Functional Proteomic Profiling of Triple-Negative Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Reverse Phase Protein Arrays (RPPA)
2.3. Antibodies
2.4. Immunohistochemistry (IHC) of Formalin-Fixed, Paraffin-Embedded (FFPE) Samples
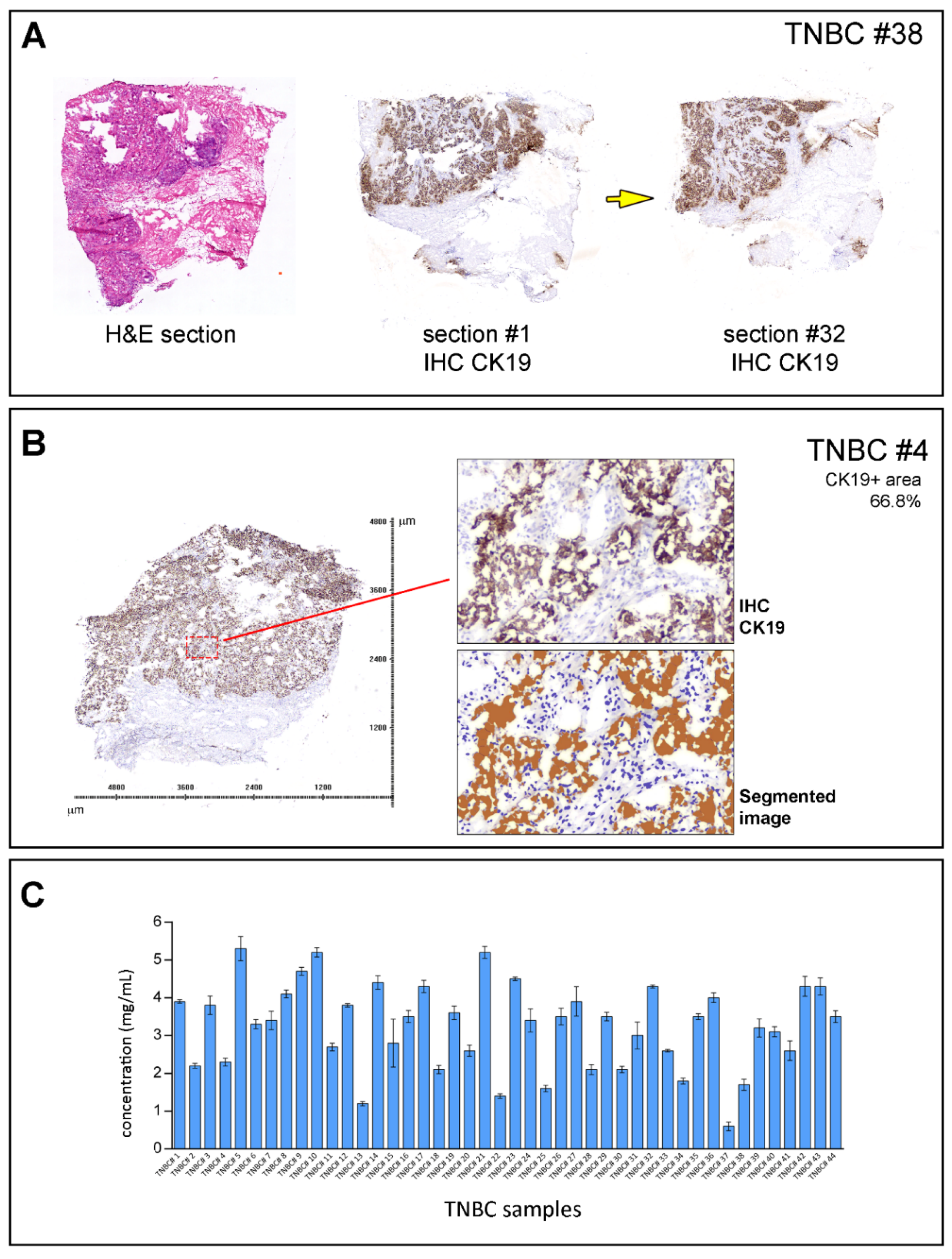
2.5. Quantitative Assessment of IHC Staining
2.6. DNA Purification and Sequencing
2.7. Data Analysis
3. Results
3.1. RPPA-Mediated Functional Profiling of Tnbcs
3.2. Pathway-Restricted Analysis of RPPA Data
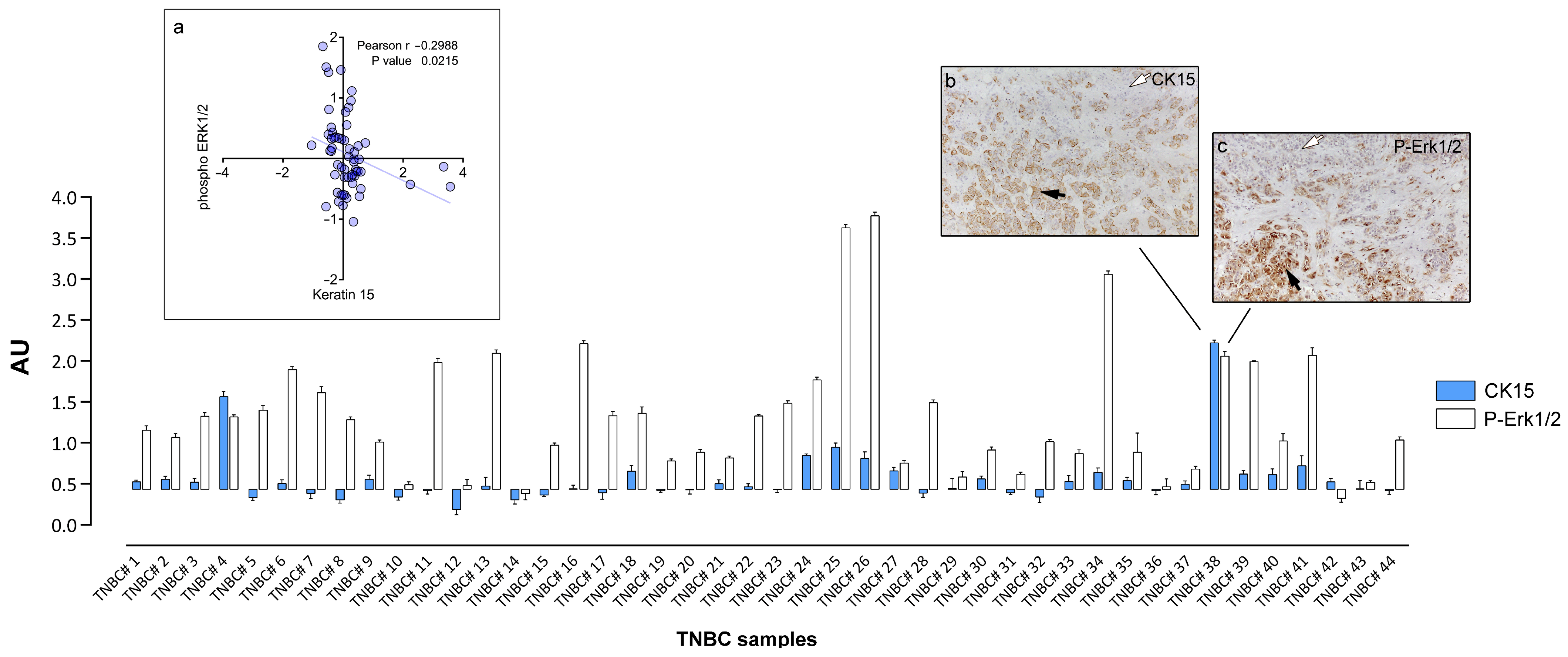
3.3. Cytokeratin 15 Expression in Tumor Cells Was Associated with Erk1/2 Signaling
3.4. Expression of Oncogenic C-Kit Defines a Subset of Tnbcs
4. Discussion
4.1. Profiling of Pathway Activation
4.2. CK15, c-Kit, and Erk1/2 Signaling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Autier, P.; Boniol, M.; La Vecchia, C.; Vatten, L.; Gavin, A.; Hery, C.; Heanue, M. Disparities in breast cancer mortality trends between 30 European countries: Retrospective trend analysis of WHO mortality database. BMJ 2010, 341, c3620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSantis, C.E.; Bray, F.; Ferlay, J.; Lortet-Tieulent, J.; Anderson, B.O.; Jemal, A. International Variation in Female Breast Cancer Incidence and Mortality Rates. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1495–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Hayat, M.J.; Howlader, N.; Reichman, M.E.; Edwards, B.K. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist 2007, 12, 20–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howlader, N.; Altekruse, S.F.; Li, C.I.; Chen, V.W.; Clarke, C.A.; Ries, L.A.; Cronin, K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [Green Version]
- Palma, G.; Frasci, G.; Chirico, A.; Esposito, E.; Siani, C.; Saturnino, C.; Arra, C.; Ciliberto, G.; Giordano, A.; D’Aiuto, M. Triple negative breast cancer: Looking for the missing link between biology and treatments. Oncotarget 2015, 6, 26560–26574. [Google Scholar] [CrossRef] [Green Version]
