Berberine Prevents Disease Progression of Nonalcoholic Steatohepatitis through Modulating Multiple Pathways
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Mice
2.3. RNA Sequencing (RNAseq) and Bioinformatic Analysis
2.4. Serum and Liver Biochemical Analysis
2.5. Histological Analysis and Immunohistochemical Staining
2.6. Bile Acid (BA) Analysis
2.7. Quantitative RT-PCR
2.8. Immunoblotting Analysis
2.9. Statistical Analysis
3. Results
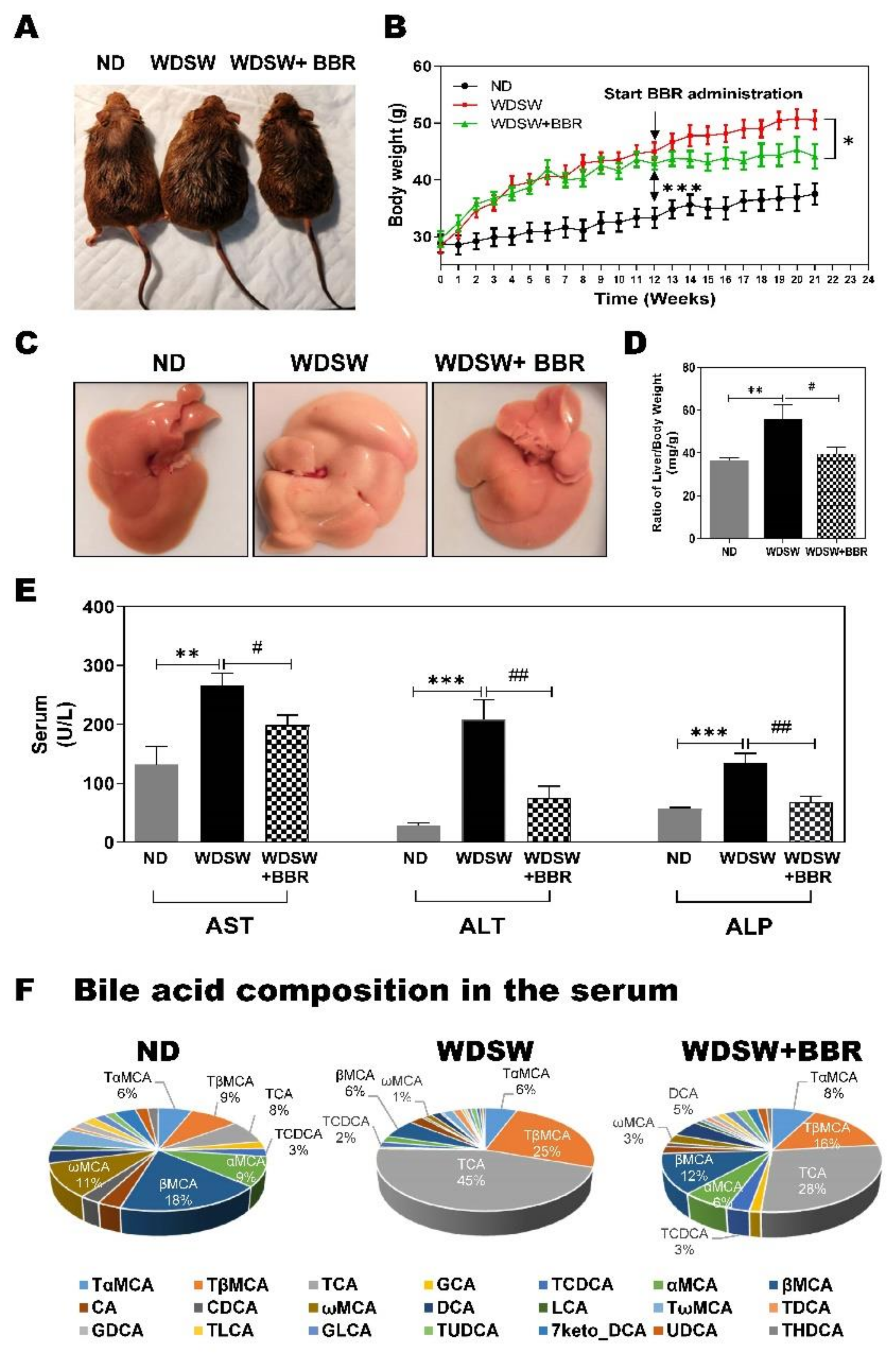
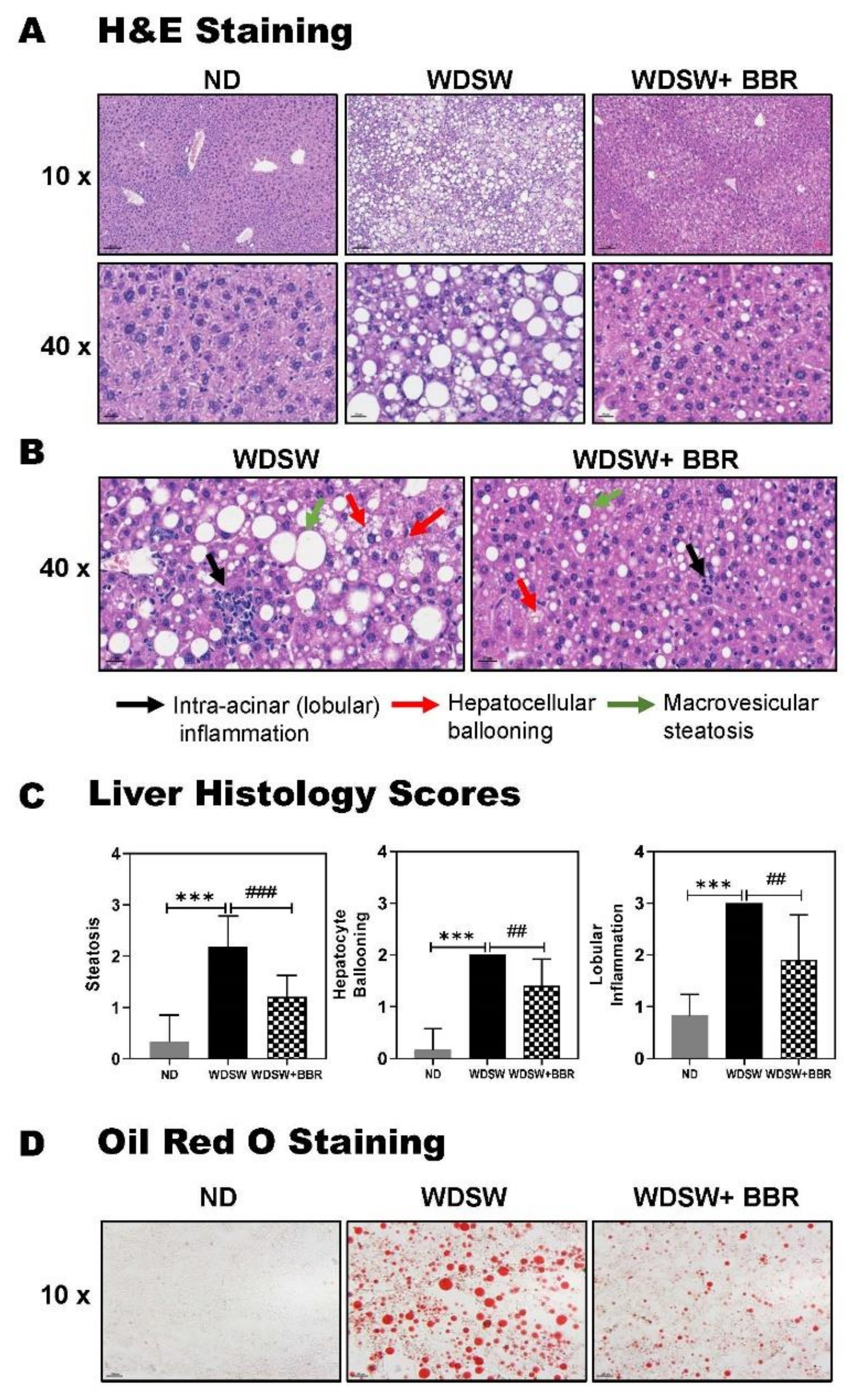
3.1. BBR Significantly Prevented NAFL to NASH Progression in WDSW-Fed Mice
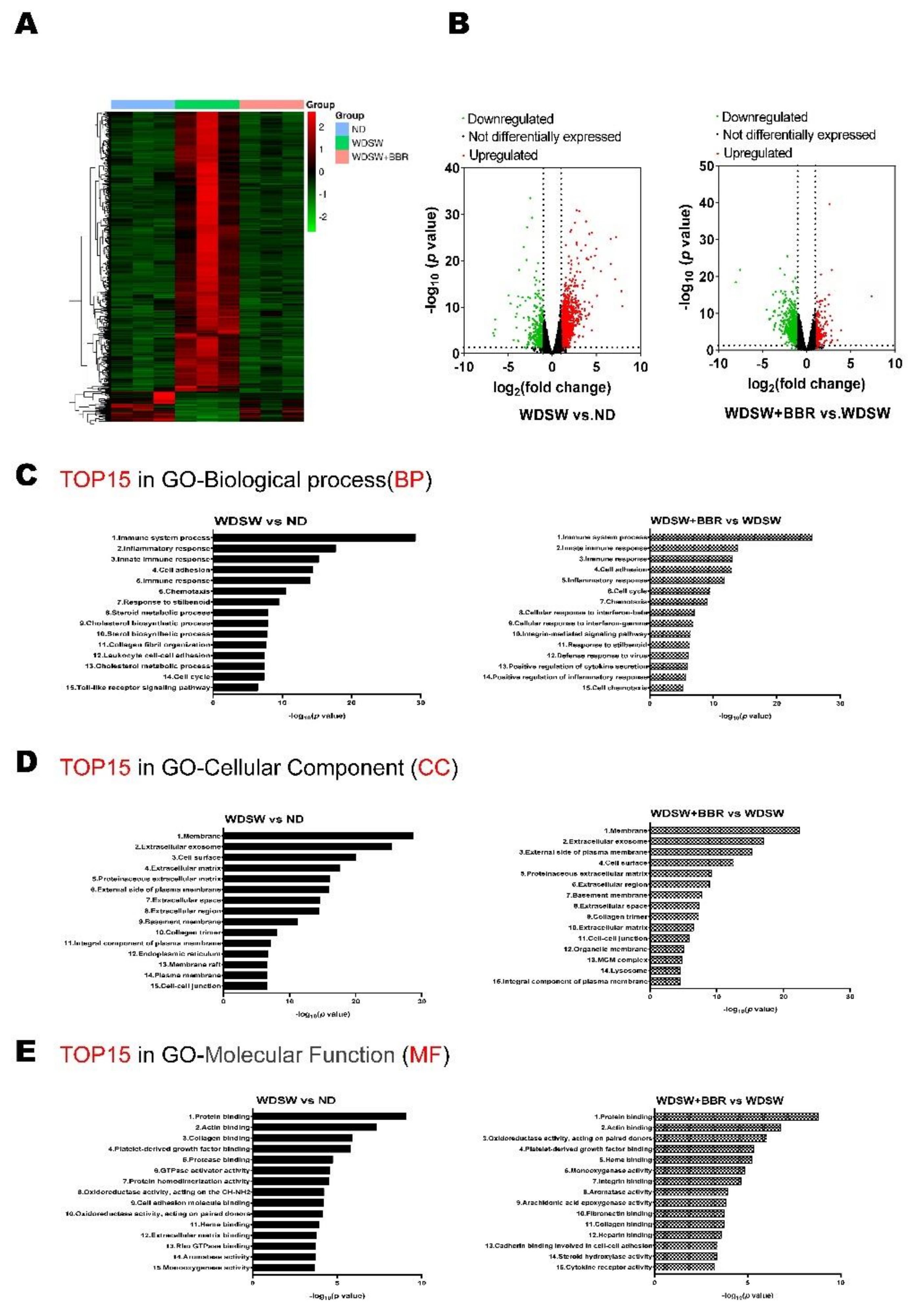
3.2. BBR Prevented NASH Progression by Modulating Global Transcriptomic Profile in WDSW-Fed Mice
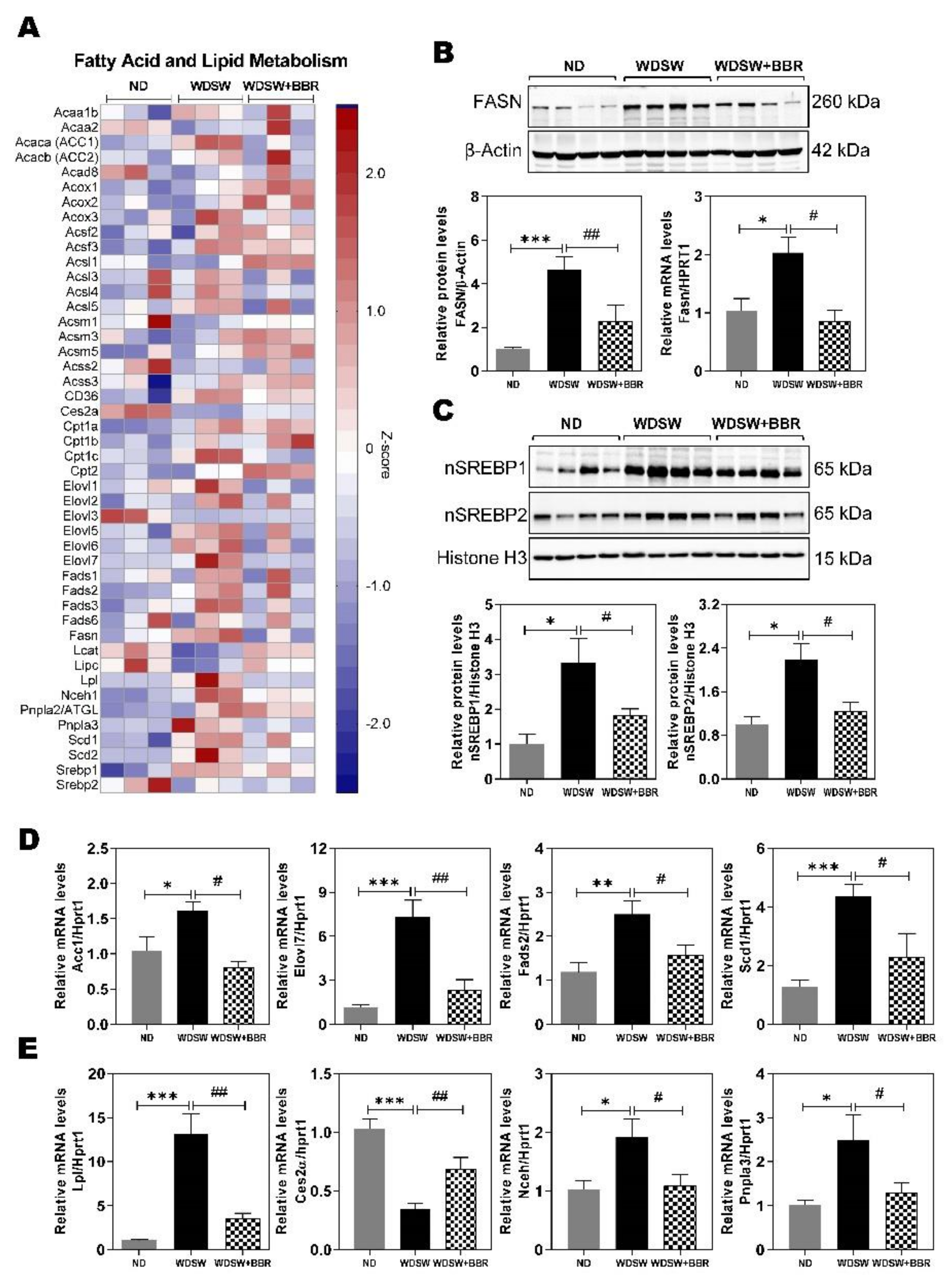
3.3. Effect of BBR on WDSW-Induced Dysregulation of Fatty Acid and Lipid Metabolism
3.4. Effect of BBR on WDSW-Induced Inflammation and Oxidative Stress
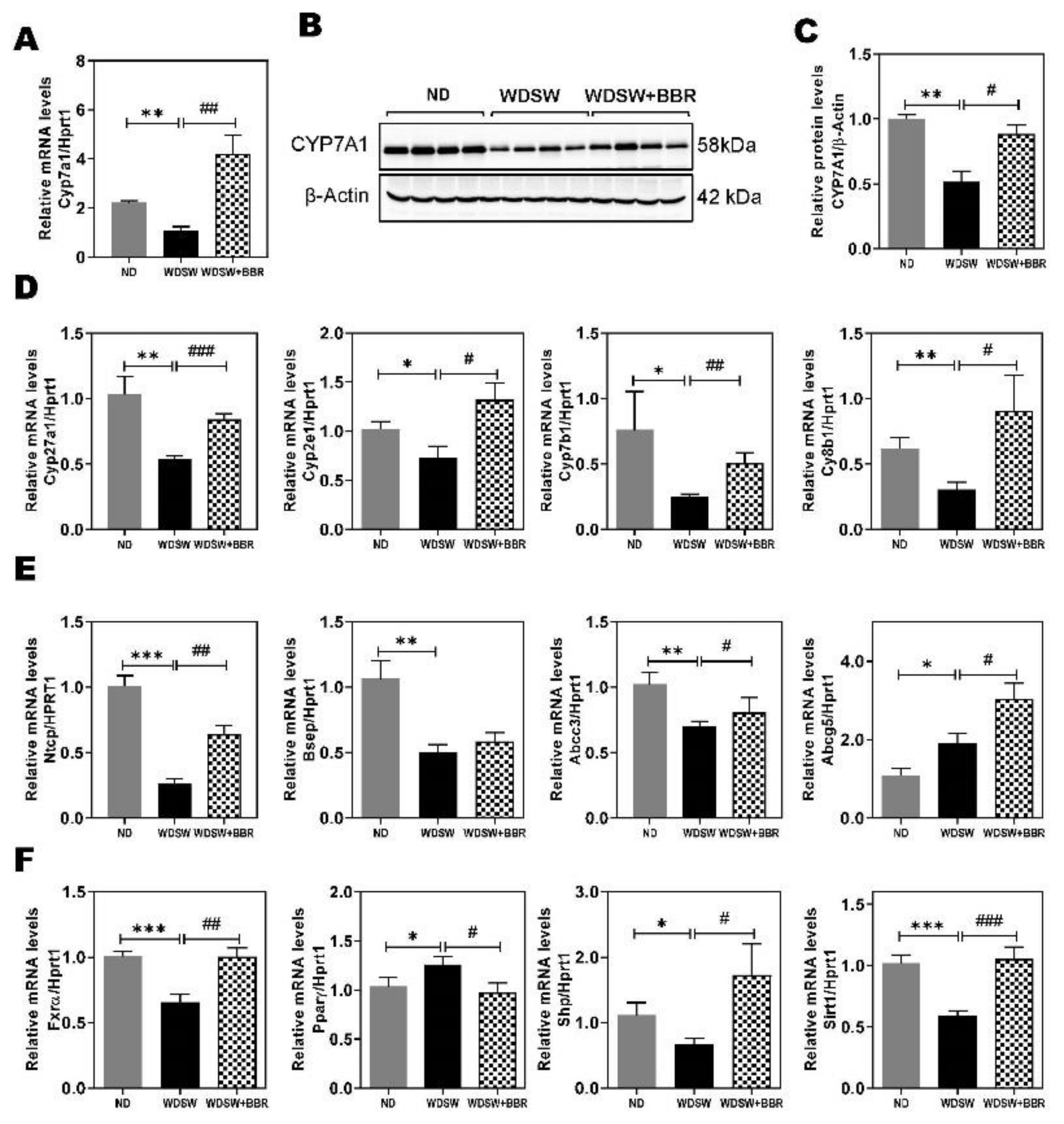
3.5. Effect of BBR on WDSW-Induced Dysregulation of Hepatic Bile Acid Metabolism
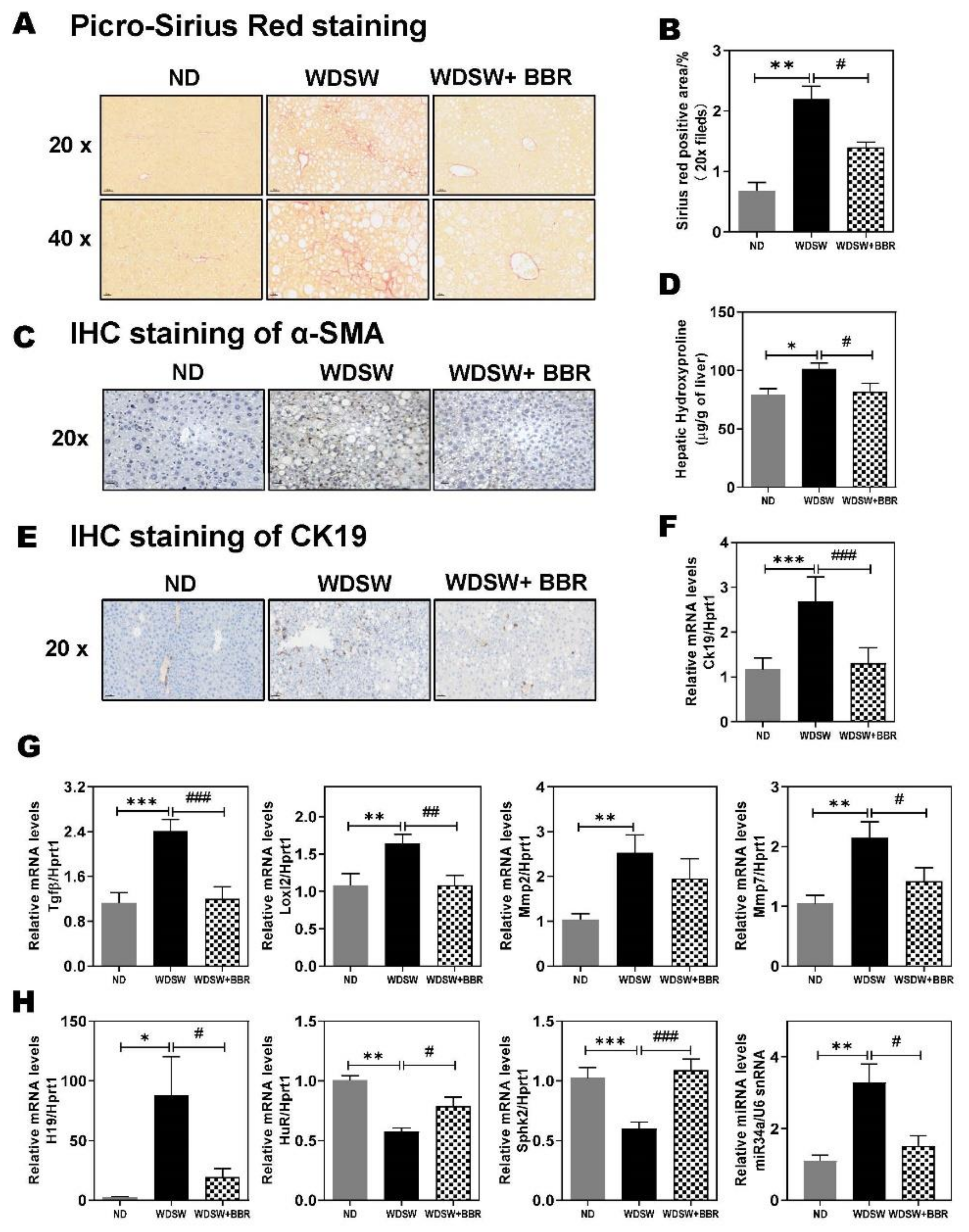
3.6. Effect of BBR on WDSW-Induced Hepatic Fibrosis
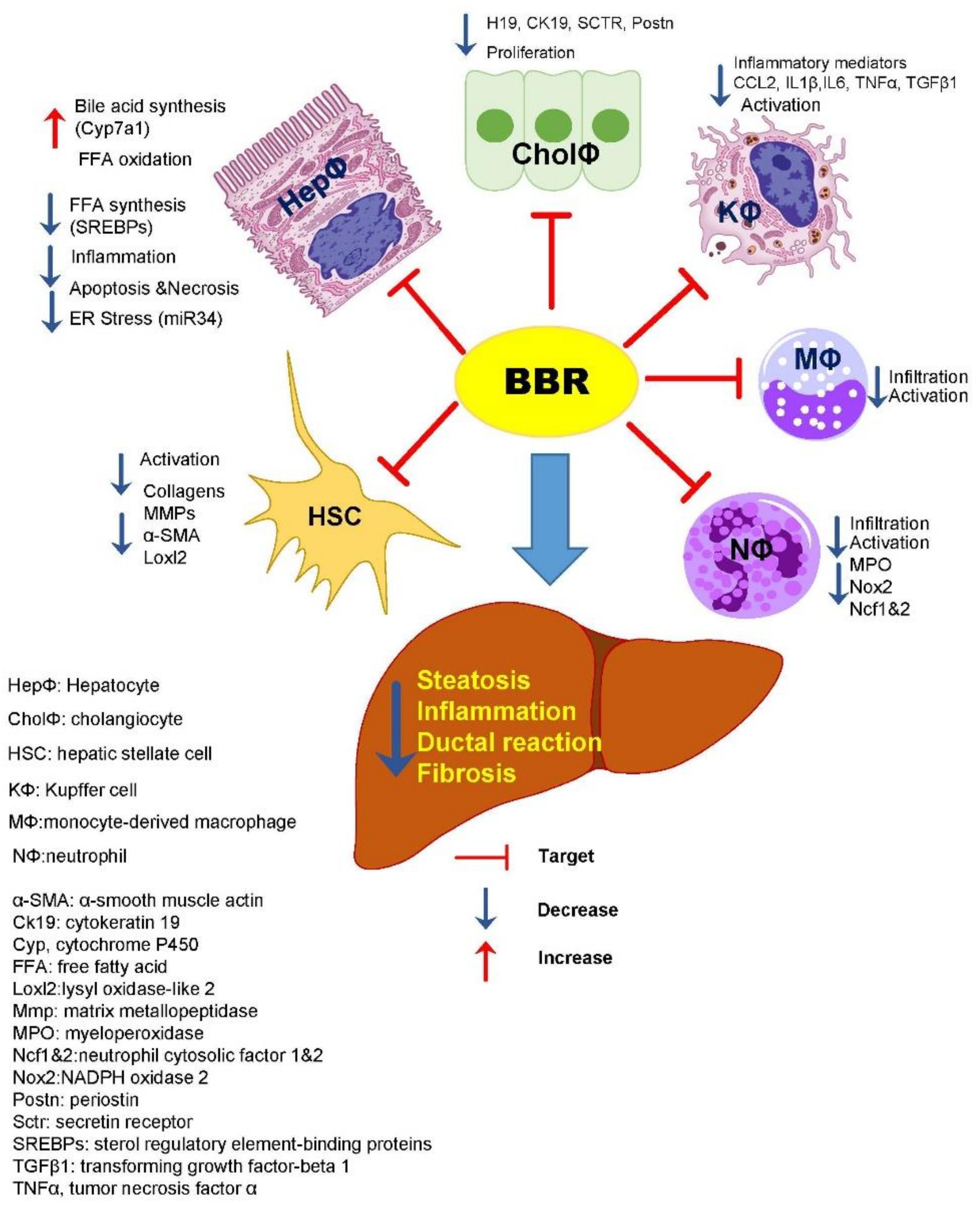
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Byrne, C.D.; Targher, G. NAFLD: A multisystem disease. J. Hepatol. 2015, 62, S47–S64. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Adolph, T.E.; Moschen, A.R. Multiple Parallel Hits Hypothesis in NAFLD – Revisited After a Decade. Hepatology 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-Z.; Li, X.-Y.; Liu, J. Recent pharmacological studies on natural products in China. Eur. J. Pharmacol. 2004, 500, 221–230. [Google Scholar] [CrossRef]
- Zhang, Y.-P.; Deng, Y.-J.; Tang, K.-R.; Chen, R.-S.; Liang, S.; Liang, Y.-J.; Han, L.; Jin, L.; Liang, Z.-E.; Chen, Y.-N.; et al. Berberine Ameliorates High-Fat Diet-Induced Non-Alcoholic Fatty Liver Disease in Rats via Activation of SIRT3/AMPK/ACC Pathway. Curr. Med. Sci. 2019, 39, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Tian, G.; Zhuang, Z.; Chen, J.; You, N.; Zhuo, L.; Liang, B.; Song, Y.; Zang, S.; Liu, J.; et al. Berberine prevents non-alcoholic steatohepatitis-derived hepatocellular carcinoma by inhib-iting inflammation and angiogenesis in mice. Am. J. Transl. Res. 2019, 11, 2668–2682. [Google Scholar]
