Overexpression of Neuroglobin Promotes Energy Metabolism and Autophagy Induction in Human Neuroblastoma SH-SY5Y Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Plasmid Construction
2.3. Sample Preparation and LC-MS/MS Analysis
2.4. LFQ Proteomics Analysis and Bioinformatics Enrichment
2.5. Bioenergetic Analysis
2.6. DNA-RNA Isolation and Real Time qPCR
2.7. Analysis of LC3-II and SQSTM1/p62 Levels by Immunoblotting
2.8. Autophagy Induction
2.9. LC3-II Immunoprecipitates
2.10. Statistical Analysis
3. Results
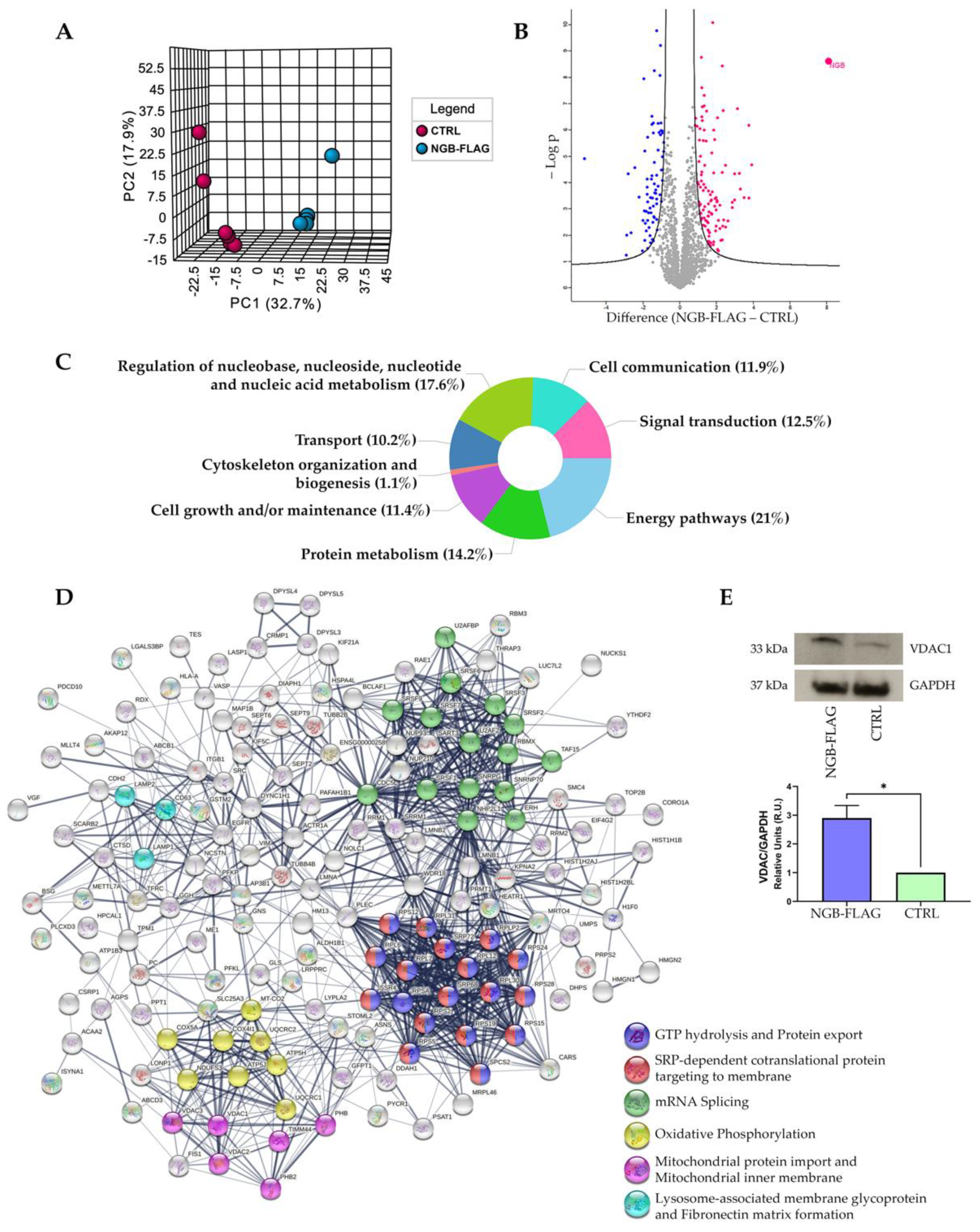
3.1. Proteomic Analysis following Genetic Overexpression of NGB in Neuroblastoma SH-SY5Y Cells
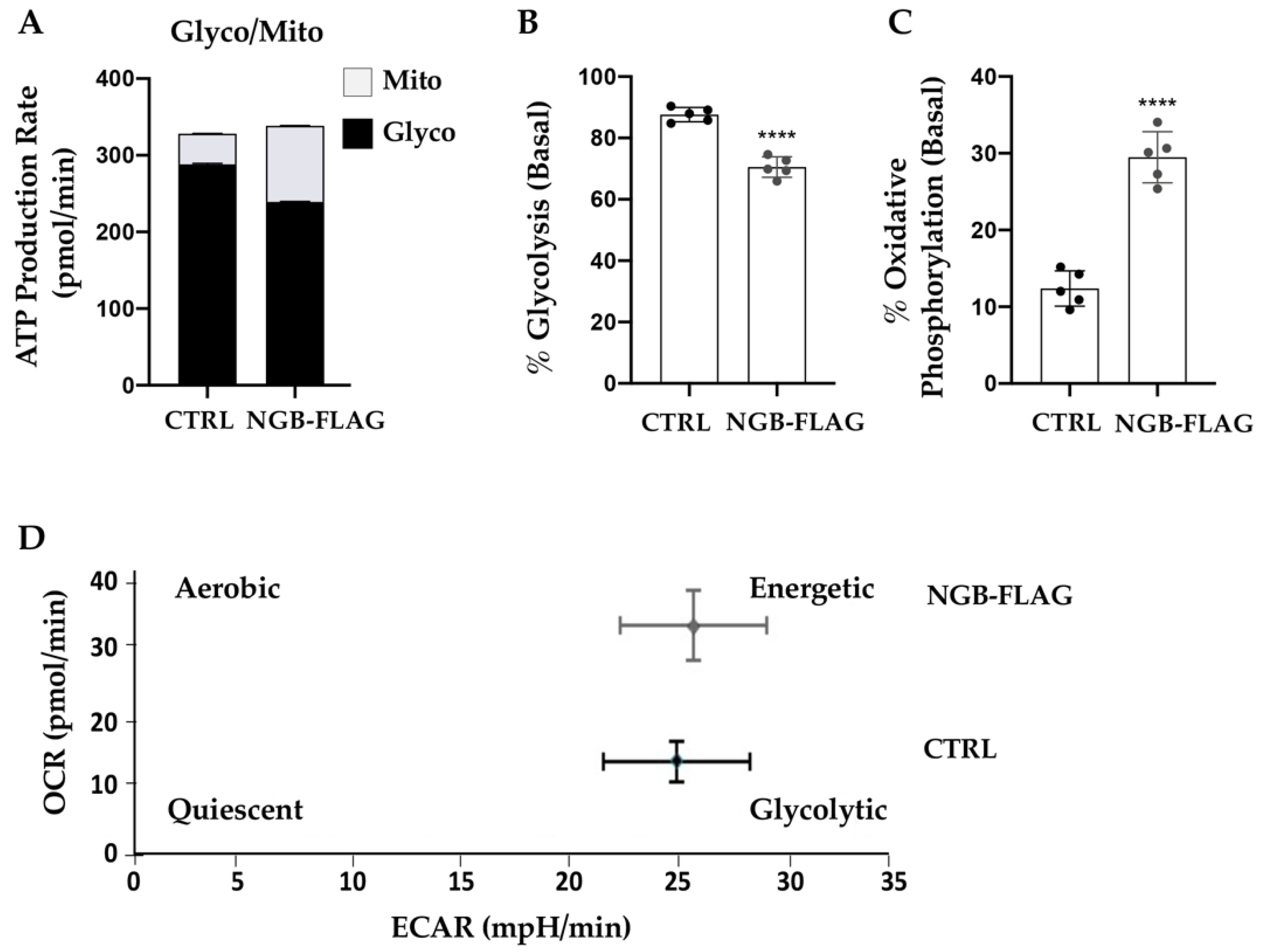
3.2. Effect of NGB Overexpression on the Energy Metabolism in Neuroblastoma SH-SY5Y Cells
3.3. NGB Overexpression Increases NRF1 mRNA and Mitochondrial DNA Levels in Neuroblastoma SH-SY5Y Cells
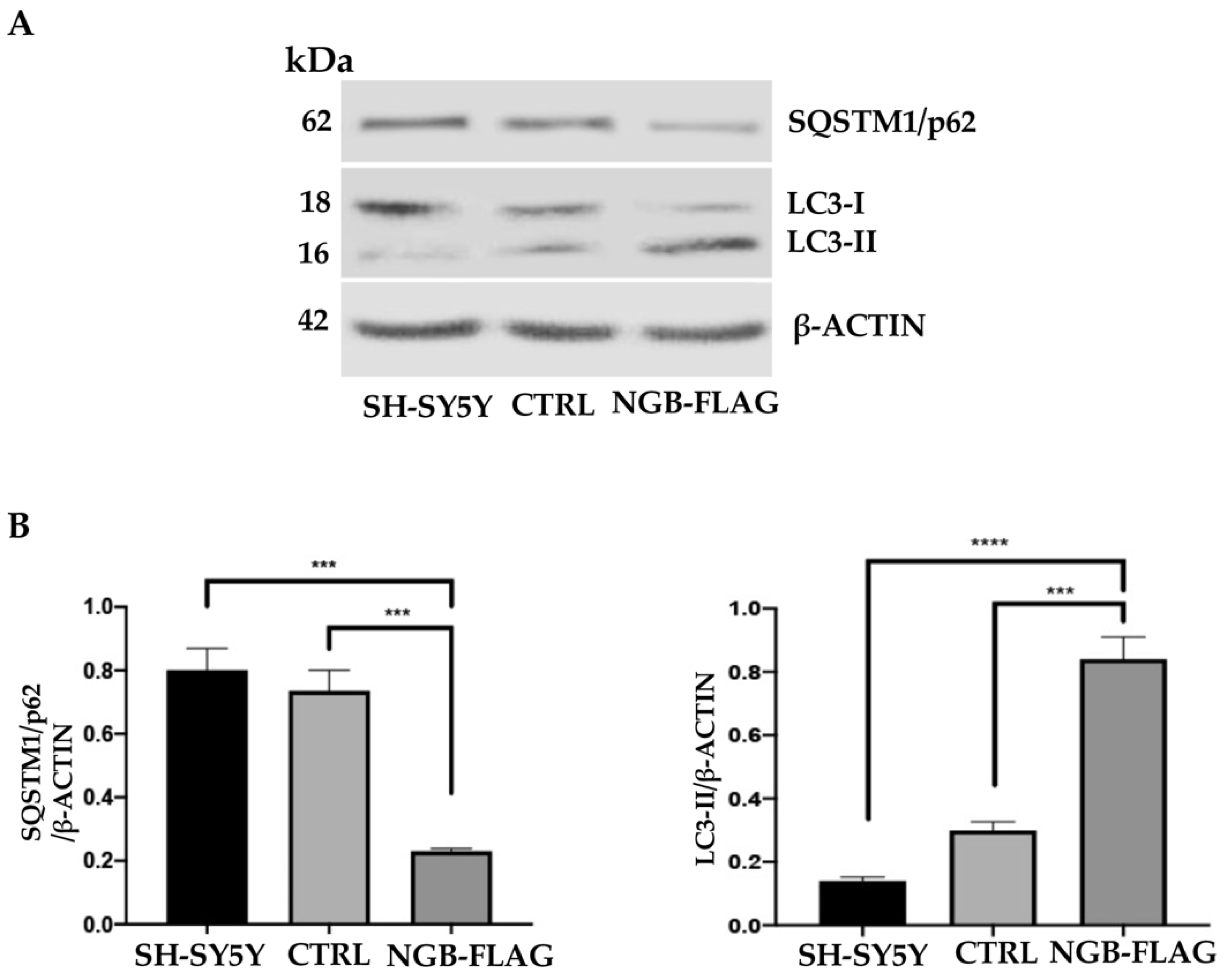
3.4. Effect of NGB Overexpression on Autophagy Induction in Neuroblastoma SH-SY5Y Cells
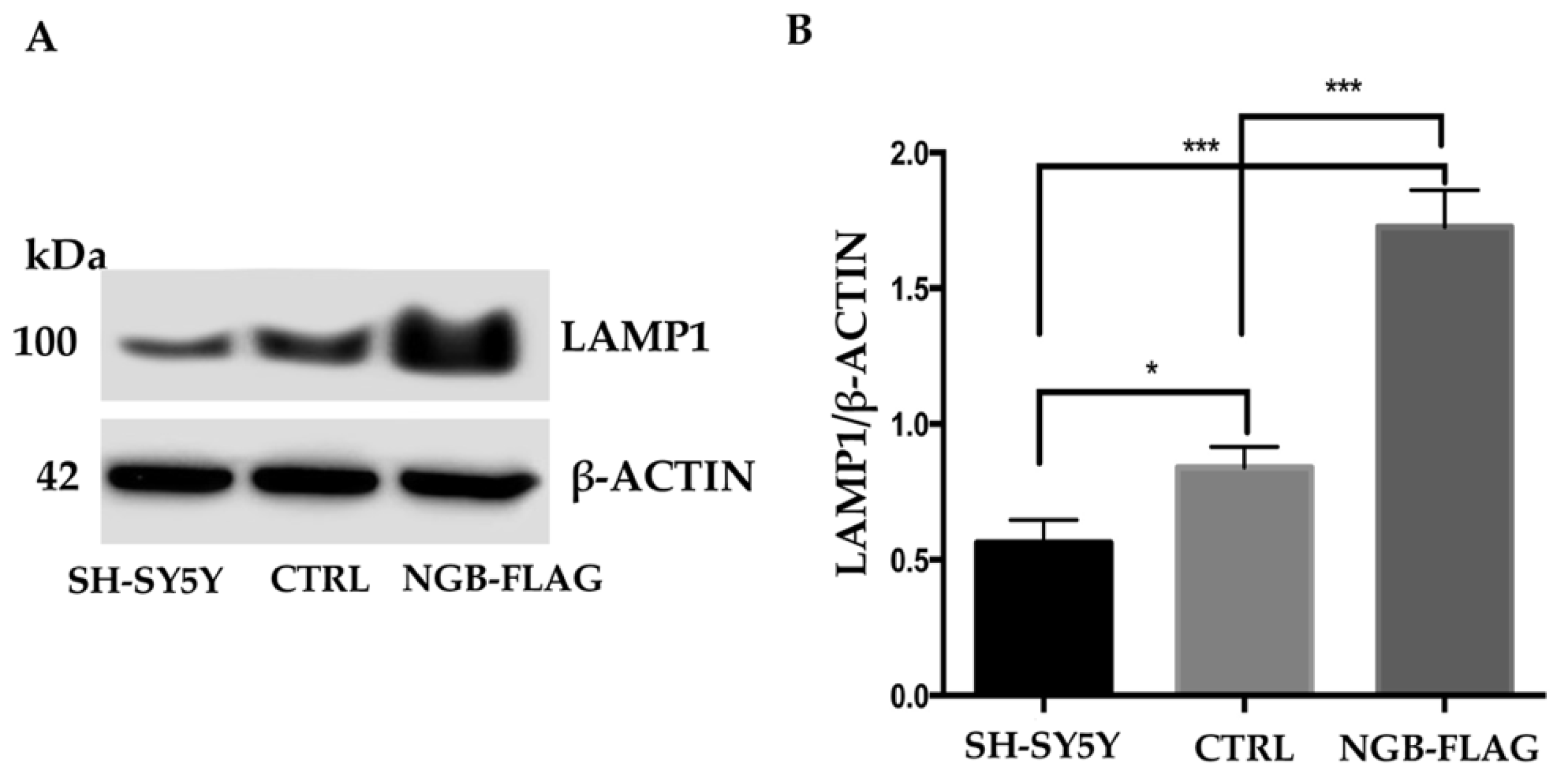
3.5. Effect of NGB Overexpression on the Lysosome Compartments of Neuroblastoma SH-SY5Y Cells
3.6. NGB Associates with LC3-II during Autolysosome Formation in Neuroblastoma SH-SY5Y-NGB-FLAG Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burmester, T.; Weich, B.; Reinhardt, S.; Hankeln, T. A vertebrate globin expressed in the brain. Nature 2000, 407, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Burmester, T.; Hankeln, T. What is the function of neuroglobin? J. Exp. Biol. 2009, 212, 1423–1428. [Google Scholar] [CrossRef] [Green Version]
- Ascenzi, P.; Gustincich, S.; Marino, M. Mammalian nerve globins in search of functions. IUBMB Life 2014, 66, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Xu, J.; Liu, N.; Wang, Y.; Li, X.; Pallast, S.; van Leyen, K.; Wang, X. Mitochondrial distribution of neuroglobin and its response to oxygen-glucose deprivation in primary-cultured mouse cortical neurons. Neuroscience 2012, 218, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Poppe, J.L.; Wang, X. Mitochondrial mechanisms of neuroglobin’s neuroprotection. Oxidative Med. Cell. Longev. 2013, 2013, 756989. [Google Scholar] [CrossRef] [Green Version]
- Raychaudhuri, S.; Skommer, J.; Henty, K.; Birch, N.; Brittain, T. Neuroglobin protects nerve cells from apoptosis by inhibiting the intrinsic pathway of cell death. Apoptosis 2010, 15, 401–411. [Google Scholar] [CrossRef] [Green Version]
- Fordel, E.; Thijs, L.; Moens, L.; Dewilde, S. Neuroglobin and cytoglobin expression in mice. Evidence for a correlation with reactive oxygen species scavenging. FEBS J. 2007, 274, 1312–1317. [Google Scholar] [CrossRef]
