Abstract
Schizophrenia is a neuropsychiatric disorder characterized by dissociation of thoughts, idea, identity, and emotions. It has no central pathophysiological mechanism and precise diagnostic markers. Despite its high heritability, there are also environmental factors implicated in the development of schizophrenia. Epigenetic factors are thought to mediate the effects of environmental factors in the development of the disorder. Epigenetic modifications like DNA methylation are a risk factor for schizophrenia. Targeted gene approach studies attempted to find candidate gene methylation, but the results are contradictory. Genome-wide methylation studies are insufficient in literature and the available data do not cover different populations like the African populations. The current genome-wide studies have limitations related to the sample and methods used. Studies are required to control for these limitations. Integration of DNA methylation, gene expression, and their effects are important in the understanding of the development of schizophrenia and search for biomarkers. There are currently no precise and functional biomarkers for the disorder. Several epigenetic markers have been reported to be common in functional and peripheral tissue. This makes the peripheral tissue epigenetic changes a surrogate of functional tissue, suggesting common epigenetic alteration can be used as biomarkers of schizophrenia in peripheral tissue.
1. Introduction
Neuropsychiatric disorders are heterogeneous disorders that occur as a result of interaction of factors like genetics, epigenetics, neurobiological and environmental factors [1]. These include neurological/neurosurgical disorders and psychiatric disorders [2]. Schizophrenia is also one psychiatric disorder which results in significant socioeconomic burdens [3].
Schizophrenia is a mental illness characterized by dissociation of thoughts, ideas, identity, and emotions [4,5,6,7]. The positive and negative symptoms of schizophrenia result from dysregulated neural pathways in the brain [5,7,8]. Schizophrenia affects approximately 20 million people globally [9]. The life expectancy of individuals with schizophrenia is reduced by 15–20 years compared to the general population [10,11]. This is exacerbated by the coexistence of other disorders like cardiovascular disease (CVD), metabolic syndrome and infectious diseases such as human immunodeficiency syndrome virus (HIV) infection and acquired immunodeficiency syndrome (AIDS) [11,12,13,14]. Understanding the pathophysiology of schizophrenia may lead to better, molecular diagnosis, which may be a key for proper therapeutic interventions. This research is of great interest in countries with large burdens of disease while having several health disparities such as in Africa.
Mental health disorders such as schizophrenia are of great concern in low and middle-income countries (LMIC) like South Africa [15,16,17,18]. Researchers demonstrate that conflicts, hunger, poverty, trauma, social inequality, and poor access to health care in LMIC attribute to the increase of mental health illnesses such as schizophrenia [17,19,20,21]. Neuropsychiatric disorders are the third contributor to the burden of disease in South Africa following HIV/AIDS and other infectious diseases [20,22]. Interestingly, HIV infection is associated with neuropsychiatric disorders like schizophrenia [23]. However, the intersection between these psychiatric disorders and HIV remains poorly investigated.
The diagnosis of schizophrenia has complex heterogeneous clinical syndrome and shares the presentation with several other psychiatric disorders [24,25,26]. Although DSM-5 provides guidelines, schizophrenia diagnosis still remains complex and imprecise [24,27,28]. Both the DSM-5 and the international classification of disease (ICD-11) have incorporated symptom specifiers in the schizophrenia clinical manifestation assessment [28,29,30]. However, the diagnosis of schizophrenia is still complex and there is great need to investigate schizophrenia in order to better understand pathophysiology. Understanding schizophrenia pathophysiology may lead to the next generation of therapeutic drug intervention and molecular biomarkers like epigenetic markers (DNA methylation).
Despite growth in the research of schizophrenia, there is still no clear central pathophysiological mechanism, molecular diagnostic, or precise biomarkers [24,25,26,31]. Epigenetic markers (DNA methylation) hold promise for better understanding of schizophrenia pathophysiology and hope for precise biomarkers. The involvement of different epigenetic markers has been investigated in schizophrenia and there are different approaches that have been undertaken. The current review is on the molecular basis of schizophrenia, with a focus on the epigenetics (DNA methylation) in schizophrenia. The review outlines the different approaches to the study of DNA methylation in schizophrenia, limitations of the current literature, and future perspectives of epigenetics in schizophrenia research.
2. The Molecular Basis of Schizophrenia: The Journey So Far
Schizophrenia has an estimated heritability ranging between 79% and 81% [32,33,34]. Many chromosomes harbor a region containing a schizophrenia risk locus [35,36] and many play a role in the development of schizophrenia each with a small to moderate effect sizes [24,37]. Owing to its high heritability and environmental risk factors, schizophrenia is considered a result of gene and environment interaction [38]. Epigenetics links genetics to environmental factors [39].
Dissecting the genetic risk of schizophrenia revealed that there is a polygenic effect in the development of the disease [40,41,42]. Many schizophrenia genetic risk loci are located in the non-coding region such as introns, promoter regions thus, suggesting that gene regulation plays a critical role in the development of the disease [43]. A large number of the schizophrenia risk loci are associated with gene expression [40,44,45], and this implicated epigenetics as a mediator of genetic risk in the pathogenesis of schizophrenia [43].
3. The Role of Epigenetic Regulations in Schizophrenia: Gene and Environment Interaction
Epigenetics is the study of genetic alterations directly affecting a gene’s expression without changing the underlying DNA sequence [46,47,48]. Epigenetic mechanisms include DNA methylation, histone modification, chromosomal remodeling, and RNA regulation through non-coding RNAs such as microRNA and long non-coding RNA [49]. Epigenetic markers may shed light to understand better the interaction between genes, the environment, and health-related quantitative traits, such as cognitive function and disease outcomes in schizophrenia [50,51,52,53]. Epigenetic mechanisms regulate the brain’s biology and cognitive process [54]. During the development of the brain, both genetic and epigenetic mechanisms play a crucial role. Hence, if the environment is not suitable for proper genetic response and the epigenetic mechanism is dysregulated, it creates a risk of developing disorders such as schizophrenia [55]. Therefore, epigenetic variations as characterized by altered DNA methylation, histone modification, and microRNA expression or epi-mutation are risks for schizophrenia [56]. Several studies have looked at different epigenetic processes in schizophrenia. However, the current review will only focus on one epigenetic variation, DNA methylation.
4. DNA Methylation and Schizophrenia
DNA methylation is the most well-studied epigenetic marker compared to others [57,58]. DNA methylation occurs when the methyl groups attach covalently to the cytosine-guanine dyads (CpG) dinucleotide [59,60]. This methyl group attachment by DNA methyltransferase (DNMT) to forms the 5-methylcytosine (5mC) [61,62,63,64]. The 5mC is then converted by the ten-eleven translocase (TET) protein family to 5-hydroxymethylcytosine (5hmC), which then initiates demethylation. The 5hmC can alternatively be converted by TET to 5-formylcytocine, which can directly be converted to unmethylated cytosine or be converted to 5-carboxycytosine (5CaC). The 5CaC can be removed by base excision repair or be directly converted to unmethylated cytosine (Figure 1) [65]. Emerging research indicates that DNA methylation also occurs at non-CpG sites. DNA methylation can either induce or suppress gene expression depending on other factors and the region where it is situated [66]. To date, several genes have been assessed for DNA methylation as candidate gene for schizophrenia. Table 1 summarizes the some of the studied candidate genes for schizophrenia.
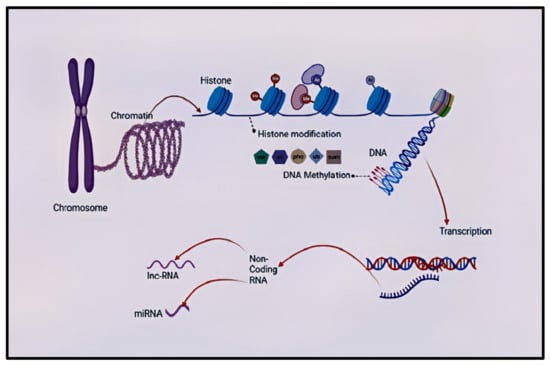
Figure 1.
Summary of epigenetic processes that can occur in the mammalian central nervous system. The DNA–protein complex in chromosomes is called a chromatin, who’s functional is a nucleosome (not shown). There is the transcriptionally accessible (found in loosely coiled chromatins) and inaccessible (found in tightly coiled chromatins) DNA. DNA interacts with the N terminal tails of the histone and this gives sites for histone modifications. Histone modification process is a covalent post-translational modification of the histone proteins, and it includes histone methylation (me), acetylation (ac), phosphorylation (pho), ubiquitination (ub), and SUMOylation (sum). Another level of epigenetic regulation is non-coding RNA. DNA methylation occurs when the methyl groups attach covalently to the cyto-sine-guanine dyads (CpG) dinucleotide and non-CpG regions of the DNA. The transcription process makes an RNA copy like mRNA (not shown) from the DNA sequence. Non-coding RNAs can be categorised into long non-coding RNAs and small non-coding RNAs (micro-RNAs). They are involved in chromatin and nuclear remodelling, gene transcription, translational repression, and degradation of messenger RNAs. DNA—deoxyribonucleic acid, RNA—ribonucleic acid, me—methylation, ac—acetylation, pho—phosphorylation, ub—ubiquitination, sum—SUMOylation, miRNA—micro RNA, lnc-RNA—long non-coding RNA.

Table 1.
Summary of some of the candidate gene methylations for schizophrenia.
There have been several genes studied as candidate genes for DNA methylation in schizophrenia patients. This review will not cover all the genes but will focus on the genes that are well covered in literature while covering all three pathways, dopaminergic, serotonergic, and GABAergic pathways. Several studies have reported changes in the DNA methylation of different gene in the functional (brain) and/or peripheral (blood) tissues of patients with schizophrenia. This studies have taken several approaches like candidate genes approach, genome wide approach and/or integrated approach. Studies have reported changes in the methylation patterns of several candidate genes. Some of the candidate gene were also reported in genome wide methylation studies. The next sections outline the changes in DNA methylation and supporting evidence from candidate gene, genome-wide and integraded approaches, respectively.
5. Gene-Specific/Candidate Gene Methylation in Schizophrenia: Changes in Candidate Gene Methylation in Schizophrenia Patients
5.1. Reelin
Reelin (RELN) gene encodes an extracellular matrix protein that is involved in cell positioning and neuronal migration during brain development [69,74]. RELN has been consistently linked to schizophrenia based on the gene function and the relationship of its variants with schizophrenia [75]. An association has been reported between the hypermethylation of RELN promoter and the down-regulation of the expression of RELN in the cortical neurons of mice [76,77], in neuroprogenitor NT2 cells [78,79] and the dorsolateral prefrontal cortex of post-mortem tissues [80]. There is contradictory evidence on methylation of RELN in the brain and peripheral tissue. Some studies have reported the hypermethylation of the RELN gene in the post-mortem brain [80,81] and in peripheral tissue [82] of patients with schizophrenia. However, other studies reported no significant difference in the methylation pattern of RELN in the brain [83] and peripheral tissue [84,85] of schizophrenia and controls. The methylation status of the RELN gene has been shown to determine the expression of RELN in different types of cells like neuronal cells [78].
The methylation of RELN gene is thought to most likely be an important factor in the regulation of RELN expression and the is also thought to be associated with psychiatric disorders [86]. This is supported by evidence of increased levels of methyl donor S-adenosylmethionine (SAM) in the prefrontal cortex of schizophrenia patients [87] and the association of RELN promoter hypermethylation with the RELN protein down-regulation [80,88,89,90,91]. A reduction in the levels of RELN gene expression and RELN protein synthesis induces both dendritic spine density deficits and cognitive impairment in an adult brain [92]
5.2. Gamma-Aminobutyric Acid
Glutamate decarboxylase 1 (GAD1) also known as glutamate decarboxylase 67 (GAD67) gene codes for an enzyme involved in the production of γ-aminobutyric acid (GABA), a major neurotransmitter of inhibitory neurons [67]. Glutamate decarboxylase 1 (GAD1) was found to be hypomethylated from the prefrontal cortex of schizophrenia cases [93]. Studies report that the downregulation of Gama-aminobutyric acid (GABA) ergic genes like glutamate decarboxylase 67 (GAD67) are mediated by hypermethylation in the frontal cortex and other brain regions. This hypermethylation occurs in the promoter region of the gene [78,80,89,90,94]. This evidence indicates the dysregulated methylation in the promoter regions of genes such as GAD67 [95].