- Honda, M.; Kubes, P. Neutrophils and neutrophil extracellular traps in the liver and gastrointestinal system. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 206–221. [Google Scholar] [CrossRef]
- Yang, J.; Ma, X.J.; Li, L.; Wang, L.; Chen, Y.G.; Liu, J.; Luo, Y.; Zhuang, Z.J.; Yang, W.J.; Zang, S.F.; et al. Berberine ameliorates non-alcoholic steatohepatitis in ApoE(-/-) mice. Exp. Ther. Med. 2017, 14, 4134–4140. [Google Scholar] [CrossRef][Green Version]
- Zha, W.; Liang, G.; Xiao, J.; Studer, E.J.; Hylemon, P.B.; Pandak, J.W.M.; Wang, G.; Li, X.; Zhou, H. Berberine Inhibits HIV Protease Inhibitor-Induced Inflammatory Response by Modulating ER Stress Signaling Pathways in Murine Macrophages. PLoS ONE 2010, 5, e9069. [Google Scholar] [CrossRef]
- Gu, S.; Cao, B.; Sun, R.; Tang, Y.; Paletta, J.L.; Wu, X.L.; Liu, L.; Zha, W.; Zhao, C.; Li, Y.; et al. A metabolomic and pharmacokinetic study on the mechanism underlying the lipid-lowering effect of orally administered berberine. Mol. Biosyst. 2015, 11, 463–474. [Google Scholar] [CrossRef]
- Asgharpour, A.; Cazanave, S.C.; Pacana, T.; Seneshaw, M.; Vincent, R.; Banini, B.A.; Kumar, D.P.; Daita, K.; Min, H.K.; Mirshahi, F.; et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepato-cellular cancer. J. Hepatol. 2016, 65, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Cazanave, S.; Podtelezhnikov, A.; Jensen, K.; Seneshaw, M.; Kumar, D.P.; Min, H.-K.; Santhekadur, P.K.; Banini, B.; Mauro, A.G.; Oseini, A.M.; et al. The Transcriptomic Signature of Disease Development and Progression of Nonalcoholic Fatty Liver Disease. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge – Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, X.; Zhu, W.; Wang, Y.; Zhao, D.; Wang, X.; Gurley, E.C.; Liang, G.; Chen, W.; Lai, G.; et al. Cholangiocyte-Derived Exosomal Long Noncoding RNA H19 Promotes Hepatic Stellate Cell Activation and Cholestatic Liver Fibrosis. Hepatology 2019, 70, 1317–1335. [Google Scholar] [CrossRef]
