDS2 | |||||
|---|---|---|---|---|---|---|
| Estimate | Std. Error | p | Estimate | Std. Error | p | |
| Primary Metabolites | ||||||
| metabolite richness | 103.229 | 5.920 | <0.001 | −8.323 | 5.838 | 0.157 |
| SLA log (m2 kg−1) | 0.145 | 0.126 | 0.255 | 0.081 | 0.102 | 0.425 |
| Cmass (%) | 0.465 | 0.587 | 0.430 | 0.295 | 0.481 | 0.541 |
| Carea log (g m−2) | −0.136 | 0.130 | 0.298 | −0.074 | 0.104 | 0.480 |
| Nmass (%) | 0.789 | 0.297 | 0.009 | 0.598 | 0.250 | 0.018 |
| Narea log (g m−2) | 0.034 | 0.164 | 0.835 | 0.015 | 0.142 | 0.917 |
| C/N ratio log | −0.357 | 0.151 | 0.020 | −0.268 | 0.126 | 0.035 |
| Specialized Metabolites | ||||||
| metabolite richness | 173.650 | 25.300 | <0.001 | 130.090 | 22.310 | <0.001 |
| SLA log (m2 kg−1) | 0.029 | 0.046 | 0.527 | 0.015 | 0.038 | 0.703 |
| Cmass (%) | 0.048 | 0.176 | 0.788 | 0.388 | 0.161 | 0.019 |
| Carea log (g m−2) | −0.029 | 0.047 | 0.537 | −0.004 | 0.039 | 0.921 |
| Nmass (%) | 0.079 | 0.079 | 0.328 | 0.031 | 0.079 | 0.698 |
| Narea log (g m−2) | −0.017 | 0.032 | 0.602 | −0.076 | 0.035 | 0.037 |
| C/N ratio log | −0.038 | 0.041 | 0.369 | 0.005 | 0.041 | 0.901 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schweiger, R.; Castells, E.; Da Sois, L.; Martínez-Vilalta, J.; Müller, C. Highly Species-Specific Foliar Metabolomes of Diverse Woody Species and Relationships with the Leaf Economics Spectrum. Cells 2021, 10, 644. https://doi.org/10.3390/cells10030644
Schweiger R, Castells E, Da Sois L, Martínez-Vilalta J, Müller C. Highly Species-Specific Foliar Metabolomes of Diverse Woody Species and Relationships with the Leaf Economics Spectrum. Cells. 2021; 10(3):644. https://doi.org/10.3390/cells10030644
Chicago/Turabian StyleSchweiger, Rabea, Eva Castells, Luca Da Sois, Jordi Martínez-Vilalta, and Caroline Müller. 2021. "Highly Species-Specific Foliar Metabolomes of Diverse Woody Species and Relationships with the Leaf Economics Spectrum" Cells 10, no. 3: 644. https://doi.org/10.3390/cells10030644
APA StyleSchweiger, R., Castells, E., Da Sois, L., Martínez-Vilalta, J., & Müller, C. (2021). Highly Species-Specific Foliar Metabolomes of Diverse Woody Species and Relationships with the Leaf Economics Spectrum. Cells, 10(3), 644. https://doi.org/10.3390/cells10030644