- Marme, F.; Schneeweiss, A. Targeted Therapies in Triple-Negative Breast Cancer. Breast Care 2015, 10, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Podo, F.; Buydens, L.M.; Degani, H.; Hilhorst, R.; Klipp, E.; Gribbestad, I.S.; Van Huffel, S.; van Laarhoven, H.W.; Luts, J.; Monleon, D.; et al. Triple-negative breast cancer: Present challenges and new perspectives. Mol. Oncol. 2010, 4, 209–229. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Prado-Vazquez, G.; Gamez-Pozo, A.; Trilla-Fuertes, L.; Arevalillo, J.M.; Zapater-Moros, A.; Ferrer-Gomez, M.; Diaz-Almiron, M.; Lopez-Vacas, R.; Navarro, H.; Main, P.; et al. A novel approach to triple-negative breast cancer molecular classification reveals a luminal immune-positive subgroup with good prognoses. Sci. Rep. 2019, 9, 1538. [Google Scholar] [CrossRef] [PubMed]
- Jezequel, P.; Loussouarn, D.; Guerin-Charbonnel, C.; Campion, L.; Vanier, A.; Gouraud, W.; Lasla, H.; Guette, C.; Valo, I.; Verriele, V.; et al. Gene-expression molecular subtyping of triple-negative breast cancer tumours: Importance of immune response. Breast Cancer Res. 2015, 17, 43. [Google Scholar] [CrossRef] [Green Version]
- Masuda, H.; Qi, Y.; Liu, S.; Hayashi, N.; Kogawa, T.; Hortobagyi, G.N.; Tripathy, D.; Ueno, N.T. Reverse phase protein array identification of triple-negative breast cancer subtypes and comparison with mRNA molecular subtypes. Oncotarget 2017, 8, 70481–70495. [Google Scholar] [CrossRef] [PubMed]
- Cabezon, T.; Gromova, I.; Gromov, P.; Serizawa, R.; Timmermans Wielenga, V.; Kroman, N.; Celis, J.E.; Moreira, J.M. Proteomic profiling of triple-negative breast carcinomas in combination with a three-tier orthogonal technology approach identifies Mage-A4 as potential therapeutic target in estrogen receptor negative breast cancer. Mol. Cell Proteomics 2013, 12, 381–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gromova, I.; Gromov, P.; Honma, N.; Kumar, S.; Rimm, D.; Talman, M.L.; Wielenga, V.T.; Moreira, J.M. High level PHGDH expression in breast is predominantly associated with keratin 5-positive cell lineage independently of malignancy. Mol. Oncol. 2015, 9, 1636–1654. [Google Scholar] [CrossRef]
- Gromov, P.; Celis, J.E.; Gromova, I.; Rank, F.; Timmermans-Wielenga, V.; Moreira, J.M. A single lysis solution for the analysis of tissue samples by different proteomic technologies. Mol. Oncol. 2008, 2, 368–379. [Google Scholar] [CrossRef] [Green Version]