- Nagahashi, M.; Ramachandran, S.; Kim, E.Y.; Allegood, J.C.; Rashid, O.M.; Yamada, A.; Zhao, R.; Milstien, S.; Zhou, H.; Spiegel, S.; et al. Sphin-gosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 2012, 72, 726–735. [Google Scholar] [CrossRef]
- Kleiner, E.D.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.-C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Kakiyama, G.; Marques, D.; Martin, R.; Takei, H.; Rodriguez-Agudo, D.; LaSalle, S.A.; Hashiguchi, T.; Liu, X.; Green, R.M.; Erickson, S.K.; et al. Insulin resistance dysregulates CYP7B1 leading to oxysterol accumulation: A pathway for NAFL to NASH transition. J. Lipid Res. 2020, 61, 1629–1644. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, K.; Janciauskiene, S.; Li, X. Endoplasmic Reticulum Stress and Lipid Metabolism. Biochem. Res. Int. 2012, 2012, 1–2. [Google Scholar] [CrossRef]
- Puri, P.; Daita, K.; Joyce, A.; Mirshahi, F.; Santhekadur, P.K.; Cazanave, S.; A Luketic, V.; Siddiqui, M.S.; Boyett, S.; Min, H.; et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology 2018, 67, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Mai, W.; Xu, Y.; Xu, J.; Zhao, D.; Ye, L.; Yu, G.; Wang, Z.; Lu, Q.; Lin, J.; Yang, T.; et al. Berberine Inhibits Nod-Like Receptor Family Pyrin Domain Containing 3 Inflammasome Activa-tion and Pyroptosis in Nonalcoholic Steatohepatitis via the ROS/TXNIP Axis. Front. Pharmacol. 2020, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Bian, H.; Wang, L.; Sun, X.; Xu, X.; Yan, H.; Xia, M.; Chang, X.; Lu, Y.; Li, Y.; et al. Berberine attenuates nonalcoholic hepatic steatosis through the AMPK-SREBP-1c-SCD1 path-way. Free Radic. Biol. Med. 2019, 141, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chen, Z.; Wang, L.; Wang, G.; Wang, Z.; Dong, X.; Wen, B.; Zhang, Z. The Pathogenesis of Diabetes Mellitus by Oxidative Stress and Inflammation: Its Inhibition by Berberine. Front. Pharmacol. 2018, 9, 782. [Google Scholar] [CrossRef]