The GAD67 promoter hypermethylation has been reported to be associated with the under expression of the gene and protein in patients with schizophrenia [89,90,96]. The altered expression of GAD67 gene is thought to lead to the impairment of working memory functions and the disturbance in cortical activity and that are evident in schizophrenia patients [97].
5.3. Catechol-O-Methyltransferase
The COMT gene codes for catechol-O-methyltransferase, which is involved in dopamine metabolism [68,98]. The COMT protein is responsible for the degradation of catecholamine like dopamine and in schizophrenia this process is disrupted [99,100]. There is evidence of an increased methylation in the promoter regions of catechol-O-methyltransferase (COMT) [101] in patients with schizophrenia. Furthermore, reports show that there is hypomethylation of membrane-bound catechol-O-methyltransferase (MB-COMT) [102,103,104], while soluble catechol-O-methyltransferase (S-COMT) is hyper-methylated [105]. However, the hypermethylation of S-COMT was not independently confirmed [106]. Other studies have reported increased methylation in the promoter regions of catechol-O-methyltransferase (COMT) [101].
An increase in COMT methylation is thought to lead a decrease in gene expression [102]. However, the potential risk of schizophrenia is thought to be related to a decreased methylation of the COMT gene [102,103,104]. It is thought that a decreased COMT methylation in schizophrenia patient leads to an increased expression and thus increased activity of COMT protein. The increased activity then causes low synaptic dopamine levels after the release of neurotransmitters and this eventually lowers dopaminergic stimulation of post-synaptic neurons [107]. When this occurs on prefrontal region (hypofrontility), it may lead to decreased executive functioning commonly seen in schizophrenia patients [108,109].
5.4. Brain-Derived Neurotrophic Factor
Brain-derived neurotrophic factor (BDNF) is a neurotrophic factor that plays a role in the inflammatory pathway in the central nervous system, and it is considered a candidate gene for the pathogenesis of schizophrenia [70,110]. BDNF is a member of the neurotrophic family growth factor supporting differentiation, maturation, and survival of neurons. It shows neuroprotective effects under adverse conditions like glutamatergic stimulation, cerebral ischemia, hypoglycemia, and neurotoxicity [111]. Studies have reported a differential methylation of the BDNF gene between patients with schizophrenia compared with controls [85,112,113]. However, other studies could not replicate the results [114,115,116]. This is thought to be due to the presence of cofounders known to affect DNA methylation like age of onset of the disease, gender and use of drugs [117,118]. This makes the standardization of samples and control of cofounders during sampling important.
Aberrant methylation of the BNDF gene has been associated with altered expression of the gene [112]. Studies reported an altered BDNF expression in the cortical areas of the brain of schizophrenia patients [112,119,120]. This alteration in turn leads to changes in protein levels known to affect dendritic growth, synaptic density, and neuronal cell size [121,122], which are implicated in schizophrenia development.
5.5. Sex Determining Region Y-Box Containing Gene 10 (SOX10)
Furthermore, genes outside of the neurotransmitter-related category have also been investigated. Sex-determining region Y (SRY)-box transcription factor 10 (SOX10) gene encodes for an oligodendrocyte-specific transcription factor [72]. The SOX10 gene is responsible oligodendrocyte differentiation [123]. The SOX10 gene regulates embryonic development and the fate of cells [124]. Iwamoto and colleagues reported the hypermethylation of SOX10 in the brain of schizophrenia patients and also found that the hypermethylation was correlated to the under expression of SOX10 [72]. Studies using post-mortem brains of schizophrenia patients have reported that the under-expression of SOX10 can lead to the dysfunction of oligodendrocytes with the downregulation of the important oligodendrocyte and myelination gene [72,125,126]. Therefore, the CpG DNA methylation status of SOX10 gene is proposed as an epigenetic sign of oligodendrocyte dysfunction in schizophrenia patients [72].
The expression of the SOX10 gene is regulated via DNA methylation [72]. Differential methylation of the SOX10 gene leads to a reduction in the expression of the gene and other oligodendrocyte-related genes [72]. The resultant under expression of SOX10 then leads to the dysfunction of the oligodendrocytes with the key oligodendrocyte and myelination genes being down-regulated has been reported in schizophrenia patients [125,126,127]. This in turn leads to the downregulation of oligodendrocyte protein evident of schizophrenia [127].
5.6. Other Genes
Several previous studies also show differential DNA methylation of genes related to the dopaminergic system, and this is in line with the dopamine hypotheses [128,129,130]. The dopamine theory proposes a hyperactive dopamine transmission in the mesolimbic regions and hypoactive dopamine transmission in the prefrontal cortex of schizophrenia patients [131,132]. Cheng and colleagues [128] have investigated differential methylation in the peripheral blood of participants with schizophrenia. This aforementioned study shows hypermethylation in the promoter region of dopamine receptor D4 (DRD4) [128]. Furthermore, Dai and colleagues have found hypermethylation in the promoter of dopamine receptor D3 (DRD3) [129]. In another line of evidence, hypomethylation of genes has also been reported. These hypomethylated genes in schizophrenia participants include dopamine receptor D2 (DRD2), DRD4 and dopamine receptor d6 (DRD6) [130]. The promoter methylation of DRD genes is thought to alter the expression of the genes and to also be involved in the development of schizophrenia [133]. Insufficient transmission of dopamine in the prefrontal cortex leads to schizophrenia-related cognitive deficits [134,135,136].
Early growth response 1 (EGR1) encodes the immediately early protein that belongs to the EGR family of cys-2-his2-type zinc-finger protein [137]. It is implicated in cell proliferation, female reproduction, immune response, cell growth, neutrophil plasticity, and memory formation [138,139]. A decrease in the EGR1 expression in the peripheral blood and prefrontal cortex of patients with schizophrenia has been reported [140,141,142]. The downregulation of EGR1 has been reported in the blood and prefrontal cortex of schizophrenia patients [140,142,143]. An upregulation of EGR1 has been found in the fibroblast and blood of schizophrenia patients [144]. Studies have reported that EGR1 gene methylation regulates the expression of the EGR1 gene [73,145]. A decreased EGR1 gene expression because of EGR1 methylation has been reported in schizophrenia patients [140,141,142]. This suggests an involvement of EGR1 gene in the development of schizophrenia [73].
Cholinergic receptor nicotinic alpha 7 (CHRNA7) gene encodes the alpha 7 nicotinic acetylcholine receptor (α7-nAChR) which is found on chromosome 15 q13.3, a region that has been identified as a schizophrenia candidate risk locus [71]. The α7-nAChR is a hemopentameric ligand-gated channel with a high permeability for calcium (Ca2+) that pre-synaptically increase neurotransmitter release from the specific terminals and post-synaptically affects gene expression [146,147]. CHRNA7 is considered a promising target for the treatment of cognitive dysfunction [148,149,150]. The CHRNA7 antagonists have been reported to improve memory and executive function in schizophrenia patients [151]. The promoter methylation of CHRNA7 gene is thought to lead to a decreased expression of the gene seen in schizophrenia patients [152]. The CHRNA7 gene is involved in the sensory processing endophenotype seen that is seen in patients with schizophrenic patients [153].However, hypermethylation of genes that regulates serotonin signaling (5-hydroxytryptamine receptor 1A (5HTR1A) and serotonin type 2A receptor (HTR2A) receptors and serotonin transporter (5-HTT) were found to be associated with schizophrenia [154,155,156]. An increase in 5-HT1A and decrease in 5-HT2A receptor densities in the dorsolateral prefrontal cortex is associated with both positive and negative symptoms of schizophrenia [157]. Brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2) is responsible for dendritic spine density abnormalities [158]. The hypomethylation of BAIAP2 has been reported in schizophrenia patients [158]. Any reduction in the level of BAIAP2 are associated with neurological disorder and memory formation deficits [159,160]. BAIAP2 participates in the proliferation, survival, and maturation of neural cells [161].
Parvalbumin (PVALB) gene encodes a high affinity calcium ion-binding protein. Deficits of brain parvalbumin (PV) are a consistent finding in schizophrenia [162]. Promoter methylation of the PVALB gene has been reported in rats that underwent schizophrenia induction using a sub chronic regime of phencyclidine (PCP) [163]. An increase in the PVALB gene promoter methylation was found to be increased in the hippocampus in schizophrenia patients’ post-mortem brain tissue. Since promoter hypermethylation can lead to a reduced gene expression, the reduced expression of PV gene in the brain of schizophrenia patients is thought to be a result of the methylation of the PV gene [162].
Peripheral blood showed hypermethylation of cytotoxic T-lymphocyte-associated protein 4 (CTLA4), which is involved in immune function, formation and maintaining peripheral tolerance of T Cells [130]. Promoter and regulatory methylation of the CTLA4 gene has been reported to alter the expression of the gene [164]. Patients with schizophrenia show alterations in cytokine production and T cell proliferation [165]. The oxytocin receptor gene (OXTR) encodes for the oxytocin receptor, which is a key element of the oxytocin system. OXTR is hypermethylated in peripheral blood [166]. Differential methylation of OXTR is thought to lead to an under-expression of oxytocin receptors [167] and oxytocin is proposed to regulate the central dopaminergic system implicated in the behavioural manifestations of schizophrenia [168,169].
Initial DNA methylation studies focused on DNA methylation alterations in candidate genes. The results from candidate gene studies are conflicted. The candidate gene approach was limited by its coverage of the genome and as such authors resorted to genome wide DNA methylation approach for a more comprehensive coverage.
6. Genome-Wide Methylation Studies in Schizophrenia: Evidence of Changes in DNA Methylation in Schizophrenia Patient
Genome-wide investigations to determine variations on methylation to provide an insight into mental health disorders like schizophrenia is insufficiently in the literature despite emerging data [170]. The first genome-wide DNA methylation study reported significant epigenetic changes associated with schizophrenia land bipolar disorder in the prefrontal cortex of patients with major psychosis using a microarray [171]. This was subsequently confirmed by Dempster and colleagues in the blood samples from 22 twin pairs discordant for SC and BP using microarray [172]. Following that, there was an increase in genome wide DNA methylation studies using different methods.
Genome-wide methylation studies by nature cover a wide range of differentially methylated patterns and due to that, they are known to have a complication with replication of results. There has been a problem in replicating the results of genome-wide studies. Despite this, there have been some common methylation patterns that were noted with genome-wide studies. The glutamate ionotropic receptor alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA) type subunit 1 (GRIA1) gene methylation was reported by two different studies [171,173]. The GRIA1 gene codes for one of the four ionotropic AMPA receptor subunits and is involved in synaptic plasticity [174]. Some of the genes reported by genome-wide studies are the known candidate genes for schizophrenia. These genes include RELN [175,176], COMT [176], DTNBP1 [177] and SOX10 [177].
Genome-wide investigations to determine variations on methylation to provide an insight into mental health disorders like schizophrenia are insufficiently reported in the literature despite emerging data [170]. Also, different populations of different demography are not yet covered, especially African populations. Furthermore, most mental health conditions like schizophrenia consist of the polygenetic effect of pleiotropic genes. Therefore, precise etiology to infer specific molecular basis like genetic or epigenetic variations is lacking [170]. To further understand the molecular basis of schizophrenia, it is essential to investigate different aspects of the molecular basis of the disease, from genes to their regulations such as epigenetics and their expression [178,179,180,181]. This line of research is critical as it may provide a direction for precision medicine for mental health disorders like schizophrenia. Therefore, genome-wide epigenetics and gene expression may shed light to understand the diseases better as well as provide insight on prognosis, diagnosis, and treatment outcomes.
Despite the use of genome-wide methylation studies, there was still a problem with identifying consistent schizophrenia specific DNA methylation patterns. This is due to the lack of reproducibility of results in genome-wide studies. This has been attributed to several limitations of studies that have been conducted thus far. Most of the genome-wide methylation studies used methylation arrays which are by design limited to the already known DMR and as such will not cover the entire genome. Most of the arrays only cover CpG regions and it has been noted that non-CpG methylation occurs. Some of the studies also did not control for the cell heterogeneity, age of diagnosis, cause of death in post-mortem brains, use of medication, and age of patients.