- Schmidt-Kastner, R.; Haberkamp, M.; Schmitz, C.; Hankeln, T.; Burmester, T. Neuroglobin mRNA expression after transient global brain ischemia and prolonged hypoxia in cell culture. Brain Res. 2006, 1103, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.Q.; Zhou, X.Y.; Lai, X.J.; Zheng, L.; Chen, X.Q. Silencing neuroglobin enhances neuronal vulnerability to oxidative injury by down-regulating 14-3-3gamma. Acta Pharmacol. Sin. 2009, 30, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Shao, G.; Gong, K.R.; Li, J.; Xu, X.J.; Gao, C.Y.; Zeng, X.Z.; Lu, G.W.; Huo, X. Antihypoxic effects of neuroglobin in hypoxia-preconditioned mice and SH-SY5Y cells. Neurosignals 2009, 17, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Hundahl, C.A.; Allen, G.C.; Hannibal, J.; Kjaer, K.; Rehfeld, J.F.; Dewilde, S.; Nyengaard, J.R.; Kelsen, J.; Hay-Schmidt, A. Anatomical characterization of cytoglobin and neuroglobin mRNA and protein expression in the mouse brain. Brain Res. 2010, 1331, 58–73. [Google Scholar] [CrossRef]
- Bentmann, A.; Schmidt, M.; Reuss, S.; Wolfrum, U.; Hankeln, T.; Burmester, T. Divergent distribution in vascular and avascular mammalian retinae links neuroglobin to cellular respiration. J. Biol. Chem. 2005, 280, 20660–20665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Marinis, E.; Fiocchetti, M.; Acconcia, F.; Ascenzi, P.; Marino, M. Neuroglobin upregulation induced by 17β-estradiol sequesters cytocrome c in the mitochondria preventing H2O2-induced apoptosis of neuroblastoma cells. Cell Death Dis. 2013, 4, e508. [Google Scholar] [CrossRef]
- Fiocchetti, M.; Cipolletti, M.; Brandi, V.; Polticelli, F.; Ascenzi, P. Neuroglobin and friends. J. Mol. Recognit. 2017, 30. [Google Scholar] [CrossRef] [PubMed]
- Fiocchetti, M.; Cipolletti, M.; Leone, S.; Naldini, A.; Carraro, F.; Giordano, D.; Verde, C.; Ascenzi, P.; Marino, M. Neuroglobin in Breast Cancer Cells: Effect of Hypoxia and Oxidative Stress on Protein Level, Localization, and Anti-Apoptotic Function. PLoS ONE 2016, 11, e0154959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garofalo, T.; Ferri, A.; Sorice, M.; Azmoon, P.; Grasso, M.; Mattei, V.; Capozzi, A.; Manganelli, V.; Misasi, R. Neuroglobin overexpression plays a pivotal role in neuroprotection through mitochondrial raft-like microdomains in neuroblastoma SK-N-BE2 cells. Mol. Cell. Neurosci. 2018, 88, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [Green Version]
- Solar Fernandez, V.; Cipolletti, M.; Ascenzi, P.; Marino, M.; Fiocchetti, M. Neuroglobin as Key Mediator in the 17β-Estradiol-Induced Antioxidant Cell Response to Oxidative Stress. Antioxid. Redox Signal. 2020, 32, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Li, R.C.; Morris, M.W.; Lee, S.K.; Pouranfar, F.; Wang, Y.; Gozal, D. Neuroglobin protects PC12 cells against oxidative stress. Brain Res. 2008, 1190, 159–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.A.; Wang, Y.; Sun, Y.; Mao, X.O.; Xie, L.; Miles, E.; Graboski, J.; Chen, S.; Ellerby, L.M.; Jin, K.; et al. Neuroglobin-overexpressing transgenic mice are resistant to cerebral and myocardial ischemia. Proc. Natl. Acad. Sci. USA 2006, 103, 17944–17948. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yu, Z.; Guo, S.; Lee, S.R.; Xing, C.; Zhang, C.; Gao, Y.; Nicholls, D.G.; Lo, E.H.; Wang, X. Effects of neuroglobin overexpression on mitochondrial function and oxidative stress following hypoxia/reoxygenation in cultured neurons. J. Neurosci. Res. 2009, 87, 164–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, S.; Takahashi, N.; Uchida, H.; Wakasugi, K. Human neuroglobin functions as an oxidative stress-responsive sensor for neuroprotection. J. Biol. Chem. 2012, 287, 30128–30138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Marinis, E.; Ascenzi, P.; Pellegrini, M.; Galluzzo, P.; Bulzomi, P.; Arevalo, M.A.; Garcia-Segura, L.M.; Marino, M. 17β-estradiol a new modulator of neuroglobin levels in neurons: Role in neuroprotection against H2O2-induced toxicity. Neurosignals 2010, 18, 223–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, W.B.; Lin, J.H.; Chen, X.W.; Wu, C.Y.; Zhong, G.X.; Zhang, L.Q.; Lin, W.P.; Liu, W.N.; Li, X.; Lin, J.L. Overexpressing neuroglobin improves functional recovery by inhibiting neuronal apoptosis after spinal cord injury. Brain Res. 2014, 1562, 100–108. [Google Scholar] [CrossRef]
- Fiocchetti, M.; De Marinis, E.; Ascenzi, P.; Marino, M. Neuroglobin and neuronal cell survival. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2013, 1834, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Liu, N.; Liu, J.; Yang, K.; Wang, X. Neuroglobin, a Novel Target for Endogenous Neuroprotection against Stroke and Neurodegenerative Disorders. Int. J. Mol. Sci. 2012, 13, 6995–7014. [Google Scholar] [CrossRef]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Dadakhujaev, S.; Noh, H.S.; Jung, E.J.; Cha, J.Y.; Baek, S.M.; Ha, J.H.; Kim, D.R. Autophagy protects the rotenone-induced cell death in alpha-synuclein overexpressing SH-SY5Y cells. Neurosci. Lett. 2010, 472, 47–52. [Google Scholar] [CrossRef]
- Fiocchetti, M.; Cipolletti, M.; Marino, M. Compensatory role of Neuroglobin in nervous and non-nervous cancer cells in response to the nutrient deprivation. PLoS ONE 2017, 12, e0189179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, Z.H.; Cai, Q. Mitochondrial transport in neurons: Impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci. 2012, 13, 77–93. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Tammineni, P. Alterations in Mitochondrial Quality Control in Alzheimer’s Disease. Front. Cell. Neurosci. 2016, 10, 24. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Zhunina, O.A.; Yabbarov, N.G.; Grechko, A.V.; Starodubova, A.V.; Ivanova, E.; Nikiforov, N.G.; Orekhov, A.N. The Role of Mitochondrial Dysfunction in Vascular Disease, Tumorigenesis, and Diabetes. Front. Mol. Biosci. 2021, 8, 671908. [Google Scholar] [CrossRef] [PubMed]
- Malpartida, A.B.; Williamson, M.; Narendra, D.P.; Wade-Martins, R.; Ryan, B.J. Mitochondrial Dysfunction and Mitophagy in Parkinson’s Disease: From Mechanism to Therapy. Trends Biochem. Sci. 2021, 46, 329–343. [Google Scholar] [CrossRef]