- Tsuchida, T.; Lee, Y.A.; Fujiwara, N.; Ybanez, M.; Allen, B.; Martins, S.; Fiel, M.I.; Goossens, N.; Chou, H.-I.; Hoshida, Y.; et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J. Hepatol. 2018, 69, 385–395. [Google Scholar] [CrossRef]
- Calvente, C.J.; Tameda, M.; Johnson, C.D.; Del Pilar, H.; Lin, Y.C.; Adronikou, N.; Jeu, X.D.M.D.; Llorente, C.; Boyer, J.; Feldstein, A.E. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. J. Clin. Investig. 2019, 129, 4091–4109. [Google Scholar] [CrossRef]
- Hwang, S.; He, Y.; Xiang, X.; Seo, W.; Kim, S.J.; Ma, J.; Ren, T.; Park, S.H.; Zhou, Z.; Feng, D.; et al. Interleukin-22 Ameliorates Neutrophil-Driven Nonalcoholic Steatohepatitis Through Multi-ple Targets. Hepatology 2020, 72, 412–429. [Google Scholar] [CrossRef]
- Hylemon, P.B.; Zhou, H.; Pandak, W.M.; Ren, S.; Gil, G.; Dent, P. Bile acids as regulatory molecules. J. Lipid Res. 2009, 50, 1509–1520. [Google Scholar] [CrossRef]
- Li, Y.; Wong, K.; Giles, A.; Jiang, J.; Lee, J.W.; Adams, A.C.; Kharitonenkov, A.; Yang, Q.; Gao, B.; Guarente, L.P.; et al. Hepatic SIRT1 Attenuates Hepatic Steatosis and Controls Energy Balance in Mice by Inducing Fibroblast Growth Factor 21. Gastroenterology 2014, 146, 539–549.e7. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A. Therapeutic Landscape for NAFLD in 2020. Gastroenterology 2020, 158, 1984–1998.e3. [Google Scholar] [CrossRef]
- Goldner, D.; Lavine, J.E. Nonalcoholic Fatty Liver Disease in Children: Unique Considerations and Challenges. Gastroenterology 2020, 158, 1967–1983.e1. [Google Scholar] [CrossRef] [PubMed]
- Doulberis, M.; Polyzos, S.A.; Papaefthymiou, A.; Katsinelos, P.; Kiosses, C.; Kountouras, J. Treatment of nonalcoholic fatty liver disease: From adult trials to perspec-tives in the management of children and adolescents. Expert Opin. Pharmacother. 2020, 21, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Cotter, T.G.; Rinella, M. Nonalcoholic Fatty Liver Disease 2020: The State of the Disease. Gastroenterology 2020, 158, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Sanyal, A.J.; George, J. International Consensus Panel: MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014.e1. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, N.; Schattenberg, J.M. Metabolic Inflammation-A Role for Hepatic Inflammatory Pathways as Drivers of Comorbidi-ties in Nonalcoholic Fatty Liver Disease? Gastroenterology 2020, 158, 1929–1947. [Google Scholar] [CrossRef]
- Affuso, F.; Mercurio, V.; Fazio, V.; Fazio, S. Cardiovascular and metabolic effects of Berberine. World J. Cardiol. 2010, 2, 71. [Google Scholar] [CrossRef]
- Bai, F.; Tao, H.; Wang, P.; Wang, L.; Zhou, X.; Wang, F.; Liu, C.; Huang, Y. Berberine hydrochloride inhibits inflammation and fibrosis after canalicular laceration repair in rabbits. Life Sci. 2020, 261, 118479. [Google Scholar] [CrossRef]
- Liang, H.; Wang, Y. Berberine alleviates hepatic lipid accumulation by increasing ABCA1 through the protein kinase C delta pathway. Biochem. Biophys. Res. Commun. 2018, 498, 473–480. [Google Scholar] [CrossRef]
- Wu, L.; Xia, M.; Duan, Y.; Zhang, L.; Jiang, H.; Hu, X.; Yan, H.; Zhang, Y.; Gu, Y.; Shi, H.; et al. Berberine promotes the recruitment and activation of brown adipose tissue in mice and humans. Cell Death Dis. 2019, 10, 468. [Google Scholar] [CrossRef]
- Xu, X.; Zhu, X.-P.; Bai, J.-Y.; Xia, P.; Li, Y.; Lu, Y.; Li, X.-Y.; Gao, X. Berberine alleviates nonalcoholic fatty liver induced by a high-fat diet in mice by activating SIRT3. FASEB J. 2019, 33, 7289–7300. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, B.; Meng, X.; Yao, S.; Jin, L.; Yang, J.; Wang, J.; Zhang, H.; Zhang, Z.; Cai, D.; et al. Berberine prevents progression from hepatic steatosis to steatohepatitis and fibrosis by reduc-ing endoplasmic reticulum stress. Sci. Rep. 2016, 6, 20848. [Google Scholar] [CrossRef] [PubMed]