7. Integrated DNA Methylation and Gene Expression: Multi-Omic Approach—Towards Precision Medicine
Based on the current data from genome-wide association studies and epigenome-wide association studies, it has become more apparent that a single omics approach might not be adequate in the search for mental health disorder development and potential biomarker discovery. This makes multi-omics/integrated studies of utmost importance in the quest to understand mental health disorders.
Zhu and colleagues reported that the DNA methylation to be correlated with gene expression in the peripheral blood of monocytes of monozygotic discordant twins [182]. Another intergraded study using whole blood DNA methylation and gene expression data from the Gene Expression Omnibus database reported the identification of blood-based signature with 46 hypo-up and 71 hyper-down genes and the authors posits that the genes may have a potential role in the diagnosis of major depressive disorder [183]. Another study by van Eijk and colleagues identified 1095 differentially methylated regions associated with 1226 differentially expressed genes. They also reported that half of the transcripts were showing differential expression in the peripheral blood of schizophrenia patients [184].
Integration of DNA methylation, gene expression, and the consideration of its global effects are essential in the understanding of the mechanism with which DNA methylation leads to the development of schizophrenia [185]. A significant increase in the expression of miRNAs has been reported in schizophrenia patients [186].
A cell-type-specific study integrating the methylome and transcriptome of distinct cells from post-mortem brain tissues showed the importance of cell and tissue type epigenetics and the importance of a whole-genome approach [43]. The study was however limited using post-mortem human brain tissue due to the cofounders related to the use of such tissues [43,187,188]. Literature also shows that the bulk of the studies on epigenetics of schizophrenia and psychosis suffer from methodological limitations. Most of the studies used pre-defined microarrays, which traditionally doesn’t cover the whole genome thus making an unbiased, whole-genome DNA methylation approach covering methylation even outside promoter region and CpG island very important in the understanding of the role of Epigenetics in mental disorders [43,189,190].
Most of the studies looking into the involvement of non-coding RNA (ncRNAs) in schizophrenia focused on microRNAs other than other ncRNAs [185,191,192]. The expression of miR137 has been reported to be altered in schizophrenia [42,193,194]. miR137 expression is essential in several signaling nodes in several gene networks that are relevant to the development and function of the brain [195,196].
An integrative study reported an overlap of intronic deferentially methylated region with miRNA. The study also reported that the aberrant DNA methylation-related miRNAs were differentially expressed thus suggesting that DNA methylation may be affecting gene expression and eventual protein expression via miRNAs [185].
Several studies have profiled the expression of miRNAs in the blood of schizophrenia patients to discover blood-based biomarkers for schizophrenia [197,198]. A recent genome-wide expression study reported that several miRs were differentially expressed in the serum of schizophrenia patients. The study reported 11 miRs that could differentiate the schizophrenia patients from controls [197].
More studies are required that intergrade different omics. Multi-omics studies will generate valuable data in the understanding of the pathophysiology of schizophrenia and possible search for biomarkers. The studies are required to account for the limitation seen in current literature. There are different limitation and cofounders seen in the current studies of DNA methylation in schizophrenia. The next section outlines the limitation and cofounders and their influence of DNA methylation. The next section also outlines the use of alternative models of schizophrenia, like animal model, in addressing limitation and controlling for cofounders.
8. Limitations of Previous Studies and Future Perspectives: Where to from Here?
Several studies have been completed in search of the potential role of epigenetics in the development of schizophrenia. The studies have yielded a lot of potential data in this regard but there is a problem with reproducibility [170]. This has been attributed to several limitations of studies that have been conducted thus far. The limitations are small sample size, the type of sample used for analysis [67], use of medication [118], smoking [117], tissue and cell-type heterogeneity [199] and methods of studying the epigenetics [54].
Epigenetics studies have widely been performed using post-mortem brain [171,177,200] and peripheral blood samples [103,173,175] this is thought to be due to the relative ease in obtaining these samples compared to brain tissue from live patients. The use of these samples introduces cofounders to the study like smoking tobacco, alcohol use, disease course, use of medication, age, infections, time, and cause of death [117,118]. Also, the epigenetic profile of the brain and peripheral tissue is different [201] and schizophrenia is a disorder of the brain [202]. All the above cofounders have been shown to influence the epigenome and transcriptome [124]. Although some of the cofounder like smoking has been shown to alter global DNA methylation [203], the control of these cofounders is required for a clear picture of the effect of methylation on schizophrenia. The use of alternative samples like samples from animal models or induced cell models may assist in the control of these cofounders.
One of the limitations is the use of medications. Studies have shown that antipsychotics affect the patterns of DNA methylation [204,205,206,207]. Olanzapine has been found to change DNA methylation patterns of the brains of mice [207], and clozapine was found to change DNA methylation in human peripheral leukocyte [208]. Studies have reported that antipsychotics may affect the promoter methylation of genes related to schizophrenia [206,207]. Antipsychotic drugs can alter the methylation patterns of schizophrenia genes and related gene expression, however, certain methylation patterns prior to the use of antipsychotics can affect the influence the efficacy of antipsychotics [105,209]. Studies are required to account for the use of medication as medication can alter the methylation patterns. However, with the use of post-mortem brain tissue, the control for use of medication is limited. This brings make the use of animal and cell model for schizophrenia of importance as they present a chance for treatment naïve schizophrenia model.
Another major limitation in previous studies is the tissue and cell-type heterogeneity in samples used for analysis [199]. It has been reported that different cell types have different epigenetic patterns [210,211,212]. There are considerations regarding the stability and biological implications of the epigenetic measurements in post-mortem tissue as changes in DNA methylation have been reported with post-mortem interval [43,187,188]. The use of mixed tissue and cell type samples limits the identification of epigenetic and transcriptomic changes in schizophrenia as the changes are masked by changes in other tissue and cell types [124]. Studies are required to control for cell and tissue type heterogeneity in the sampling process to give a clear picture of DNA methylation in different parts and cell of the brain schizophrenia. This will help with the pathophysiology of schizophrenia and with evidence needed for the development of new therapeutic targets and diagnostic markers.
Despite the different approaches used, there still is limited reproducibility and this can also be attributed to the method used to assess DNA methylation in different studies [54]. The methodological limitations are related to the different coverages of the available methods. Targeted approaches are by definition limited in their coverage and will only pick up methylation in targeted genes [213]. To deal with this limitation, genome-wide approaches are used as they cover the whole genome methylation [171,172]. This however is also limited based on the coverage of the platform used. Most genome-wide studies are using methylation arrays, which use predefined probes for the detection of methylation, which is biased in their analysis of methylation [43,189,190]. An unbiased genome-wide methylation approach has revealed methylation outside the promoter regions and non-CpG methylation are important [43]. For an unbiased picture of DNA methylation in schizophrenia patients, studies are required to look at CpG and non-CpG genome wide methylation. Such studies will provide a holistic picture thus enabling further understanding of the pathophysiology of schizophrenia and assisting in the search for diagnostic biomarkers.
To understand the potential involvement of the epigenome in the development of schizophrenia, studies that control for the above-mentioned limitation are needed. The studies need to control for, in sampling, the sample type, tissue, and cell-type heterogeneity. Studies should also use methods that have a broader coverage like sequencing [43]. To complete the search for potential biomarkers for schizophrenia, studies with defined tissue and cell types should compare differentially methylated region in brain tissue and peripheral blood tissue as the common epigenetic pattern in both brain and blood could serve as a potential biomarker [140,177,214].
Data from schizophrenia animal models shows added evidence of the effect of environmental factors on epigenetic [215]. Different animal models have been used and are based on factors implicated in schizophrenia like maternal immune activation during pregnancy, inhibitors of foetal neurogenesis, pre- and post-natal stress, nutritional deficiencies, drug abuse, exposure to toxicants, reduced postpartum maternal care, and cannabis use in adolescence. The models capture a wide spectrum of neurological and behavioural changes related to schizophrenia and shows difference in several epigenetic markers [124,216]. The use of schizophrenia animal models enables the comparison of epigenetic markers like DNA methylation in functional and peripheral tissues and different stages of development [213] and testing target sequences that are influenced by changes in the chromatin and to also test their effects on the development and functioning of the brain [217]. The use of animal models enables researchers to study mental health using samples that are free from cofounders like use of medication, smoking, alcohol use, cause of death, infections, cannabis use and disease course.
Several limitations have been noted with current studies and new studies are required to control for these limitations. The use of animal models for schizophrenia is of importance in addressing some of the limitations. Control of cofounders like age, use of medications, smoking can be relatively challenging with the use of post-mortem brain tissue. The use of animal models is necessary for the control of these cofounders.
9. Possibility of Blood-Based Biomarker: Towards a Laboratory Screening, Diagnosis, and/or Monitoring Tool
A biological marker, or biomarker, is a trait that can be evaluated and measured as an indicator of biological process, pathological process, or treatment response [218]. There are currently no precise and functional biomarkers for neuropsychiatric disorders like schizophrenia [219]. Studies of epigenetic changes in schizophrenia hold promise for the development of diagnostic and prognostic biomarkers for schizophrenia and a therapeutic target [177]. To achieve this goal, studies are required that will identify aberrant DNA methylation profiles in functional tissue and determine if the results are translatable to diagnostically feasible tissue [177].
Methylation status changes in peripheral tissue like blood are thought to mirror methylation changes in the brain [220]. Some methylation markers were found to be similarly altered in both brain and peripheral tissue. Several studies have attempted to compare the methylation patterns among brain and peripheral tissue and these studies identified about 2–7% of CpG sites that showed a correlation between the brain and peripheral tissue [201,221,222,223]. This suggests that such common epigenetic alterations may be used as potential biomarkers for schizophrenia. Peripheral epigenetics are important in the identification of biomarkers, but the epigenetics signature of the brain isn’t a mirror image of the peripheral tissue. This means that common changes must be found and replicated between the brain and peripheral tissue.
Memories are stored at the molecular level and in the functional organization of the brain and mind, combining to alter cognition, behavior, and clinical symptoms [224,225]. Since all memories are controlled by molecular mechanisms, they require gene expression changes and protein synthesis. These proteins are encoded by genes which can be regulated via epigenetic modification [226,227,228]. The diagnostic procedure of schizophrenia is difficult, and this is because it’s a heterogeneous clinical syndrome [24,27]. Although the current diagnosis incorporated symptom specifiers [28,29,30], the diagnosis of schizophrenia is still difficult and imprecise [24,27]. Thus, making it necessary to discover new, better ways, including biomarkers for diagnosis of schizophrenia. Studies are required that will compare DNA methylation in functional tissue with peripheral tissues to assist in the search for biomarkers.
10. Conclusions
Based on the results and limitations from the current literature, we envisage the need for a tissue and cell type specific, unbiased whole genome DNA methylation study integrated with gene expression, considering global effects of DNA methylation (miRNAs). The study will need to control for confounders related to the use of post-mortem tissue and compare the epigenetics of the CNS tissue and non-CNS tissue for biomarker discovery. Peripheral epigenetics are important in the identification of biomarkers, but the epigenetics signature of the brain is not a mirror image of the peripheral tissue. This means that the common changes must be found and replicated between brain and peripheral tissue.