- Elkamhawy, A.; Lee, J.; Park, B.G.; Park, I.; Pae, A.N.; Roh, E.J. Novel quinazoline-urea analogues as modulators for Aβ-induced mitochondrial dysfunction: Design, synthesis, and molecular docking study. Eur. J. Med. Chem. 2014, 84, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Elkamhawy, A.; Park, J.E.; Hassan, A.H.E.; Pae, A.N.; Lee, J.; Paik, S.; Park, B.G.; Roh, E.J. Pyrazinyl ureas revisited: 1-(3-(Benzyloxy)pyrazin-2-yl)-3-(3,4-dichlorophenyl)urea, a new blocker of Aβ-induced mPTP opening for Alzheimer’s disease. Eur. J. Med. Chem. 2018, 157, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.H.; Kwon, Y.T. Crosstalk and Interplay between the Ubiquitin-Proteasome System and Autophagy. Mol. Cells 2017, 40, 441–449. [Google Scholar] [CrossRef] [Green Version]
- Elkamhawy, A.; Kim, H.J.; Elsherbeny, M.H.; Paik, S.; Park, J.H.; Gotina, L.; Abdellattif, M.H.; Gouda, N.A.; Cho, J.; Lee, K.; et al. Discovery of 3,4-dichloro-N-(1H-indol-5-yl)benzamide: A highly potent, selective, and competitive hMAO-B inhibitor with high BBB permeability profile and neuroprotective action. Bioorganic Chem. 2021, 116, 105352. [Google Scholar] [CrossRef]
- Elkamhawy, A.; Woo, J.; Gouda, N.A.; Kim, J.; Nada, H.; Roh, E.J.; Park, K.D.; Cho, J.; Lee, K. Melatonin Analogues Potently Inhibit MAO-B and Protect PC12 Cells against Oxidative Stress. Antioxidants 2021, 10, 1604. [Google Scholar] [CrossRef] [PubMed]
- Elsherbeny, M.H.; Kim, J.; Gouda, N.A.; Gotina, L.; Cho, J.; Pae, A.N.; Lee, K.; Park, K.D.; Elkamhawy, A.; Roh, E.J. Highly Potent, Selective, and Competitive Indole-Based MAO-B Inhibitors Protect PC12 Cells against 6-Hydroxydopamine- and Rotenone-Induced Oxidative Stress. Antioxidants 2021, 10, 1641. [Google Scholar] [CrossRef]
- Strother, L.; Miles, G.B.; Holiday, A.R.; Cheng, Y.; Doherty, G.H. Long-term culture of SH-SY5Y neuroblastoma cells in the absence of neurotrophins: A novel model of neuronal ageing. J. Neurosci. Methods 2021, 362, 109301. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, D.; Huang, X.; Huang, H. Astragaloside IV Protects 6-Hydroxydopamine-Induced SH-SY5Y Cell Model of Parkinson’s Disease via Activating the JAK2/STAT3 Pathway. Front. Neurosci. 2021, 15, 631501. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Fiocchetti, M.; Ascenzi, P.; Marino, M.; Caterino, M.; Ruoppolo, M. Proteomic and Bioinformatic Investigation of Altered Pathways in Neuroglobin-Deficient Breast Cancer Cells. Molecules 2021, 26, 2397. [Google Scholar] [CrossRef]
- Cozzolino, M.; Amori, I.; Pesaresi, M.G.; Ferri, A.; Nencini, M.; Carrì, M.T. Cysteine 111 affects aggregation and cytotoxicity of mutant Cu,Zn-superoxide dismutase associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008, 283, 866–874. [Google Scholar] [CrossRef] [Green Version]