- Arrese, M.; Cabrera, D.; Kalergis, A.M.; Feldstein, A.E. Innate Immunity and Inflammation in NAFLD/NASH. Dig. Dis. Sci. 2016, 61, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, X.; Zhao, D.; Wang, X.; Gurley, E.C.; Liu, R.; Li, X.; Hylemon, P.B.; Chen, W.; Zhou, H. Berberine inhibits free fatty acid and LPS-induced inflammation via modulating ER stress response in macrophages and hepatocytes. PLoS ONE 2020, 15, e0232630. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Du, X.; Ma, H.; Yao, J. The Anti-Cancer Mechanisms of Berberine: A Review. Cancer Manag. Res. 2020, 12, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, S. Berberine pharmacology and the gut microbiota: A hidden therapeutic link. Pharmacol. Res. 2020, 155, 104722. [Google Scholar] [CrossRef] [PubMed]
- Van Der Windt, D.J.; Sud, V.; Zhang, H.; Varley, P.R.; Goswami, J.; Yazdani, H.O.; Tohme, S.; Loughran, P.; O’Doherty, R.M.; Minervini, M.I.; et al. Neutrophil extracellular traps promote inflammation and development of hepato-cellular carcinoma in nonalcoholic steatohepatitis. Hepatology 2018, 68, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, H.; Ma, L.; Zhou, J.; Guo, X.; Woo, S.L.; Pei, Y.; Knight, L.R.; Deveau, M.; Chen, Y.; et al. Expression of STING Is Increased in Liver Tissues From Patients With NAFLD and Promotes Mac-rophage-Mediated Hepatic Inflammation and Fibrosis in Mice. Gastroenterology 2018, 155, 1971–1984. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Targeting bile acids and lipotoxicity for NASH treatment. Hepatol. Commun. 2017, 1, 1002–1004. [Google Scholar] [CrossRef]
- Chen, J.Y.; Levy-Wilson, B.; Goodart, S.; Cooper, A.D. Mice expressing the human CYP7A1 gene in the mouse CYP7A1 knock-out back-ground lack induction of CYP7A1 expression by cholesterol feeding and have increased hypercholesterolemia when fed a high fat diet. J. Biol. Chem. 2002, 277, 42588–42595. [Google Scholar] [CrossRef]
- Venetsanaki, V.; Karabouta, Z.; Polyzos, S.A. Farnesoid X nuclear receptor agonists for the treatment of nonalcoholic steato-hepatitis. Eur. J. Pharmacol. 2019, 863, 172661. [Google Scholar] [CrossRef]
- Repa, J.J.; Mangelsdorf, D.J. Nuclear receptor regulation of cholesterol and bile acid metabolism. Curr. Opin. Biotechnol. 1999, 10, 557–563. [Google Scholar] [CrossRef]