Author Contributions
T.M. (Thabo Magwai), K.R.X. and T.M. (Thabisile Mpofana) conceived the concept. T.M. (Thabo Magwai) conducted literature review, and wrote the manuscript draft. K.R.X., T.M. (Thabisile Mpofana), B.C., K.B.S. and F.O.O. revised and added a part. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Research Foundation Thuthuka grant number [118049], National Research Foundation Thuthuka grant number [129827] and Developing Research Innovation, Localisation and Leadership in South Africa (DRILL), grant number [D43TW010131]. The APC was funded by [D43TW010131].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lin, C.C.; Huang, T.L. Epigenetic Biomarkers in Neuropsychiatric Disorders. In Neuropsychiatric Disorders and Epigenetics; Yasui, D.H., Peedicayil, J., Grayson, D.R., Eds.; Academic Press: Boston, MA, USA, 2017; Chapter 3; pp. 35–66. [Google Scholar] [CrossRef]
- Peedicayil, J.; Grayson, D.R.; Yasui, D.H. Introduction to Neuropsychiatric Disorders and Epigenetics. In Neuropsychiatric Disorders and Epigenetics; Yasui, D.H., Peedicayil, J., Grayson, D.R., Eds.; Academic Press: Boston, MA, USA, 2017; Chapter 1; pp. 3–8. [Google Scholar] [CrossRef]
- Stuchlik, A.; Sumiyoshi, T. Cognitive Deficits in Schizophrenia and Other Neuropsychiatric Disorders: Convergence of Preclinical and Clinical Evidence. Front. Behav. Neurosci. 2014, 8, 444. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Birnbaum, R.; Weinberger, D.R. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci. 2017, 18, 727–740. [Google Scholar] [CrossRef]
- Kim, J.S.; Shin, K.S.; Jung, W.H.; Kim, S.N.; Kwon, J.S.; Chung, C.K. Power spectral aspects of the default mode network in schizophrenia: An MEG study. BMC Neurosci. 2014, 15, 104. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, A.; Heim, G. Eugen Bleuler’s Dementia Praecox or the Group of Schizophrenias (1911): A Centenary Appreciation and Reconsideration. Schizophr. Bull. 2011, 37, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Owen, M.J.; Sawa, A.; Mortensen, P.B. Schizophrenia. Lancet 2016, 388, 86–97. [Google Scholar] [CrossRef]
- Howes, O.D.; Murray, R.M. Schizophrenia: An integrated sociodevelopmental-cognitive model. Lancet 2014, 383, 1677–1687. [Google Scholar] [CrossRef]
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- Laursen, T.M.; Wahlbeck, K.; Hällgren, J.; Westman, J.; Ösby, U.; Alinaghizadeh, H.; Gissler, M.; Nordentoft, M. Life expectancy and death by diseases of the circulatory system in patients with bipolar disorder or schizophrenia in the Nordic countries. PLoS ONE 2013, 8, e67133. [Google Scholar] [CrossRef]
- Laursen, T.M.; Nordentoft, M.; Mortensen, P.B. Excess early mortality in schizophrenia. Ann. Rev. Clin. Psychol. 2014, 10, 425–448. [Google Scholar] [CrossRef]
- Mere, S.M.; Paruk, S. A chart review of human immunodeficiency virus status in patients admitted with psychosis in Durban, South Africa. S. Afr. J. Psychiatry 2018, 24, 1129. [Google Scholar] [CrossRef]
- Saloojee, S.; Burns, J.K.; Motala, A.A. Very low rates of screening for metabolic syndrome among patients with severe mental illness in Durban, South Africa. BMC Psychiatry 2014, 14, 228. [Google Scholar] [CrossRef][Green Version]
- Saloojee, S.; Burns, J.K.; Motala, A.A. Metabolic Syndrome in South African Patients with Severe Mental Illness: Prevalence and Associated Risk Factors. PLoS ONE 2016, 11, e0149209. [Google Scholar] [CrossRef]
- Alloh, F.T.; Regmi, P.; Onche, I.; Van Teijlingen, E.; Trenoweth, S. Mental Health in low-and middle income countries (LMICs): Going beyond the need for funding. Health Prospect J. Public Health 2018, 17, 12–17. [Google Scholar] [CrossRef]
- Mackenzie, J.; Kesner, C. Mental Health Funding and the SDGs: What Now and Who Pays? 2016. Available online: https://cdn.odi.org/media/documents/Mental_health_funding_and_the_SDGs_what_next_and_who_pays.pdf (accessed on 12 August 2021).
- Rathod, S.; Pinninti, N.; Irfan, M.; Gorczynski, P.; Rathod, P.; Gega, L.; Naeem, F. Mental Health Service Provision in Low- and Middle-Income Countries. Health Serv. Insights 2017, 10, 1–7. [Google Scholar] [CrossRef]
- Wang, P.S.; Aguilar-Gaxiola, S.; Alonso, J.; Angermeyer, M.C.; Borges, G.; Bromet, E.J.; Bruffaerts, R.; De Girolamo, G.; De Graaf, R.; Gureje, O. Use of mental health services for anxiety, mood, and substance disorders in 17 countries in the WHO world mental health surveys. Lancet 2007, 370, 841–850. [Google Scholar] [CrossRef]
- Alloh, F.T.; Regmi, P.R. Effect of economic and security challenges on the Nigerian health sector. Afr. Health Sci. 2017, 17, 591–592. [Google Scholar] [CrossRef]
- Department of Health. National Mental Health Policy Framework and Strategic Plan 2013–2020; Department of Health Pretoria: Pretoria, South Africa, 2013. [Google Scholar]
- Mugagga, F.; Nabaasa, B.B. The centrality of water resources to the realization of Sustainable Development Goals (SDG). A review of potentials and constraints on the African continent. Int. Soil Water Conserv. Res. 2016, 4, 215–223. [Google Scholar] [CrossRef]
- Bradshaw, D.; Norman, R.; Schneider, M. A clarion call for action based on refined DALY estimates for South Africa. S. Afr. Med. J. 2007, 97, 438–440. [Google Scholar] [CrossRef]
- Nebhinani, N.; Mattoo, S.K. Psychotic disorders with HIV infection: A review. Ger. J. Psychiatry 2013, 16, 43–48. [Google Scholar]
- Wong, A.H.C.; Van Tol, H.H. Schizophrenia: From phenomenology to neurobiology. Neurosci. Biobehav. Rev. 2003, 27, 269–306. [Google Scholar] [CrossRef]
- Kaur Multani, P.; Saini, N.; Kaur, R.; Sharma, P. Biomarkers for Drugs of Abuse and Neuropsychiatric Disorders: Models and Mechanisms. In Biomarkers in Toxicology; Gupta, R.C., Ed.; Academic Press: Boston, MA, USA, 2014; Chapter 59; pp. 983–1001. [Google Scholar] [CrossRef]
- Oh, S.L.; Vicnesh, J.; Ciaccio, E.J.; Yuvaraj, R.; Acharya, U.R. Deep Convolutional Neural Network Model for Automated Diagnosis of Schizophrenia Using EEG Signals. Appl. Sci. 2019, 9, 2870. [Google Scholar] [CrossRef]
- Keller, W.R.; Fischer, B.A.; Carpenter, J.; William, T. Revisiting the diagnosis of schizophrenia: Where have we been and where are we going? CNS Neurosci. Ther. 2011, 17, 83–88. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®), 5th ed.; American Psychiatric Pub: Washington, DC, USA, 2013. [Google Scholar]
- Valle, R. Schizophrenia in ICD-11: Comparison of ICD-10 and DSM-5. Rev. Psiquiatr. Salud Ment. 2020, 13, 95–104. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. International Classification of Diseases for Mortality and Morbidity Statistics (11th Revision); World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Perkovic, M.N.; Erjavec, G.N.; Strac, D.S.; Uzun, S.; Kozumplik, O.; Pivac, N. Theranostic Biomarkers for Schizophrenia. Int. J. Mol. Sci. 2017, 18, 733. [Google Scholar] [CrossRef] [PubMed]
- Hilker, R.; Helenius, D.; Fagerlund, B.; Skytthe, A.; Christensen, K.; Werge, T.M.; Nordentoft, M.; Glenthøj, B. Heritability of schizophrenia and schizophrenia spectrum based on the nationwide Danish twin register. Biol. Psychiatry 2018, 83, 492–498. [Google Scholar] [CrossRef]
- Lichtenstein, P.; Yip, B.H.; Björk, C.; Pawitan, Y.; Cannon, T.D.; Sullivan, P.F.; Hultman, C.M. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: A population-based study. Lancet 2009, 373, 234–239. [Google Scholar] [CrossRef]
- Sullivan, P.F.; Kendler, K.S.; Neale, M.C. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 2003, 60, 1187–1192. [Google Scholar] [CrossRef]
- Henriksen, M.G.; Nordgaard, J.; Jansson, L.B. Genetics of schizophrenia: Overview of methods, findings and limitations. Front. Hum. Neurosci. 2017, 11, 322. [Google Scholar] [CrossRef]
- Ng, M.Y.; Levinson, D.F.; Faraone, S.V.; Suárez, B.K.; DeLisi, L.E.; Arinami, T.; Riley, B.; Paunio, T.; Pulver, A.E.; Holmans, P.A. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol. Psychiatry 2009, 14, 774–785. [Google Scholar] [CrossRef]
- Modinos, G.; Iyegbe, C.; Prata, D.; Rivera, M.; Kempton, M.J.; Valmaggia, L.R.; Sham, P.C.; van Os, J.; McGuire, P. Molecular genetic gene–environment studies using candidate genes in schizophrenia: A systematic review. Schizophr. Res. 2013, 150, 356–365. [Google Scholar] [CrossRef]
- Karl, T.; Arnold, J.C. Schizophrenia: A consequence of gene-environment interactions? Front. Behav. Neurosci. 2014, 8, 435. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Gusev, A.; Mancuso, N.; Won, H.; Kousi, M.; Finucane, H.K.; Reshef, Y.; Song, L.; Safi, A.; McCarroll, S.; Neale, B.M. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018, 50, 538–548. [Google Scholar] [CrossRef]
- Loh, P.R.; Bhatia, G.; Gusev, A.; Finucane, H.K.; Bulik-Sullivan, B.K.; Pollack, S.J.; de Candia, T.R.; Lee, S.H.; Wray, N.R.; Kendler, K.S.; et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015, 47, 1385–1392. [Google Scholar] [CrossRef]
- Ripke, S.; Neale, B.M.; Corvin, A.; Walters, J.T.R.; Farh, K.-H.; Holmans, P.A.; Lee, P.; Bulik-Sullivan, B.; Collier, D.A.; Huang, H.; et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef]
- Mendizabal, I.; Berto, S.; Usui, N.; Toriumi, K.; Chatterjee, P.; Douglas, C.; Huh, I.; Jeong, H.; Layman, T.; Tamminga, C.A.; et al. Cell type-specific epigenetic links to schizophrenia risk in the brain. Genome Biol. 2019, 20, 135. [Google Scholar] [CrossRef]
- Fromer, M.; Roussos, P.; Sieberts, S.K.; Johnson, J.S.; Kavanagh, D.H.; Perumal, T.M.; Ruderfer, D.M.; Oh, E.C.; Topol, A.; Shah, H.R. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016, 19, 1442–1453. [Google Scholar] [CrossRef]
- Jaffe, A.E.; Straub, R.E.; Shin, J.H.; Tao, R.; Gao, Y.; Collado-Torres, L.; Kam-Thong, T.; Xi, H.S.; Quan, J.; Chen, Q. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci. 2018, 21, 1117–1125. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Gayon, J. From Mendel to epigenetics: History of genetics. Comptes Rendus Biol. 2016, 339, 225–230. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A Landscape Takes Shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Kuehner, J.N.; Bruggeman, E.C.; Wen, Z.; Yao, B. Epigenetic Regulations in Neuropsychiatric Disorders. Front. Genet. 2019, 10, 268. [Google Scholar] [CrossRef] [PubMed]
