Comparative Mutational Profiling of Hematopoietic Progenitor Cells and Circulating Endothelial Cells (CECs) in Patients with Primary Myelofibrosis
Abstract
:1. Introduction
2. Patients and Methods
2.1. Patients and Healthy Controls
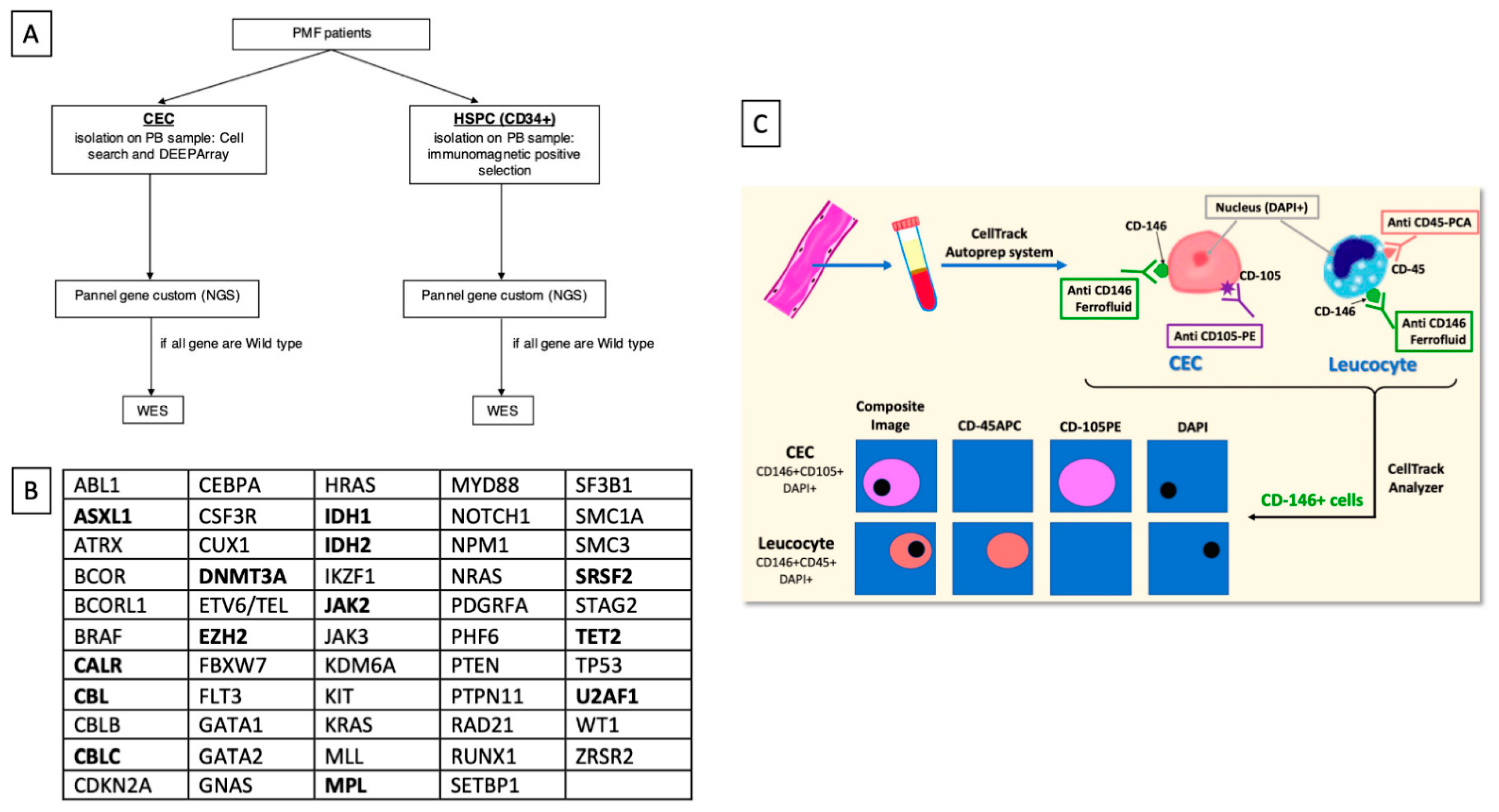
2.2. Study Plan