- Winters, M.; Dabir, B.; Yu, M.; Kohn, E.C. Constitution and quantity of lysis buffer alters outcome of reverse phase protein microarrays. Proteomics 2007, 7, 4066–4068. [Google Scholar] [CrossRef]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar] [CrossRef]
- van Oostrum, J.; Calonder, C.; Rechsteiner, D.; Ehrat, M.; Mestan, J.; Fabbro, D.; Voshol, H. Tracing pathway activities with kinase inhibitors and reverse phase protein arrays. Proteomics Clin. Appl. 2009, 3, 412–422. [Google Scholar] [CrossRef]
- Voshol, H.; Ehrat, M.; Traenkle, J.; Bertrand, E.; van Oostrum, J. Antibody-based proteomics: Analysis of signaling networks using reverse protein arrays. FEBS J. 2009, 276, 6871–6879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ummanni, R.; Mannsperger, H.A.; Sonntag, J.; Oswald, M.; Sharma, A.K.; Konig, R.; Korf, U. Evaluation of reverse phase protein array (RPPA)-based pathway-activation profiling in 84 non-small cell lung cancer (NSCLC) cell lines as platform for cancer proteomics and biomarker discovery. Biochim. Biophys. Acta 2014, 1844, 950–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.; Savage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef] [Green Version]
- Prat, A.; Adamo, B.; Cheang, M.C.; Anders, C.K.; Carey, L.A.; Perou, C.M. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013, 18, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celis, J.E.; Gromova, I.; Cabezon, T.; Gromov, P.; Shen, T.; Timmermans-Wielenga, V.; Rank, F.; Moreira, J.M. Identification of a subset of breast carcinomas characterized by expression of cytokeratin 15: Relationship between CK15+ progenitor/amplified cells and pre-malignant lesions and invasive disease. Mol. Oncol. 2007, 1, 321–349. [Google Scholar] [CrossRef] [Green Version]
- Soady, K.J.; Kendrick, H.; Gao, Q.; Tutt, A.; Zvelebil, M.; Ordonez, L.D.; Quist, J.; Tan, D.W.; Isacke, C.M.; Grigoriadis, A.; et al. Mouse mammary stem cells express prognostic markers for triple-negative breast cancer. Breast Cancer Res. 2015, 17, 31. [Google Scholar] [CrossRef] [Green Version]
- Asselin-Labat, M.L.; Shackleton, M.; Stingl, J.; Vaillant, F.; Forrest, N.C.; Eaves, C.J.; Visvader, J.E.; Lindeman, G.J. Steroid hormone receptor status of mouse mammary stem cells. J. Natl. Cancer Inst. 2006, 98, 1011–1014. [Google Scholar] [CrossRef] [Green Version]