- Bakulski, K.M.; Fallin, M.D. Epigenetic epidemiology: Promises for public health research. Environ. Mol. Mutagenesis 2014, 55, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Kular, L.; Kular, S. Epigenetics applied to psychiatry: Clinical opportunities and future challenges. Psychiatry Clin. Neurosci. 2018, 72, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Down, T.A.; Balding, D.J.; Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011, 12, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Wiechmann, T.; Gassen, N.C.; Binder, E.B. Gene-Stress-Epigenetic Regulation of FKBP5: Clinical and Translational Implications. Neuropsychopharmacology 2016, 41, 261–274. [Google Scholar] [CrossRef]
- Dempster, E.; Viana, J.; Pidsley, R.; Mill, J. Epigenetic studies of schizophrenia: Progress, predicaments, and promises for the future. Schizophr. Bull. 2013, 39, 11–16. [Google Scholar] [CrossRef]
- Hoffmann, A.; Sportelli, V.; Ziller, M.; Spengler, D. Epigenomics of major depressive disorders and schizophrenia: Early life decides. Int. J. Mol. Sci. 2017, 18, 1711. [Google Scholar] [CrossRef]
- Nestler, E.J.; Peña, C.J.; Kundakovic, M.; Mitchell, A.; Akbarian, S. Epigenetic basis of mental illness. Neuroscientist 2016, 22, 447–463. [Google Scholar] [CrossRef]
- Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007, 447, 425–432. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Zeng, Y.; Chen, T. DNA Methylation Reprogramming during Mammalian Development. Genes 2019, 10, 257. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Grayson, D.R. Schizophrenia and the epigenetic hypothesis. Epigenomics 2010, 2, 341–344. [Google Scholar] [CrossRef]
- Grayson, D.R.; Guidotti, A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology 2013, 38, 138–166. [Google Scholar] [CrossRef]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and function of mammalian DNA methyltransferases. Chembiochem 2011, 12, 206–222. [Google Scholar] [CrossRef]
- Adachi, M.; Monteggia, L.M. Decoding transcriptional repressor complexes in the adult central nervous system. Neuropharmacology 2014, 80, 45–52. [Google Scholar] [CrossRef][Green Version]
- Gavin, D.P.; Chase, K.A.; Sharma, R.P. Active DNA demethylation in post-mitotic neurons: A reason for optimism. Neuropharmacology 2013, 75, 233–245. [Google Scholar] [CrossRef]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef]
- Nishioka, M.; Bundo, M.; Kasai, K.; Iwamoto, K. DNA methylation in schizophrenia: Progress and challenges of epigenetic studies. Genome Med. 2012, 4, 96. [Google Scholar] [CrossRef]
- Martinez-Jauand, M.; Sitges, C.; Rodriguez, V.; Picornell, A.; Ramon, M.; Buskila, D.; Montoya, P. Pain sensitivity in fibromyalgia is associated with catechol-O-methyltransferase (COMT) gene. Eur. J. Pain 2013, 17, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Sozuguzel, M.D.; Sazci, A.; Yildiz, M. Female gender specific association of the Reelin (RELN) gene rs7341475 variant with schizophrenia. Mol. Biol. Rep. 2019, 46, 3411–3416. [Google Scholar] [CrossRef]
- Janicijevic, S.M.; Dejanovic, S.D.; Borovcanin, M. Interplay of brain-derived neurotrophic factor and cytokines in schizophrenia. Serb. J. Exp. Clin. Res. 2018, 108, 110–117. [Google Scholar] [CrossRef]
- Leonard, S.; Freedman, R. Genetics of chromosome 15q13–q14 in schizophrenia. Biol. Psychiatry 2006, 60, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, K.; Bundo, M.; Yamada, K.; Takao, H.; Iwayama-Shigeno, Y.; Yoshikawa, T.; Kato, T. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J. Neurosci. 2005, 25, 5376–5381. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.-M.; Chen, S.-J.; Hsu, S.-H.; Cheng, M.-C. Functional analyses and effect of DNA methylation on the EGR1 gene in patients with schizophrenia. Psychiatry Res. 2019, 275, 276–282. [Google Scholar] [CrossRef]
- Niu, S.; Yabut, O.; D’Arcangelo, G. The Reelin signaling pathway promotes dendritic spine development in hippocampal neurons. J. Neurosci. 2008, 28, 10339–10348. [Google Scholar] [CrossRef]
- Li, W.; Guo, X.; Xiao, S. Evaluating the relationship between reelin gene variants (rs7341475 and rs262355) and schizophrenia: A meta-analysis. Neurosci. Lett. 2015, 609, 42–47. [Google Scholar] [CrossRef]
- Dong, E.; Agis-Balboa, R.; Simonini, M.; Grayson, D.; Costa, E.; Guidotti, A. Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc. Natl. Acad. Sci. USA 2005, 102, 12578–12583. [Google Scholar] [CrossRef]
- Noh, J.S.; Sharma, R.P.; Veldic, M.; Salvacion, A.A.; Jia, X.; Chen, Y.; Costa, E.; Guidotti, A.; Grayson, D.R. DNA methyltransferase 1 regulates reelin mRNA expression in mouse primary cortical cultures. Proc. Natl. Acad. Sci. USA 2005, 102, 1749–1754. [Google Scholar] [CrossRef]
- Chen, Y.; Sharma, R.P.; Costa, R.H.; Costa, E.; Grayson, D.R. On the epigenetic regulation of the human reelin promoter. Nucleic Acids Res. 2002, 30, 2930–2939. [Google Scholar] [CrossRef]
- Mitchell, C.P.; Chen, Y.; Kundakovic, M.; Costa, E.; Grayson, D.R. Histone deacetylase inhibitors decrease reelin promoter methylation in vitro. J. Neurochem. 2005, 93, 483–492. [Google Scholar] [CrossRef]
- Abdolmaleky, H.M.; Cheng, K.H.; Russo, A.; Smith, C.L.; Faraone, S.V.; Wilcox, M.; Shafa, R.; Glatt, S.J.; Nguyen, G.; Ponte, J.F. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: A preliminary report. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2005, 134, 60–66. [Google Scholar] [CrossRef]
- Eastwood, S.; Harrison, P. Interstitial white matter neurons express less reelin and are abnormally distributed in schizophrenia: Towards an integration of molecular and morphologic aspects of the neurodevelopmental hypothesis. Mol. Psychiatry 2003, 8, 821–831. [Google Scholar] [CrossRef]
- Fikri, R.M.N.; Norlelawati, A.T.; El-Huda, A.R.N.; Hanisah, M.N.; Kartini, A.; Norsidah, K.; Zamzila, A.N. Reelin (RELN) DNA methylation in the peripheral blood of schizophrenia. J. Psychiatr. Res. 2017, 88, 28–37. [Google Scholar] [CrossRef]
- Tochigi, M.; Iwamoto, K.; Bundo, M.; Komori, A.; Sasaki, T.; Kato, N.; Kato, T. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol. Psychiatry 2008, 63, 530–533. [Google Scholar] [CrossRef]
- Bönsch, D.; Wunschel, M.; Lenz, B.; Janssen, G.; Weisbrod, M.; Sauer, H. Methylation matters? Decreased methylation status of genomic DNA in the blood of schizophrenic twins. Psychiatry Res. 2012, 198, 533–537. [Google Scholar] [CrossRef]
- Ikegame, T.; Bundo, M.; Sunaga, F.; Asai, T.; Nishimura, F.; Yoshikawa, A.; Kawamura, Y.; Hibino, H.; Tochigi, M.; Kakiuchi, C. DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci. Res. 2013, 77, 208–214. [Google Scholar] [CrossRef]
- Tamura, Y.; Kunugi, H.; Ohashi, J.; Hohjoh, H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol. Psychiatry 2007, 12, 593–600. [Google Scholar] [CrossRef]
- Guidotti, A.; Ruzicka, W.; Grayson, D.R.; Veldic, M.; Pinna, G.; Davis, J.M.; Costa, E. S-adenosyl methionine and DNA methyltransferase-1 mRNA overexpression in psychosis. NeuroReport 2007, 18, 57–60. [Google Scholar] [CrossRef]
- Grayson, D.R.; Chen, Y.; Costa, E.; Dong, E.; Guidotti, A.; Kundakovic, M.; Sharma, R.P. The human reelin gene: Transcription factors (+), repressors (−) and the methylation switch (+/−) in schizophrenia. Pharmacol. Ther. 2006, 111, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Grayson, D.R.; Jia, X.; Chen, Y.; Sharma, R.P.; Mitchell, C.P.; Guidotti, A.; Costa, E. Reelin promoter hypermethylation in schizophrenia. Proc. Natl. Acad. Sci. USA 2005, 102, 9341–9346. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, A.; Auta, J.; Davis, J.M.; Gerevini, V.D.; Dwivedi, Y.; Grayson, D.R.; Impagnatiello, F.; Pandey, G.; Pesold, C.; Sharma, R. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: A postmortem brain study. Arch. Gen. Psychiatry 2000, 57, 1061–1069. [Google Scholar] [CrossRef]
- Lintas, C.; Persico, A.M. Neocortical RELN promoter methylation increases significantly after puberty. NeuroReport 2010, 21, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, A.; Grayson, D.R.; Caruncho, H.J. Epigenetic RELN Dysfunction in Schizophrenia and Related Neuropsychiatric Disorders. Front. Cell. Neurosci. 2016, 10, 89. [Google Scholar] [CrossRef]
- Huang, H.-S.; Akbarian, S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS ONE 2007, 2, e809. [Google Scholar] [CrossRef]
- Fatemi, S.H.; Stary, J.M.; Earle, J.A.; Araghi-Niknam, M.; Eagan, E. GABAergic dysfunction in schizophrenia and mood disorders as reflected by decreased levels of glutamic acid decarboxylase 65 and 67 kDa and Reelin proteins in cerebellum. Schizophr. Res. 2005, 72, 109–122. [Google Scholar] [CrossRef]
- Gavin, D.P.; Sharma, R.P. Histone modifications, DNA methylation, and schizophrenia. Neurosci. Biobehav. Rev. 2010, 34, 882–888. [Google Scholar] [CrossRef]
- Guidotti, A.; Auta, J.; Chen, Y.; Davis, J.M.; Dong, E.; Gavin, D.P.; Grayson, D.R.; Matrisciano, F.; Pinna, G.; Satta, R.; et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology 2011, 60, 1007–1016. [Google Scholar] [CrossRef]
- Lewis, D.A.; Hashimoto, T.; Volk, D.W. Cortical inhibitory neurons and schizophrenia. Nat. Rev. Neurosci. 2005, 6, 312–324. [Google Scholar] [CrossRef]
- Tunbridge, E.M. The catechol-O-methyltransferase gene: Its regulation and polymorphisms. Int. Rev. Neurobiol. 2010, 95, 7–27. [Google Scholar] [CrossRef]
- Axelrod, J.; Tomchick, R. Enzymatic O-Methylation of Epinephrine and Other Catechols. J. Biol. Chem. 1958, 233, 702–705. [Google Scholar] [CrossRef]
- Shifman, S.; Bronstein, M.; Sternfeld, M.; Pisanté-Shalom, A.; Lev-Lehman, E.; Weizman, A.; Reznik, I.; Spivak, B.; Grisaru, N.; Karp, L.; et al. A Highly Significant Association between a COMT Haplotype and Schizophrenia. Am. J. Hum. Genet. 2002, 71, 1296–1302. [Google Scholar] [CrossRef]
- Mill, J.; Dempster, E.; Caspi, A.; Williams, B.; Moffitt, T.; Craig, I. Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2006, 141, 421–425. [Google Scholar] [CrossRef]
- Abdolmaleky, H.M.; Cheng, K.-h.; Faraone, S.V.; Wilcox, M.; Glatt, S.J.; Gao, F.; Smith, C.L.; Shafa, R.; Aeali, B.; Carnevale, J. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum. Mol. Genet. 2006, 15, 3132–3145. [Google Scholar] [CrossRef]