- Gottlieb, A.; Canbay, A. Why Bile Acids Are So Important in Non-Alcoholic Fatty Liver Disease (NAFLD) Progression. Cells 2019, 8, 1358. [Google Scholar] [CrossRef] [PubMed]
- Bechmann, L.P.; Kocabayoglu, P.; Sowa, J.P.; Sydor, S.; Best, J.; Schlattjan, M.; Beilfuss, A.; Schmitt, J.; Hannivoort, R.A.; Kilicarslan, A.; et al. Free fatty acids repress small heterodimer partner (SHP) activation and adi-ponectin counteracts bile acid-induced liver injury in superobese patients with nonalcoholic steatohepatitis. Hepatology 2013, 57, 1394–1406. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, F.; Claudel, T.; Sturm, E.; Staels, B. The Farnesoid X Receptor (FXR) as Modulator of Bile Acid Metabolism. Rev. Endocr. Metab. Disord. 2004, 5, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Wang, Y. Oxysterols as lipid mediators: Their biosynthetic genes, enzymes and metabolites. Prostaglandins Other Lipid Mediat. 2020, 147, 106381. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, R.; Huang, Z.; Gurley, E.C.; Wang, X.; Wang, J.; He, H.; Yang, H.; Lai, G.; Zhang, L.; et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology 2018, 68, 599–615. [Google Scholar] [CrossRef]
- Wang, Y.; Aoki, H.; Yang, J.; Peng, K.; Liu, R.; Li, X.; Qiang, X.; Sun, L.; Gurley, E.C.; Lai, G.; et al. The role of sphingosine 1-phosphate receptor 2 in bile-acid-induced cholangiocyte prolifera-tion and cholestasis-induced liver injury in mice. Hepatology 2017, 65, 2005–2018. [Google Scholar] [CrossRef]
- Nagahashi, M.; Takabe, K.; Liu, R.; Peng, K.; Wang, X.; Wang, Y.; Hait, N.C.; Wang, X.; Allegood, J.C.; Yamada, A.; et al. Conjugated bile acid-activated S1P receptor 2 is a key regulator of sphingosine kinase 2 and hepatic gene expression. Hepatology 2015, 61, 1216–1226. [Google Scholar] [CrossRef]