- Villadsen, R.; Fridriksdottir, A.J.; Ronnov-Jessen, L.; Gudjonsson, T.; Rank, F.; LaBarge, M.A.; Bissell, M.J.; Petersen, O.W. Evidence for a stem cell hierarchy in the adult human breast. J. Cell Biol. 2007, 177, 87–101. [Google Scholar] [CrossRef]
- Moreira, J.M.; Cabezon, T.; Gromova, I.; Gromov, P.; Timmermans-Wielenga, V.; Machado, I.; Llombart-Bosch, A.; Kroman, N.; Rank, F.; Celis, J.E. Tissue proteomics of the human mammary gland: Towards an abridged definition of the molecular phenotypes underlying epithelial normalcy. Mol. Oncol. 2010, 4, 539–561. [Google Scholar] [CrossRef] [Green Version]
- Lennartsson, J.; Jelacic, T.; Linnekin, D.; Shivakrupa, R. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells 2005, 23, 16–43. [Google Scholar] [CrossRef] [PubMed]
- Millis, S.Z.; Gatalica, Z.; Winkler, J.; Vranic, S.; Kimbrough, J.; Reddy, S.; O’Shaughnessy, J.A. Predictive Biomarker Profiling of >6000 Breast Cancer Patients Shows Heterogeneity in TNBC, With Treatment Implications. Clin. Breast Cancer 2015, 15, 473–481 e473. [Google Scholar] [CrossRef] [Green Version]
- Coombs, C.C.; Tallman, M.S.; Levine, R.L. Molecular therapy for acute myeloid leukaemia. Nat. Rev. Clin. Oncol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, Y.; Akbani, R.; Ju, Z.; Roebuck, P.L.; Liu, W.; Yang, J.Y.; Broom, B.M.; Verhaak, R.G.; Kane, D.W.; et al. TCPA: A resource for cancer functional proteomics data. Nat. Methods 2013, 10, 1046–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, K.; Tsuda, H.; Koizumi, F.; Shimizu, C.; Yonemori, K.; Ando, M.; Kodaira, M.; Yunokawa, M.; Fujiwara, Y.; Tamura, K. Activated PI3K/AKT and MAPK pathways are potential good prognostic markers in node-positive, triple-negative breast cancer. Ann. Oncol. 2014, 25, 1973–1979. [Google Scholar] [CrossRef]
- Aleskandarany, M.A.; Negm, O.H.; Green, A.R.; Ahmed, M.A.; Nolan, C.C.; Tighe, P.J.; Ellis, I.O.; Rakha, E.A. Epithelial mesenchymal transition in early invasive breast cancer: An immunohistochemical and reverse phase protein array study. Breast Cancer Res. Treat 2014, 145, 339–348. [Google Scholar] [CrossRef]