- Nohesara, S.; Ghadirivasfi, M.; Mostafavi, S.; Eskandari, M.-R.; Ahmadkhaniha, H.; Thiagalingam, S.; Abdolmaleky, H.M. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J. Psychiatr. Res. 2011, 45, 1432–1438. [Google Scholar] [CrossRef]
- Walton, E.; Liu, J.; Hass, J.; White, T.; Scholz, M.; Roessner, V.; Gollub, R.; Calhoun, V.D.; Ehrlich, S. MB-COMT promoter DNA methylation is associated with working-memory processing in schizophrenia patients and healthy controls. Epigenetics 2014, 9, 1101–1107. [Google Scholar] [CrossRef]
- Melas, P.A.; Rogdaki, M.; Ösby, U.; Schalling, M.; Lavebratt, C.; Ekström, T.J. Epigenetic aberrations in leukocytes of patients with schizophrenia: Association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J. 2012, 26, 2712–2718. [Google Scholar] [CrossRef]
- Murphy, B.C.; O’Reilly, R.L.; Singh, S.M. Site-specific cytosine methylation in S-COMT promoter in 31 brain regions with implications for studies involving schizophrenia. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2005, 133, 37–42. [Google Scholar] [CrossRef]
- Sesack, S.R.; Hawrylak, V.A.; Matus, C.; Guido, M.A.; Levey, A.I. Dopamine Axon Varicosities in the Prelimbic Division of the Rat Prefrontal Cortex Exhibit Sparse Immunoreactivity for the Dopamine Transporter. J. Neurosci. 1998, 18, 2697–2708. [Google Scholar] [CrossRef]
- Glahn, D.C.; Ragland, J.D.; Abramoff, A.; Barrett, J.; Laird, A.R.; Bearden, C.E.; Velligan, D.I. Beyond hypofrontality: A quantitative meta-analysis of functional neuroimaging studies of working memory in schizophrenia. Hum. Brain Mapp. 2005, 25, 60–69. [Google Scholar] [CrossRef]
- Karlsgodt, K.H.; Sanz, J.; van Erp, T.G.M.; Bearden, C.E.; Nuechterlein, K.H.; Cannon, T.D. Re-evaluating dorsolateral prefrontal cortex activation during working memory in schizophrenia. Schizophr. Res. 2009, 108, 143–150. [Google Scholar] [CrossRef]
- Wynn, J.K.; Green, M.F.; Hellemann, G.; Karunaratne, K.; Davis, M.C.; Marder, S.R. The effects of curcumin on brain-derived neurotrophic factor and cognition in schizophrenia: A randomized controlled study. Schizophr. Res. 2018, 195, 572–573. [Google Scholar] [CrossRef]
- Bathina, S.; Das, U.N. Brain-derived neurotrophic factor and its clinical implications. Arch. Med. Sci. AMS 2015, 11, 1164–1189. [Google Scholar] [CrossRef]
- Kordi-Tamandani, D.M.; Sahranavard, R.; Torkamanzehi, A. DNA methylation and expression profiles of the brain-derived neurotrophic factor (BDNF) and dopamine transporter (DAT1) genes in patients with schizophrenia. Mol. Biol. Rep. 2012, 39, 10889–10893. [Google Scholar] [CrossRef]
- Ovenden, E.S.; McGregor, N.W.; Emsley, R.A.; Warnich, L. DNA methylation and antipsychotic treatment mechanisms in schizophrenia: Progress and future directions. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 81, 38–49. [Google Scholar] [CrossRef]
- Amoli, M.M.; Khatami, F.; Arzaghi, S.M.; Enayati, S.; Nejatisafa, A.-A. Over-expression of TGF-β1 gene in medication free Schizophrenia. Psychoneuroendocrinology 2019, 99, 265–270. [Google Scholar] [CrossRef]
- Huang, T.-L.; Lee, C.-T. Associations between serum brain-derived neurotrophic factor levels and clinical phenotypes in schizophrenia patients. J. Psychiatr. Res. 2006, 40, 664–668. [Google Scholar] [CrossRef]
- Shimizu, E.; Hashimoto, K.; Watanabe, H.; Komatsu, N.; Okamura, N.; Koike, K.; Shinoda, N.; Nakazato, M.; Kumakiri, C.; Okada, S.-I. Serum brain-derived neurotrophic factor (BDNF) levels in schizophrenia are indistinguishable from controls. Neurosci. Lett. 2003, 351, 111–114. [Google Scholar] [CrossRef]
- Marzi, S.J.; Sugden, K.; Arseneault, L.; Belsky, D.W.; Burrage, J.; Corcoran, D.L.; Danese, A.; Fisher, H.L.; Hannon, E.; Moffitt, T.E. Analysis of DNA methylation in young people: Limited evidence for an association between victimization stress and epigenetic variation in blood. Am. J. Psychiatry 2018, 175, 517–529. [Google Scholar] [CrossRef]
- Swathy, B.; Banerjee, M. Understanding epigenetics of schizophrenia in the backdrop of its antipsychotic drug therapy. Epigenomics 2017, 9, 721–736. [Google Scholar] [CrossRef]
- Hashimoto, T.; Bergen, S.L.; Nguyen, Q.L.; Xu, B.; Monteggia, L.M.; Pierri, J.N.; Sun, Z.; Sampson, A.R.; Lewis, D.A. Relationship of Brain-Derived Neurotrophic Factor and Its Receptor TrkB to Altered Inhibitory Prefrontal Circuitry in Schizophrenia. J. Neurosci. 2005, 25, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Weickert, C.S.; Hyde, T.M.; Lipska, B.K.; Herman, M.M.; Weinberger, D.R.; Kleinman, J.E. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol. Psychiatry 2003, 8, 592–610. [Google Scholar] [CrossRef] [PubMed]
- Causing, C.G.; Gloster, A.; Aloyz, R.; Bamji, S.X.; Chang, E.; Fawcett, J.; Kuchel, G.; Miller, F.D. Synaptic Innervation Density Is Regulated by Neuron-Derived BDNF. Neuron 1997, 18, 257–267. [Google Scholar] [CrossRef]
- Katoh-Semba, R.; Takeuchi, I.K.; Semba, R.; Kato, K. Distribution of Brain-Derived Neurotrophic Factor in Rats and Its Changes with Development in the Brain. J. Neurochem. 1997, 69, 34–42. [Google Scholar] [CrossRef]
- Stolt, C.C.; Rehberg, S.; Ader, M.; Lommes, P.; Riethmacher, D.; Schachner, M.; Bartsch, U.; Wegner, M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002, 16, 165–170. [Google Scholar] [CrossRef]
- Richetto, J.; Meyer, U. Epigenetic Modifications in Schizophrenia and Related Disorders: Molecular Scars of Environmental Exposures and Source of Phenotypic Variability. Biol. Psychiatry 2021, 89, 215–226. [Google Scholar] [CrossRef]
- Tkachev, D.; Mimmack, M.L.; Ryan, M.M.; Wayland, M.; Freeman, T.; Jones, P.B.; Starkey, M.; Webster, M.J.; Yolken, R.H.; Bahn, S. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet 2003, 362, 798–805. [Google Scholar] [CrossRef]
- Iwamoto, K.; Bundo, M.; Yamada, K.; Takao, H.; Iwayama, Y.; Yoshikawa, T.; Kato, T. A family-based and case-control association study of SOX10 in schizophrenia. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2006, 141B, 477–481. [Google Scholar] [CrossRef]
- Flynn, S.W.; Lang, D.J.; Mackay, A.L.; Goghari, V.; Vavasour, I.M.; Whittall, K.P.; Smith, G.N.; Arango, V.; Mann, J.J.; Dwork, A.J.; et al. Abnormalities of myelination in schizophrenia detected in vivo with MRI, and post-mortem with analysis of oligodendrocyte proteins. Mol. Psychiatry 2003, 8, 811–820. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, Y.; Zhou, K.; Wang, L.; Li, J.; Zhuang, Q.; Xu, X.; Xu, L.; Zhang, K.; Dai, D. Male-specific association between dopamine receptor D4 gene methylation and schizophrenia. PLoS ONE 2014, 9, 2930–2939. [Google Scholar] [CrossRef]
- Dai, D.; Cheng, J.; Zhou, K.; Lv, Y.; Zhuang, Q.; Zheng, R.; Zhang, K.; Jiang, D.; Gao, S.; Duan, S. Significant association between DRD3 gene body methylation and schizophrenia. Psychiatry Res. 2014, 220, 772–777. [Google Scholar] [CrossRef]
- Kordi-Tamandani, D.M.; Dahmardeh, N.; Torkamanzehi, A. Evaluation of hypermethylation and expression pattern of GMR2, GMR5, GMR8, and GRIA3 in patients with schizophrenia. Gene 2013, 515, 163–166. [Google Scholar] [CrossRef]
- Pogarell, O.; Koch, W.; Karch, S.; Dehning, S.; Müller, N.; Tatsch, K.; Poepperl, G.; Möller, H.J. Dopaminergic neurotransmission in patients with schizophrenia in relation to positive and negative symptoms. Pharmacopsychiatry 2012, 45 (Suppl. S1), S36–S41. [Google Scholar] [CrossRef]
- Walter, H.; Kammerer, H.; Frasch, K.; Spitzer, M.; Abler, B. Altered reward functions in patients on atypical antipsychotic medication in line with the revised dopamine hypothesis of schizophrenia. Psychopharmacology 2009, 206, 121–132. [Google Scholar] [CrossRef]
- Kordi-Tamandani, D.M.; Sahranavard, R.; Torkamanzehi, A. Analysis of association between dopamine receptor genes’ methylation and their expression profile with the risk of schizophrenia. Psychiatr. Genet. 2013, 23, 183–187. [Google Scholar] [CrossRef]
- Abi-Dargham, A.; Mawlawi, O.; Lombardo, I.; Gil, R.; Martinez, D.; Huang, Y.; Hwang, D.-R.; Keilp, J.; Kochan, L.; Van Heertum, R.; et al. Prefrontal Dopamine D1 Receptors and Working Memory in Schizophrenia. J. Neurosci. 2002, 22, 3708–3719. [Google Scholar] [CrossRef]
- Rybakowski, J.K.; Borkowska, A.; Czerski, P.M.; Kapelski, P.; Dmitrzak-Weglarz, M.; Hauser, J. An association study of dopamine receptors polymorphisms and the Wisconsin Card Sorting Test in schizophrenia. J. Neural Transm. 2005, 112, 1575–1582. [Google Scholar] [CrossRef]
- Lee, K.Y.; Joo, E.-J.; Ji, Y.I.; Kim, D.-H.; Park, J.; Chung, I.-W.; Lee, S.I.; Joo, Y.H.; Ahn, Y.M.; Song, J.Y.; et al. Associations between DRDs and schizophrenia in a Korean population: Multi-stage association analyses. Exp. Mol. Med. 2011, 43, 44–52. [Google Scholar] [CrossRef]
- Sukhatme, V.P. The Egr family of nuclear signal transducers. Am. J. Kidney Dis. 1991, 17, 615–618. [Google Scholar] [CrossRef]
- Poirier, R.; Cheval, H.; Mailhes, C.; Garel, S.; Charnay, P.; Davis, S.; Laroche, S. Distinct functions of egr gene family members in cognitive processes. Front. Neurosci. 2008, 2, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Vawter, M.P.; Weickert, C.S.; Ferran, E.; Matsumoto, M.; Overman, K.; Hyde, T.M.; Weinberger, D.R.; Bunney, W.E.; Kleinman, J.E. Gene expression of metabolic enzymes and a protease inhibitor in the prefrontal cortex are decreased in schizophrenia. Neurochem. Res. 2004, 29, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, F.; Shugart, Y.; Yang, L.; Li, X.; Liu, Z.; Sun, N.; Yang, C.; Guo, X.; Shi, J. The early growth response protein 1-miR-30a-5p-neurogenic differentiation factor 1 axis as a novel biomarker for schizophrenia diagnosis and treatment monitoring. Transl. Psychiatry 2017, 7, e998. [Google Scholar] [CrossRef] [PubMed]
- Ramaker, R.C.; Bowling, K.M.; Lasseigne, B.N.; Hagenauer, M.H.; Hardigan, A.A.; Davis, N.S.; Gertz, J.; Cartagena, P.M.; Walsh, D.M.; Vawter, M.P. Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017, 9, 72. [Google Scholar] [CrossRef]
- Xu, Y.; Yue, W.; Yao Shugart, Y.; Li, S.; Cai, L.; Li, Q.; Cheng, Z.; Wang, G.; Zhou, Z.; Jin, C. Exploring transcription factors-microRNAs co-regulation networks in schizophrenia. Schizophr. Bull. 2016, 42, 1037–1045. [Google Scholar] [CrossRef]