- De Pasquale, V.; Costanzo, M.; Siciliano, R.A.; Mazzeo, M.F.; Pistorio, V.; Bianchi, L.; Marchese, E.; Ruoppolo, M.; Pavone, L.M.; Caterino, M. Proteomic Analysis of Mucopolysaccharidosis IIIB Mouse Brain. Biomolecules 2020, 10, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pasquale, V.; Caterino, M.; Costanzo, M.; Fedele, R.; Ruoppolo, M.; Pavone, L.M. Targeted Metabolomic Analysis of a Mucopolysaccharidosis IIIB Mouse Model Reveals an Imbalance of Branched-Chain Amino Acid and Fatty Acid Metabolism. Int. J. Mol. Sci. 2020, 21, 4211. [Google Scholar] [CrossRef]
- Giacco, A.; Delli Paoli, G.; Senese, R.; Cioffi, F.; Silvestri, E.; Moreno, M.; Ruoppolo, M.; Caterino, M.; Costanzo, M.; Lombardi, A.; et al. The saturation degree of fatty acids and their derived acylcarnitines determines the direct effect of metabolically active thyroid hormones on insulin sensitivity in skeletal muscle cells. FASEB J. 2019, 33, 1811–1823. [Google Scholar] [CrossRef] [Green Version]
- Costanzo, M.; Cevenini, A.; Marchese, E.; Imperlini, E.; Raia, M.; Del Vecchio, L.; Caterino, M.; Ruoppolo, M. Label-Free Quantitative Proteomics in a Methylmalonyl-CoA Mutase-Silenced Neuroblastoma Cell Line. Int. J. Mol. Sci. 2018, 19, 3580. [Google Scholar] [CrossRef] [Green Version]
- Costanzo, M.; Caterino, M.; Cevenini, A.; Jung, V.; Chhuon, C.; Lipecka, J.; Fedele, R.; Guerrera, I.C.; Ruoppolo, M. Proteomics Reveals that Methylmalonyl-CoA Mutase Modulates Cell Architecture and Increases Susceptibility to Stress. Int. J. Mol. Sci. 2020, 21, 4998. [Google Scholar] [CrossRef]
- Melo, M.G.; Remacle, N.; Cudré-Cung, H.P.; Roux, C.; Poms, M.; Cudalbu, C.; Barroso, M.; Gersting, S.W.; Feichtinger, R.G.; Mayr, J.A.; et al. The first knock-in rat model for glutaric aciduria type I allows further insights into pathophysiology in brain and periphery. Mol. Genet. Metab. 2021, 133, 157–181. [Google Scholar] [CrossRef]
- Costanzo, M.; Caterino, M.; Cevenini, A.; Jung, V.; Chhuon, C.; Lipecka, J.; Fedele, R.; Guerrera, I.C.; Ruoppolo, M. Dataset of a comparative proteomics experiment in a methylmalonyl-CoA mutase knockout HEK 293 cell model. Data Brief 2020, 33, 06453. [Google Scholar] [CrossRef]
- Caterino, M.; Ruoppolo, M.; Villani, G.R.D.; Marchese, E.; Costanzo, M.; Sotgiu, G.; Dore, S.; Franconi, F.; Campesi, I. Influence of Sex on Urinary Organic Acids: A Cross-Sectional Study in Children. Int. J. Mol. Sci. 2020, 21, 582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caterino, M.; Costanzo, M.; Fedele, R.; Cevenini, A.; Gelzo, M.; Di Minno, A.; Andolfo, I.; Capasso, M.; Russo, R.; Annunziata, A.; et al. The Serum Metabolome of Moderate and Severe COVID-19 Patients Reflects Possible Liver Alterations Involving Carbon and Nitrogen Metabolism. Int. J. Mol. Sci. 2021, 22, 9548. [Google Scholar] [CrossRef] [PubMed]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2021, 433, 166747. [Google Scholar] [CrossRef]
- Caterino, M.; Ruoppolo, M.; Mandola, A.; Costanzo, M.; Orrù, S.; Imperlini, E. Protein-protein interaction networks as a new perspective to evaluate distinct functional roles of voltage-dependent anion channel isoforms. Mol. Biosyst. 2017, 13, 2466–2476. [Google Scholar] [CrossRef]