- Rieger-Christ, K.M.; Lee, P.; Zagha, R.; Kosakowski, M.; Moinzadeh, A.; Stoffel, J.; Ben-Ze’ev, A.; Libertino, J.A.; Summerhayes, I.C. Novel expression of N-cadherin elicits in vitro bladder cell invasion via the Akt signaling pathway. Oncogene 2004, 23, 4745–4753. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.H.; Jo, U.; Kohrman, A.; Rezaeian, A.H.; Chou, P.C.; Logothetis, C.; Lin, H.K. Posttranslational regulation of Akt in human cancer. Cell Biosci. 2014, 4, 59. [Google Scholar] [CrossRef] [Green Version]
- TCGA-Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Stemke-Hale, K.; Gonzalez-Angulo, A.M.; Lluch, A.; Neve, R.M.; Kuo, W.L.; Davies, M.; Carey, M.; Hu, Z.; Guan, Y.; Sahin, A.; et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008, 68, 6084–6091. [Google Scholar] [CrossRef] [Green Version]
- Schneider, B.P.; Winer, E.P.; Foulkes, W.D.; Garber, J.; Perou, C.M.; Richardson, A.; Sledge, G.W.; Carey, L.A. Triple-negative breast cancer: Risk factors to potential targets. Clin. Cancer Res. 2008, 14, 8010–8018. [Google Scholar] [CrossRef] [Green Version]
- Swanton, C.; Caldas, C. Molecular classification of solid tumours: Towards pathway-driven therapeutics. Br. J. Cancer 2009, 100, 1517–1522. [Google Scholar] [CrossRef] [Green Version]
- Cossu-Rocca, P.; Orru, S.; Muroni, M.R.; Sanges, F.; Sotgiu, G.; Ena, S.; Pira, G.; Murgia, L.; Manca, A.; Uras, M.G.; et al. Analysis of PIK3CA Mutations and Activation Pathways in Triple Negative Breast Cancer. PLoS ONE 2015, 10, e0141763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, M.L.; Gong, C.; Chen, C.H.; Hu, H.; Huang, P.; Zheng, M.; Yao, Y.; Wei, S.; Wulf, G.; Lieberman, J.; et al. The Rab2A GTPase promotes breast cancer stem cells and tumorigenesis via Erk signaling activation. Cell Rep. 2015, 11, 111–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.J.; Yang, J.Y.; Xia, W.; Chen, C.T.; Xie, X.; Chao, C.H.; Woodward, W.A.; Hsu, J.M.; Hortobagyi, G.N.; Hung, M.C. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell 2011, 19, 86–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashiwagi, S.; Yashiro, M.; Takashima, T.; Aomatsu, N.; Kawajiri, H.; Ogawa, Y.; Onoda, N.; Ishikawa, T.; Wakasa, K.; Hirakawa, K. c-Kit expression as a prognostic molecular marker in patients with basal-like breast cancer. Br. J. Surg. 2013, 100, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Y.; Guan, B.; Rao, Q.; Wang, J.; Ma, H.; Zhang, Z.; Zhou, X. C-kit and PDGFRA gene mutations in triple negative breast cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 4280–4285. [Google Scholar]


| Patient | c-KIT | |||
|---|---|---|---|---|
| Exon 9 | Exon 11 | Exon 13 | Exon 17 | |
| TNBC #15 | wild-type | wild-type | wild-type | c.2394 G > T (silent/polymorphism) |
| TNBC #17 | wild-type | wild-type | wild-type | wild-type |
| TNBC #18 | c.1414 G > A (silent/polymorphism) | c.1678_1680 del3 p.Val560 del | wild-type | wild-type |
| TNBC #21 | wild-type | c.1660_1662 del3 p.Glu1554 del | wild-type | wild-type |
| TNBC #26 | wild-type | wild-type | wild-type | wild-type |
| TNBC #27 | wild-type | wild-type | wild-type | wild-type |
| TNBC #31 | wild-type | wild-type | wild-type | wild-type |
| TNBC #32 | wild-type | wild-type | wild-type | wild-type |
| TNBC #34 | wild-type | c.1678_1680 del3 p.Val560 del | wild-type | wild-type |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gromova, I.; Espinoza, J.A.; Grauslund, M.; Santoni-Rugiu, E.; Møller Talman, M.-L.; van Oostrum, J.; Moreira, J.M.A. Functional Proteomic Profiling of Triple-Negative Breast Cancer. Cells 2021, 10, 2768. https://doi.org/10.3390/cells10102768
Gromova I, Espinoza JA, Grauslund M, Santoni-Rugiu E, Møller Talman M-L, van Oostrum J, Moreira JMA. Functional Proteomic Profiling of Triple-Negative Breast Cancer. Cells. 2021; 10(10):2768. https://doi.org/10.3390/cells10102768
Chicago/Turabian StyleGromova, Irina, Jaime A. Espinoza, Morten Grauslund, Eric Santoni-Rugiu, Maj-Lis Møller Talman, Jan van Oostrum, and José M. A. Moreira. 2021. "Functional Proteomic Profiling of Triple-Negative Breast Cancer" Cells 10, no. 10: 2768. https://doi.org/10.3390/cells10102768
APA StyleGromova, I., Espinoza, J. A., Grauslund, M., Santoni-Rugiu, E., Møller Talman, M.-L., van Oostrum, J., & Moreira, J. M. A. (2021). Functional Proteomic Profiling of Triple-Negative Breast Cancer. Cells, 10(10), 2768. https://doi.org/10.3390/cells10102768