- Pérez-Santiago, J.; Diez-Alarcia, R.; Callado, L.F.; Zhang, J.X.; Chana, G.; White, C.H.; Glatt, S.J.; Tsuang, M.T.; Everall, I.P.; Meana, J.J. A combined analysis of microarray gene expression studies of the human prefrontal cortex identifies genes implicated in schizophrenia. J. Psychiatr. Res. 2012, 46, 1464–1474. [Google Scholar] [CrossRef]
- Cattane, N.; Minelli, A.; Milanesi, E.; Maj, C.; Bignotti, S.; Bortolomasi, M.; Chiavetto, L.B.; Gennarelli, M. Altered gene expression in schizophrenia: Findings from transcriptional signatures in fibroblasts and blood. PLoS ONE 2015, 10, e116686. [Google Scholar] [CrossRef]
- Penner, M.R.; Parrish, R.R.; Hoang, L.T.; Roth, T.L.; Lubin, F.D.; Barnes, C.A. Age-related changes in Egr1 transcription and DNA methylation within the hippocampus. Hippocampus 2016, 26, 1008–1020. [Google Scholar] [CrossRef]
- Albuquerque, E.X.; Pereira, E.F.; Alkondon, M.; Rogers, S.W. Mammalian nicotinic acetylcholine receptors: From structure to function. Physiol. Rev. 2009, 89, 73–120. [Google Scholar] [CrossRef]
- Araud, T.; Graw, S.; Berger, R.; Lee, M.; Neveu, E.; Bertrand, D.; Leonard, S. The chimeric gene CHRFAM7A, a partial duplication of the CHRNA7 gene, is a dominant negative regulator of α7* nAChR function. Biochem. Pharmacol. 2011, 82, 904–914. [Google Scholar] [CrossRef]
- Jones, C. α7 nicotinic acetylcholine receptor: A potential target in treating cognitive decline in schizophrenia. J. Clin. Psychopharmacol. 2018, 38, 247–249. [Google Scholar] [CrossRef]
- Thomsen, M.S.; Hansen, H.H.; Timmerman, M.B.; Mikkelsen, J.D. Cognitive improvement by activation of α7 nicotinic acetylcholine receptors: From animal models to human pathophysiology. Curr. Pharm. Des. 2010, 16, 323–343. [Google Scholar] [CrossRef]
- Wallace, T.L.; Porter, R.H. Targeting the nicotinic alpha7 acetylcholine receptor to enhance cognition in disease. Biochem. Pharmacol. 2011, 82, 891–903. [Google Scholar] [CrossRef]
- Preskorn, S.H.; Gawryl, M.; Dgetluck, N.; Palfreyman, M.; Bauer, L.O.; Hilt, D.C. Normalizing effects of EVP-6124, an alpha-7 nicotinic partial agonist, on event-related potentials and cognition: A proof of concept, randomized trial in patients with schizophrenia. J. Psychiatr. Pract. 2014, 20, 12–24. [Google Scholar] [CrossRef]
- Canastar, A.; Logel, J.; Graw, S.; Finlay-Schultz, J.; Osborne, C.; Palionyte, M.; Drebing, C.; Plehaty, M.; Wilson, L.; Eyeson, R.; et al. Promoter Methylation and Tissue-Specific Transcription of the α7 Nicotinic Receptor Gene, CHRNA7. J. Mol. Neurosci. 2012, 47, 389–400. [Google Scholar] [CrossRef]
- Leonard, S.; Gault, J.; Adams, C.; Breese, C.R.; Rollins, Y.; Adler, L.E.; Olincy, A.; Freedman, R. Nicotinic Receptors, Smoking and Schizophrenia. Restor. Neurol. Neurosci. 1998, 12, 195–201. [Google Scholar]
- Abdolmaleky, H.M.; Nohesara, S.; Ghadirivasfi, M.; Lambert, A.W.; Ahmadkhaniha, H.; Ozturk, S.; Wong, C.K.; Shafa, R.; Mostafavi, A.; Thiagalingam, S. DNA hypermethylation of serotonin transporter gene promoter in drug naive patients with schizophrenia. Schizophr. Res. 2014, 152, 373–380. [Google Scholar] [CrossRef]
- Carrard, A.; Salzmann, A.; Malafosse, A.; Karege, F. Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. J. Affect. Disord. 2011, 132, 450–453. [Google Scholar] [CrossRef]
- Cheah, S.-Y.; Lawford, B.R.; Young, R.M.; Morris, C.P.; Voisey, J. mRNA expression and DNA methylation analysis of serotonin receptor 2A (HTR2A) in the human schizophrenic brain. Genes 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Ngan, E.T.C.; Lakshmi, N.Y.; Thomas, J.R.; Peter, F.L. Decreased Serotonin 2A Receptor Densities in Neuroleptic-Naive Patients With Schizophrenia: A PET Study Using [18F] Setoperone. Am. J. Psychiatry 2000, 157, 1016–1018. [Google Scholar] [CrossRef]
- McKinney, B.; Ding, Y.; Lewis, D.; Sweet, R. DNA methylation as a putative mechanism for reduced dendritic spine density in the superior temporal gyrus of subjects with schizophrenia. Transl. Psychiatry 2017, 7, e1032. [Google Scholar] [CrossRef] [PubMed]
- Fromer, M.; Pocklington, A.J.; Kavanagh, D.H.; Williams, H.J.; Dwyer, S.; Gormley, P.; Georgieva, L.; Rees, E.; Palta, P.; Ruderfer, D.M.; et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014, 506, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.M.; Moran, J.L.; Fromer, M.; Ruderfer, D.; Solovieff, N.; Roussos, P.; O’Dushlaine, C.; Chambert, K.; Bergen, S.E.; Kähler, A.; et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014, 506, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Russo, S.J.; Bolanos, C.A.; Theobald, D.E.; DeCarolis, N.A.; Renthal, W.; Kumar, A.; Winstanley, C.A.; Renthal, N.E.; Wiley, M.D.; Self, D.W.; et al. IRS2-Akt pathway in midbrain dopamine neurons regulates behavioral and cellular responses to opiates. Nat. Neurosci. 2007, 10, 93–99. [Google Scholar] [CrossRef]
- Fachim, H.A.; Srisawat, U.; Dalton, C.F.; Reynolds, G.P. Parvalbumin promoter hypermethylation in postmortem brain in schizophrenia. Epigenomics 2018, 10, 519–524. [Google Scholar] [CrossRef]
- Fachim, H.A.; Srisawat, U.; Dalton, C.F.; Harte, M.K.; Marsh, S.; Neill, J.C.; Reynolds, G.P. Subchronic administration of phencyclidine produces hypermethylation in the parvalbumin gene promoter in rat brain. Epigenomics 2016, 8, 1179–1183. [Google Scholar] [CrossRef]
- Connor, C.M.; Akbarian, S. DNA methylation changes in schizophrenia and bipolar disorder. Epigenetics 2008, 3, 55–58. [Google Scholar] [CrossRef]
- Lei, C.; Yu-Hong, Y.; Lin, H.; Kuo-Chen, C. Modulation of Cytokine Network in the Comorbidity of Schizophrenia and Tuberculosis. Curr. Top. Med. Chem. 2016, 16, 655–665. [Google Scholar]
- Rubin, L.H.; Connelly, J.J.; Reilly, J.L.; Carter, C.S.; Drogos, L.L.; Pournajafi-Nazarloo, H.; Ruocco, A.C.; Keedy, S.K.; Matthew, I.; Tandon, N. Sex and diagnosis-specific associations between DNA methylation of the oxytocin receptor gene with emotion processing and temporal-limbic and prefrontal brain volumes in psychotic disorders. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2016, 1, 141–151. [Google Scholar] [CrossRef]
- Kusui, C.; Kimura, T.; Ogita, K.; Nakamura, H.; Matsumura, Y.; Koyama, M.; Azuma, C.; Murata, Y. DNA Methylation of the Human Oxytocin Receptor Gene Promoter Regulates Tissue-Specific Gene Suppression. Biochem. Biophys. Res. Commun. 2001, 289, 681–686. [Google Scholar] [CrossRef]
- Baskerville, T.A.; Douglas, A.J. Dopamine and oxytocin interactions underlying behaviors: Potential contributions to behavioral disorders. CNS Neurosci. Ther. 2010, 16, e92–e123. [Google Scholar] [CrossRef]
- Feifel, D.; Shilling, P.D.; MacDonald, K. A Review of Oxytocin’s Effects on the Positive, Negative, and Cognitive Domains of Schizophrenia. Biol. Psychiatry 2016, 79, 222–233. [Google Scholar] [CrossRef]
- Smigielski, L.; Jagannath, V.; Rössler, W.; Walitza, S.; Grünblatt, E. Epigenetic mechanisms in schizophrenia and other psychotic disorders: A systematic review of empirical human findings. Mol. Psychiatry 2020, 25, 1718–1748. [Google Scholar] [CrossRef]
- Mill, J.; Tang, T.; Kaminsky, Z.; Khare, T.; Yazdanpanah, S.; Bouchard, L.; Jia, P.; Assadzadeh, A.; Flanagan, J.; Schumacher, A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008, 82, 696–711. [Google Scholar] [CrossRef]
- Dempster, E.L.; Pidsley, R.; Schalkwyk, L.C.; Owens, S.; Georgiades, A.; Kane, F.; Kalidindi, S.; Picchioni, M.; Kravariti, E.; Toulopoulou, T.; et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum. Mol. Genet. 2011, 20, 4786–4796. [Google Scholar] [CrossRef]
- Aberg, K.A.; McClay, J.L.; Nerella, S.; Xie, L.Y.; Clark, S.L.; Hudson, A.D.; Bukszár, J.; Adkins, D.; Consortium, S.S.; Hultman, C.M. MBD-seq as a cost-effective approach for methylome-wide association studies: Demonstration in 1500 case–control samples. Epigenomics 2012, 4, 605–621. [Google Scholar] [CrossRef]
- Kavanagh, D.; Tansey, K.; O’Donovan, M.C.; Owen, M.J. Schizophrenia genetics: Emerging themes for a complex disorder. Mol. Psychiatry 2015, 20, 72–76. [Google Scholar] [CrossRef]
- Aberg, K.A.; McClay, J.L.; Nerella, S.; Clark, S.; Kumar, G.; Chen, W.; Khachane, A.N.; Xie, L.; Hudson, A.; Gao, G. Methylome-wide association study of schizophrenia: Identifying blood biomarker signatures of environmental insults. JAMA Psychiatry 2014, 71, 255–264. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, C.; Cheng, L.; Reilly, J.L.; Bishop, J.R.; Sweeney, J.A.; Chen, H.Y.; Gershon, E.S.; Liu, C. Correlation between DNA methylation and gene expression in the brains of patients with bipolar disorder and schizophrenia. Bipolar Disord. 2014, 16, 790–799. [Google Scholar] [CrossRef]
- Wockner, L.F.; Noble, E.P.; Lawford, B.R.; Young, R.M.; Morris, C.P.; Whitehall, V.L.; Voisey, J. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl. Psychiatry 2014, 4, e339. [Google Scholar] [CrossRef]
- Ota, V.K.; Moretti, P.N.; Santoro, M.L.; Talarico, F.; Spindola, L.M.; Xavier, G.; Carvalho, C.M.; Marques, D.F.; Costa, G.O.; Pellegrino, R.; et al. Gene expression over the course of schizophrenia: From clinical high-risk for psychosis to chronic stages. NPJ Schizophrenia 2019, 5, 5. [Google Scholar] [CrossRef]
- Ota, V.K.; Noto, C.; Gadelha, A.; Santoro, M.L.; Spindola, L.M.; Gouvea, E.S.; Stilhano, R.S.; Ortiz, B.B.; Silva, P.N.; Sato, J.R.; et al. Changes in gene expression and methylation in the blood of patients with first-episode psychosis. Schizophr. Res. 2014, 159, 358–364. [Google Scholar] [CrossRef]
- Pesce, M.; Ferrone, A.; Rizzuto, A.; Tatangelo, R.; Iezzi, I.; Ladu, S.; Franceschelli, S.; Speranza, L.; Patruno, A.; Felaco, M.; et al. The SHP-1 expression is associated with cytokines and psychopathological status in unmedicated first episode Schizophrenia patients. Brain Behav. Immun. 2014, 41, 251–260. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, J.; Tang, X.; Feng, X.; Yu, M.; Sha, W.; Wang, X.; Zhang, X.; Yi, H.; Zhang, X. DNA methylation and gene expression of the chemokine (C-X-C motif) ligand 1 in patients with deficit and non-deficit schizophrenia. Psychiatry Res. 2018, 268, 82–86. [Google Scholar] [CrossRef]
- Zhu, Y.; Strachan, E.; Fowler, E.; Bacus, T.; Roy-Byrne, P.; Zhao, J. Genome-wide profiling of DNA methylome and transcriptome in peripheral blood monocytes for major depression: A Monozygotic Discordant Twin Study. Transl. Psychiatry 2019, 9, 215. [Google Scholar] [CrossRef]