- Miller, F.J.; Rosenfeldt, F.L.; Zhang, C.; Linnane, A.W.; Nagley, P. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: Lack of change of copy number with age. Nucleic Acids Res. 2003, 31, e61. [Google Scholar] [CrossRef]
- Bosc, C.; Broin, N.; Fanjul, M.; Saland, E.; Farge, T.; Courdy, C.; Batut, A.; Masoud, R.; Larrue, C.; Skuli, S.; et al. Autophagy regulates fatty acid availability for oxidative phosphorylation through mitochondria-endoplasmic reticulum contact sites. Nat. Commun. 2020, 11, 4056. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Furuta, N.; Yoshimori, T.; Amano, A. Mediatory molecules that fuse autophagosomes and lysosomes. Autophagy 2010, 6, 417–418. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Zhuo, M.; Gorgun, F.M.; Englander, E.W. Overexpressed neuroglobin raises threshold for nitric oxide-induced impairment of mitochondrial respiratory activities and stress signaling in primary cortical neurons. Nitric Oxide 2013, 32, 21–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbat, C.; Trucy, M.; Sorice, M.; Garofalo, T.; Manganelli, V.; Fischer, A.; Mazerolles, F. p56lck, LFA-1 and PI3K but not SHP-2 interact with GM1- or GM3-enriched microdomains in a CD4-p56lck association-dependent manner. Biochem. J. 2007, 402, 471–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciarlo, L.; Manganelli, V.; Matarrese, P.; Garofalo, T.; Tinari, A.; Gambardella, L.; Marconi, M.; Grasso, M.; Misasi, R.; Sorice, M.; et al. Raft-like microdomains play a key role in mitochondrial impairment in lymphoid cells from patients with Huntington’s disease. J. Lipid Res. 2012, 53, 2057–2068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorice, M.; Mattei, V.; Tasciotti, V.; Manganelli, V.; Garofalo, T.; Misasi, R. Trafficking of PrPc to mitochondrial raft-like microdomains during cell apoptosis. Prion 2012, 6, 354–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iessi, E.; Marconi, M.; Manganelli, V.; Sorice, M.; Malorni, W.; Garofalo, T.; Matarrese, P. On the role of sphingolipids in cell survival and death. Int. Rev. Cell. Mol. Biol. 2020, 351, 149–195. [Google Scholar] [CrossRef]
- Matarrese, P.; Garofalo, T.; Manganelli, V.; Gambardella, L.; Marconi, M.; Grasso, M.; Tinari, A.; Misasi, R.; Malorni, W.; Sorice, M. Evidence for the involvement of GD3 ganglioside in autophagosome formation and maturation. Autophagy 2014, 10, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, T.; Matarrese, P.; Manganelli, V.; Marconi, M.; Tinari, A.; Gambardella, L.; Faggioni, A.; Misasi, R.; Sorice, M.; Malorni, W. Evidence for the involvement of lipid rafts localized at the ER-mitochondria associated membranes in autophagosome formation. Autophagy 2016, 12, 917–935. [Google Scholar] [CrossRef] [Green Version]