- Zhang, Z.; Zong, C.; Jiang, M.; Hu, H.; Cheng, X.; Ni, J.; Yi, X.; Jiang, B.; Tian, F.; Chang, M.-W.; et al. Hepatic HuR modulates lipid homeostasis in response to high-fat diet. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Lin, B.; Ma, Y.; Wu, S.; Liu, Y.; Liu, L.; Wu, L. Novel Serum Biomarkers for Noninvasive Diagnosis and Screening of Nonalcoholic Fatty Liver Disease-Related Hepatic Fibrosis. OMICS J. Integr. Biol. 2019, 23, 181–189. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, Y.; Ren, H.; Wang, S.; Zhong, H.; Zhao, X.; Ma, J.; Gu, X.; Xue, Y.; Huang, S.; et al. Gut microbiome-related effects of berberine and probiotics on type 2 diabetes (the PREMOTE study). Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Yang, N.; Kong, B.; Cao, B.; Feng, D.; Yu, X.; Ge, C.; Huang, J.; Shen, J.; Wang, P.; et al. Orally Administered Berberine Modulates Hepatic Lipid Metabolism by Altering Microbial Bile Acid Metabolism and the Intestinal FXR Signaling Pathway. Mol. Pharmacol. 2017, 91, 110–122. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Tai, Y.-L.; Zhao, D.; Zhang, Y.; Yan, J.; Kakiyama, G.; Wang, X.; Gurley, E.C.; Liu, J.; Liu, J.; et al. Berberine Prevents Disease Progression of Nonalcoholic Steatohepatitis through Modulating Multiple Pathways. Cells 2021, 10, 210. https://doi.org/10.3390/cells10020210
Wang Y, Tai Y-L, Zhao D, Zhang Y, Yan J, Kakiyama G, Wang X, Gurley EC, Liu J, Liu J, et al. Berberine Prevents Disease Progression of Nonalcoholic Steatohepatitis through Modulating Multiple Pathways. Cells. 2021; 10(2):210. https://doi.org/10.3390/cells10020210
Chicago/Turabian StyleWang, Yanyan, Yun-Ling Tai, Derrick Zhao, Yuan Zhang, Junkai Yan, Genta Kakiyama, Xuan Wang, Emily C. Gurley, Jinze Liu, Jinpeng Liu, and et al. 2021. "Berberine Prevents Disease Progression of Nonalcoholic Steatohepatitis through Modulating Multiple Pathways" Cells 10, no. 2: 210. https://doi.org/10.3390/cells10020210
APA StyleWang, Y., Tai, Y.-L., Zhao, D., Zhang, Y., Yan, J., Kakiyama, G., Wang, X., Gurley, E. C., Liu, J., Liu, J., Liu, J., Lai, G., Hylemon, P. B., Pandak, W. M., Chen, W., & Zhou, H. (2021). Berberine Prevents Disease Progression of Nonalcoholic Steatohepatitis through Modulating Multiple Pathways. Cells, 10(2), 210. https://doi.org/10.3390/cells10020210