- Xie, Y.; Xiao, L.; Chen, L.; Zheng, Y.; Zhang, C.; Wang, G. Integrated Analysis of Methylomic and Transcriptomic Data to Identify Potential Diagnostic Biomarkers for Major Depressive Disorder. Genes 2021, 12, 178. [Google Scholar] [CrossRef]
- Van Eijk, K.R.; De Jong, S.; Strengman, E.; Buizer-Voskamp, J.E.; Kahn, R.S.; Boks, M.P.; Horvath, S.; Ophoff, R.A. Identification of schizophrenia-associated loci by combining DNA methylation and gene expression data from whole blood. Eur. J. Hum. Genet. 2015, 23, 1106–1110. [Google Scholar] [CrossRef]
- Zhao, H.; Xu, J.; Pang, L.; Zhang, Y.; Fan, H.; Liu, L.; Liu, T.; Yu, F.; Zhang, G.; Lan, Y. Genome-wide DNA methylome reveals the dysfunction of intronic microRNAs in major psychosis. BMC Med. Genom. 2015, 8, 1–12. [Google Scholar] [CrossRef]
- Beveridge, N.J.; Gardiner, E.; Carroll, A.; Tooney, P.; Cairns, M. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry 2010, 15, 1176–1189. [Google Scholar] [CrossRef]
- Pidsley, R.; Mill, J. Epigenetic studies of psychosis: Current findings, methodological approaches, and implications for postmortem research. Biol. Psychiatry 2011, 69, 146–156. [Google Scholar] [CrossRef]
- Sjöholm, L.K.; Ransome, Y.; Ekström, T.J.; Karlsson, O. Evaluation of post-mortem effects on global brain DNA methylation and hydroxymethylation. Basic Clin. Pharmacol. Toxicol. 2018, 122, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Mendizabal, I.; Yi, S.V. Whole-genome bisulfite sequencing maps from multiple human tissues reveal novel CpG islands associated with tissue-specific regulation. Hum. Mol. Genet. 2016, 25, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Gu, H.; Müller, F.; Donaghey, J.; Tsai, L.T.-Y.; Kohlbacher, O.; De Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chang, X.; Hahn, C.-G.; Gur, R.E.; Sleiman, P.A.; Hakonarson, H. Non-coding RNA dysregulation in the amygdala region of schizophrenia patients contributes to the pathogenesis of the disease. Transl. Psychiatry 2018, 8, 44. [Google Scholar] [CrossRef]
- Ragan, C.; Patel, K.; Edson, J.; Zhang, Z.-H.; Gratten, J.; Mowry, B. Small non-coding RNA expression from anterior cingulate cortex in schizophrenia shows sex specific regulation. Schizophr. Res. 2017, 183, 82–87. [Google Scholar] [CrossRef]
- Ripke, S.; Sanders, A.R.; Kendler, K.S.; Levinson, D.F.; Sklar, P.; Holmans, P.A.; Lin, D.-Y.; Duan, J.; Ophoff, R.A.; Andreassen, O.A. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011, 43, 969–976. [Google Scholar] [CrossRef]
- Sakamoto, K.; Crowley, J.J. A comprehensive review of the genetic and biological evidence supports a role for MicroRNA-137 in the etiology of schizophrenia. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2018, 177, 242–256. [Google Scholar] [CrossRef]
- Collins, A.; Kim, Y.; Bloom, R.; Kelada, S.; Sethupathy, P.; Sullivan, P. Transcriptional targets of the schizophrenia risk gene MIR137. Transl. Psychiatry 2014, 4, e404. [Google Scholar] [CrossRef]
- Wright, C.; Calhoun, V.D.; Ehrlich, S.; Wang, L.; Turner, J.A.; Bizzozero, N.I.P. Meta gene set enrichment analyses link miR-137-regulated pathways with schizophrenia risk. Front. Genet. 2015, 6, 147. [Google Scholar] [CrossRef]
- Du, Y.; Yu, Y.; Hu, Y.; Li, X.-W.; Wei, Z.-X.; Pan, R.-Y.; Li, X.-S.; Zheng, G.-E.; Qin, X.-Y.; Liu, Q.-S. Genome-wide, integrative analysis implicates exosome-derived microRNA dysregulation in schizophrenia. Schizophr. Bull. 2019, 45, 1257–1266. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Yu, S.-L.; Hsieh, M.H.; Chen, C.-H.; Chen, H.-Y.; Wen, C.-C.; Huang, Y.-H.; Hsiao, P.-C.; Hsiao, C.K.; Liu, C.-M. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE 2011, 6, e21635. [Google Scholar] [CrossRef]
- Jaffe, A.E.; Irizarry, R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014, 15, 1–9. [Google Scholar] [CrossRef]
- Alelú-Paz, R.; González-Corpas, A.; Ashour, N.; Escanilla, A.; Monje, A.; Guerrero Marquez, C.; Algora Weber, M.; Ropero, S. DNA methylation pattern of gene promoters of major neurotransmitter systems in older patients with schizophrenia with severe and mild cognitive impairment. Int. J. Geriatr. Psychiatry 2015, 30, 558–565. [Google Scholar] [CrossRef]
- Davies, M.N.; Volta, M.; Pidsley, R.; Lunnon, K.; Dixit, A.; Lovestone, S.; Coarfa, C.; Harris, R.A.; Milosavljevic, A.; Troakes, C. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012, 13, 1–14. [Google Scholar] [CrossRef]
- Karlsgodt, K.H.; Sun, D.; Cannon, T.D. Structural and Functional Brain Abnormalities in Schizophrenia. Curr. Dir. Psychol. Sci. 2010, 19, 226–231. [Google Scholar] [CrossRef]
- Zong, D.; Liu, X.; Li, J.; Ouyang, R.; Chen, P. The role of cigarette smoke-induced epigenetic alterations in inflammation. Epigenetics Chromatin 2019, 12, 65. [Google Scholar] [CrossRef]
- Guidotti, A.; Grayson, D.R. DNA methylation and demethylation as targets for antipsychotic therapy. Dialogues Clin. Neurosci. 2014, 16, 419–429. [Google Scholar] [CrossRef]
- Tang, H.; Dalton, C.F.; Srisawat, U.; Zhang, Z.J.; Reynolds, G.P. Methylation at a transcription factor-binding site on the 5-HT1A receptor gene correlates with negative symptom treatment response in first episode schizophrenia. Int. J. Neuropsychopharmacol. 2014, 17, 645–649. [Google Scholar] [CrossRef]
- Dong, E.; Tueting, P.; Matrisciano, F.; Grayson, D.R.; Guidotti, A. Behavioral and molecular neuroepigenetic alterations in prenatally stressed mice: Relevance for the study of chromatin remodeling properties of antipsychotic drugs. Transl. Psychiatry 2016, 6, e711. [Google Scholar] [CrossRef]
- Melka, M.G.; Castellani, C.A.; Laufer, B.I.; Rajakumar, N.; O’Reilly, R.; Singh, S.M. Olanzapine induced DNA methylation changes support the dopamine hypothesis of psychosis. J. Mol. Psychiatry 2013, 1, 19. [Google Scholar] [CrossRef]
- Kinoshita, M.; Numata, S.; Tajima, A.; Yamamori, H.; Yasuda, Y.; Fujimoto, M.; Watanabe, S.; Umehara, H.; Shimodera, S.; Nakazawa, T.; et al. Effect of Clozapine on DNA Methylation in Peripheral Leukocytes from Patients with Treatment-Resistant Schizophrenia. Int. J. Mol. Sci. 2017, 18, 632. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, G.P.; Fachim, H.A. Does DNA methylation influence the effects of psychiatric drugs? Epigenomics 2016, 8, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Sloan, S.A.; Zhang, Y.; Enge, M.; Caneda, C.; Shuer, L.M.; Hayden Gephart, M.G.; Barres, B.A.; Quake, S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 2015, 112, 7285–7290. [Google Scholar] [CrossRef] [PubMed]
- Lake, B.B.; Ai, R.; Kaeser, G.E.; Salathia, N.S.; Yung, Y.C.; Liu, R.; Wildberg, A.; Gao, D.; Fung, H.-L.; Chen, S.; et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 2016, 352, 1586–1590. [Google Scholar] [CrossRef]
- Ecker, J.R.; Geschwind, D.H.; Kriegstein, A.R.; Ngai, J.; Osten, P.; Polioudakis, D.; Regev, A.; Sestan, N.; Wickersham, I.R.; Zeng, H. The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron 2017, 96, 542–557. [Google Scholar] [CrossRef]
- Richetto, J.; Massart, R.; Weber-Stadlbauer, U.; Szyf, M.; Riva, M.A.; Meyer, U. Genome-wide DNA Methylation Changes in a Mouse Model of Infection-Mediated Neurodevelopmental Disorders. Biol. Psychiatry 2017, 81, 265–276. [Google Scholar] [CrossRef]
- Khavari, B.; Cairns, M.J. Epigenomic Dysregulation in Schizophrenia: In Search of Disease Etiology and Biomarkers. Cells 2020, 9, 1837. [Google Scholar] [CrossRef]
- Grayson, D.R.; Guidotti, A. DNA Methylation in Animal Models of Psychosis. In Progress in Molecular Biology and Translational Science; Grayson, D.R., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 157, pp. 105–132. [Google Scholar]
- Meyer, U.; Feldon, J. Epidemiology-driven neurodevelopmental animal models of schizophrenia. Prog. Neurobiol. 2010, 90, 285–326. [Google Scholar] [CrossRef]
- Javidfar, B.; Park, R.; Kassim, B.S.; Bicks, L.K.; Akbarian, S. The epigenomics of schizophrenia, in the mouse. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2017, 174, 631–640. [Google Scholar] [CrossRef]
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharm. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- Peedicayil, J. Identification of Biomarkers in Neuropsychiatric Disorders Based on Systems Biology and Epigenetics. Front. Genet. 2019, 10, 985. [Google Scholar] [CrossRef]
- Aberg, K.A.; Xie, L.Y.; McClay, J.L.; Nerella, S.; Vunck, S.; Snider, S.; Beardsley, P.M.; van den Oord, E.J.C.G. Testing two models describing how methylome-wide studies in blood are informative for psychiatric conditions. Epigenomics 2013, 5, 367–377. [Google Scholar] [CrossRef]
- Hannon, E.; Lunnon, K.; Schalkwyk, L.; Mill, J. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 2015, 10, 1024–1032. [Google Scholar] [CrossRef]
- Walton, E.; Hass, J.; Liu, J.; Roffman, J.L.; Bernardoni, F.; Roessner, V.; Kirsch, M.; Schackert, G.; Calhoun, V.; Ehrlich, S. Correspondence of DNA Methylation Between Blood and Brain Tissue and Its Application to Schizophrenia Research. Schizophr. Bull. 2016, 42, 406–414. [Google Scholar] [CrossRef]
- Horvath, S.; Zhang, Y.; Langfelder, P.; Kahn, R.S.; Boks, M.P.; van Eijk, K.; van den Berg, L.H.; Ophoff, R.A. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012, 13, R97. [Google Scholar] [CrossRef]
- Flores, G.; Morales-Medina, J.C.; Diaz, A. Neuronal and brain morphological changes in animal models of schizophrenia. Behav. Brain Res. 2016, 301, 190–203. [Google Scholar] [CrossRef]
- Tomoda, T.; Sumitomo, A.; Jaaro-Peled, H.; Sawa, A. Utility and validity of DISC1 mouse models in biological psychiatry. Neuroscience 2016, 321, 99–107. [Google Scholar] [CrossRef]
- Katche, C.; Cammarota, M.; Medina, J.H. Molecular signatures and mechanisms of long-lasting memory consolidation and storage. Neurobiol. Learn. Mem. 2013, 106, 40–47. [Google Scholar] [CrossRef]
- Miranda, M.; Kent, B.A.; Morici, J.F.; Gallo, F.; Weisstaub, N.V.; Saksida, L.M.; Bussey, T.J.; Bekinschtein, P. Molecular Mechanisms in Perirhinal Cortex Selectively Necessary for Discrimination of Overlapping Memories, but Independent of Memory Persistence. eNeuro 2017, 4, 1–20. [Google Scholar] [CrossRef]
- Roesler, R. Molecular mechanisms controlling protein synthesis in memory reconsolidation. Neurobiol. Learn. Mem. 2017, 142, 30–40. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).