- Manganelli, V.; Capozzi, A.; Recalchi, S.; Riitano, G.; Mattei, V.; Longo, A.; Misasi, R.; Garofalo, T.; Sorice, M. The Role of Cardiolipin as a Scaffold Mitochondrial Phospholipid in Autophagosome Formation: In Vitro Evidence. Biomolecules 2021, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Manganelli, V.; Matarrese, P.; Antonioli, M.; Gambardella, L.; Vescovo, T.; Gretzmeier, C.; Longo, A.; Capozzi, A.; Recalchi, S.; Riitano, G.; et al. Raft-like lipid microdomains drive autophagy initiation via AMBRA1-ERLIN1 molecular association within MAMs. Autophagy 2021, 17, 2528–2548. [Google Scholar] [CrossRef]
- Jain, K.; Paranandi, K.S.; Sridharan, S.; Basu, A. Autophagy in breast cancer and its implications for therapy. Am. J. Cancer Res. 2013, 3, 251–265. [Google Scholar]
- Karantza-Wadsworth, V.; White, E. Role of autophagy in breast cancer. Autophagy 2007, 3, 610–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nat. Rev. Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; Niso-Santano, M.; Baehrecke, E.H.; Kroemer, G. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell. Biol. 2014, 15, 81–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vomero, M.; Manganelli, V.; Barbati, C.; Colasanti, T.; Capozzi, A.; Finucci, A.; Spinelli, F.R.; Ceccarelli, F.; Perricone, C.; Truglia, S.; et al. Reduction of autophagy and increase in apoptosis correlates with a favorable clinical outcome in patients with rheumatoid arthritis treated with anti-TNF drugs. Arthritis Res. Ther. 2019, 21, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manganelli, V.; Longo, A.; Mattei, V.; Recalchi, S.; Riitano, G.; Caissutti, D.; Capozzi, A.; Sorice, M.; Misasi, R.; Garofalo, T. Role of ERLINs in the Control of Cell Fate through Lipid Rafts. Cells 2021, 10, 2408. [Google Scholar] [CrossRef]
- Fiocchetti, M.; Solar Fernandez, V.; Segatto, M.; Leone, S.; Cercola, P.; Massari, A.; Cavaliere, F.; Marino, M. Extracellular Neuroglobin as a Stress-Induced Factor Activating Pre-Adaptation Mechanisms against Oxidative Stress and Chemotherapy-Induced Cell Death in Breast Cancer. Cancers 2020, 12, 2451. [Google Scholar] [CrossRef] [PubMed]
- Roca-Portoles, A.; Tait, S.W.G. Mitochondrial quality control: From molecule to organelle. Cell. Mol. Life Sci. 2021, 78, 3853–3866. [Google Scholar] [CrossRef] [PubMed]
- Panchal, K.; Tiwari, A.K. Mitochondrial dynamics, a key executioner in neurodegenerative diseases. Mitochondrion 2019, 47, 151–173. [Google Scholar] [CrossRef]
- Han, S.; Zhang, M.; Jeong, Y.Y.; Margolis, D.J.; Cai, Q. The role of mitophagy in the regulation of mitochondrial energetic status in neurons. Autophagy 2021. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, V.; Menzies, K.J.; Auwerx, J. Repairing Mitochondrial Dysfunction in Disease. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 353–389. [Google Scholar] [CrossRef] [PubMed]




| Target | Forward Sequence | Reverse Sequence |
|---|---|---|
| ND5 | 5′-AGCATTCGGAAGCATCTTTG-3′ | 3′-TCGGATGTCTTGTTCGTCTG-5′ |
| ND2 | 5′-CTACCGCATTCCTACTACTCAACTT-3′ | 3′-GCTTGTTTCAGGTGCGAGAT-5′ |
| NRF1 | 5′-CAGCCGCTCTGAGAACTTCA- 3′ | 3′-CGGTGTAAGTAGCCACATGGA-5′ |
| IL2 | 5′TAGGCCACAGAATTGAAAGATCT- 3′ | 3′GTAGGTGGAAATTCTAGCATCATCC-5′ |
| UniProt ID | Gene Name | Protein Description | Difference | Cluster |
|---|---|---|---|---|
| P13073 | COX4I1 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | 2.3 | Oxidative Phosphorylation |
| P56134 | ATP5J2 | ATP synthase subunit f, mitochondrial | 2.2 | |
| P31930 | UQCRC1 | Cytochrome b-c1 complex subunit 1, mitochondrial | 2.0 | |
| O75947 | ATP5H | ATP synthase subunit d, mitochondrial | 1.9 | |
| P22695 | UQCRC2 | Cytochrome b-c1 complex subunit 2, mitochondrial | 1.7 | |
| P20674 | COX5A | Cytochrome c oxidase subunit 5A, mitochondrial | 1.6 | |
| O75489 | NDUFS3 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | 1.4 | |
| P00403 | MT-CO2 | Cytochrome c oxidase subunit 2 | 1.3 | |
| Q99623 | PHB2 | Prohibitin-2 | 1.8 | Mitochondrial protein import, and integral component of mitochondrial inner membrane |
| P35232 | PHB | Prohibitin | 1.7 | |
| P21796 | VDAC1 | Voltage-dependent anion-selective channel protein 1 | 1.7 | |
| Q9Y277 | VDAC3 | Voltage-dependent anion-selective channel protein 3 | 1.5 | |
| O43615 | TIMM44 | Mitochondrial import inner membrane translocase subunit TIM44 | 1.5 | |
| P45880 | VDAC2 | Voltage-dependent anion-selective channel protein 2 | 1.4 | |
| P08962 | CD63 | CD63 antigen | 3.9 | Lysosome-associated membrane glycoprotein, and Fibronectin matrix formation |
| P11279 | LAMP1 | Lysosome-associated membrane glycoprotein 1 | 2.3 | |
| P13473 | LAMP2 | Lysosome-associated membrane glycoprotein 2 | 2.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manganelli, V.; Salvatori, I.; Costanzo, M.; Capozzi, A.; Caissutti, D.; Caterino, M.; Valle, C.; Ferri, A.; Sorice, M.; Ruoppolo, M.; et al. Overexpression of Neuroglobin Promotes Energy Metabolism and Autophagy Induction in Human Neuroblastoma SH-SY5Y Cells. Cells 2021, 10, 3394. https://doi.org/10.3390/cells10123394
Manganelli V, Salvatori I, Costanzo M, Capozzi A, Caissutti D, Caterino M, Valle C, Ferri A, Sorice M, Ruoppolo M, et al. Overexpression of Neuroglobin Promotes Energy Metabolism and Autophagy Induction in Human Neuroblastoma SH-SY5Y Cells. Cells. 2021; 10(12):3394. https://doi.org/10.3390/cells10123394
Chicago/Turabian StyleManganelli, Valeria, Illari Salvatori, Michele Costanzo, Antonella Capozzi, Daniela Caissutti, Marianna Caterino, Cristiana Valle, Alberto Ferri, Maurizio Sorice, Margherita Ruoppolo, and et al. 2021. "Overexpression of Neuroglobin Promotes Energy Metabolism and Autophagy Induction in Human Neuroblastoma SH-SY5Y Cells" Cells 10, no. 12: 3394. https://doi.org/10.3390/cells10123394
APA StyleManganelli, V., Salvatori, I., Costanzo, M., Capozzi, A., Caissutti, D., Caterino, M., Valle, C., Ferri, A., Sorice, M., Ruoppolo, M., Garofalo, T., & Misasi, R. (2021). Overexpression of Neuroglobin Promotes Energy Metabolism and Autophagy Induction in Human Neuroblastoma SH-SY5Y Cells. Cells, 10(12), 3394. https://doi.org/10.3390/cells10123394