2.3. CD34 + HSPC Detection and Selection
2.4. CellSearch CECs Identification and Collection
2.5. NGS Analysis
2.6. Statistical Analysis
3. Results
3.1. Patients and Healthy Controls Characteristics
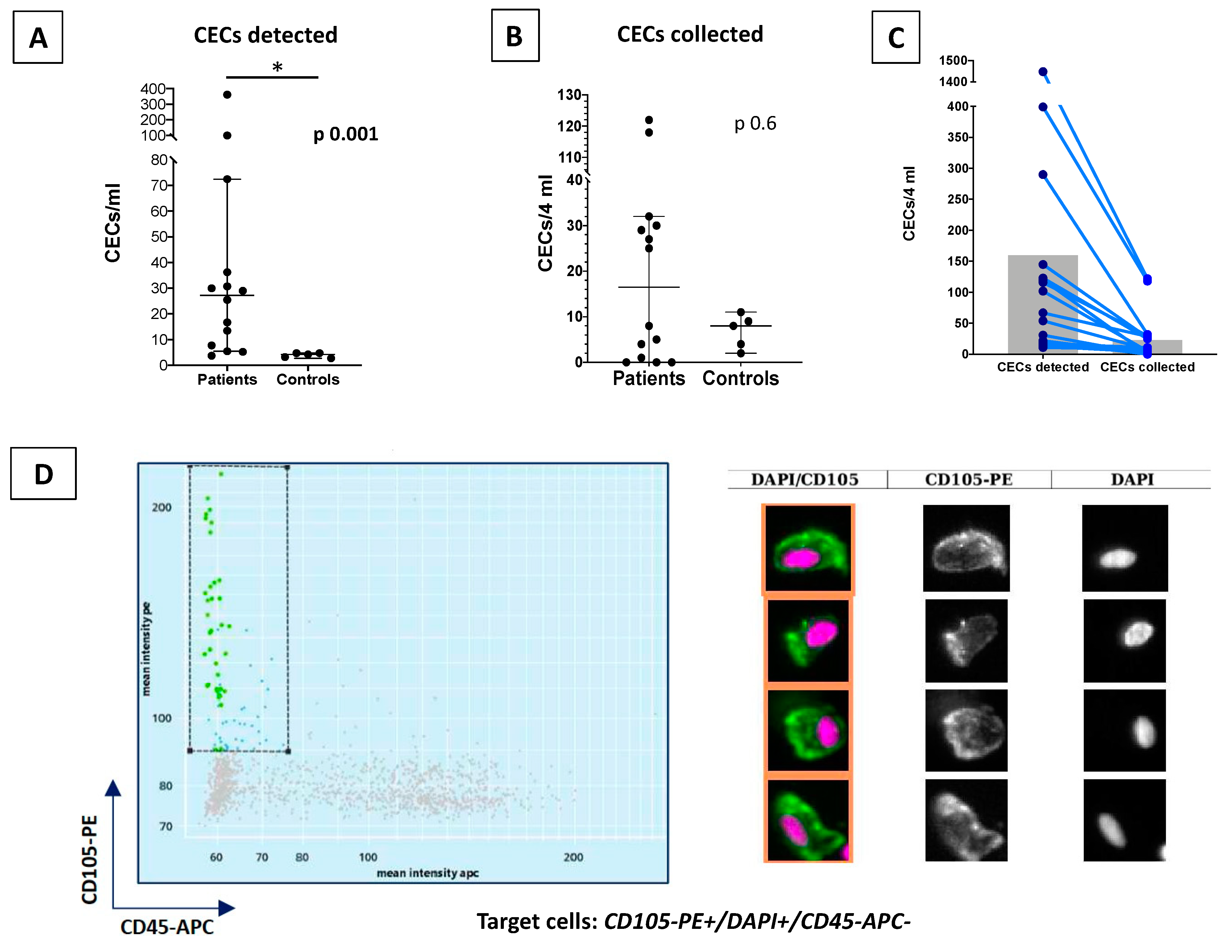
3.2. CEC and HSPCs Enumeration and Collection
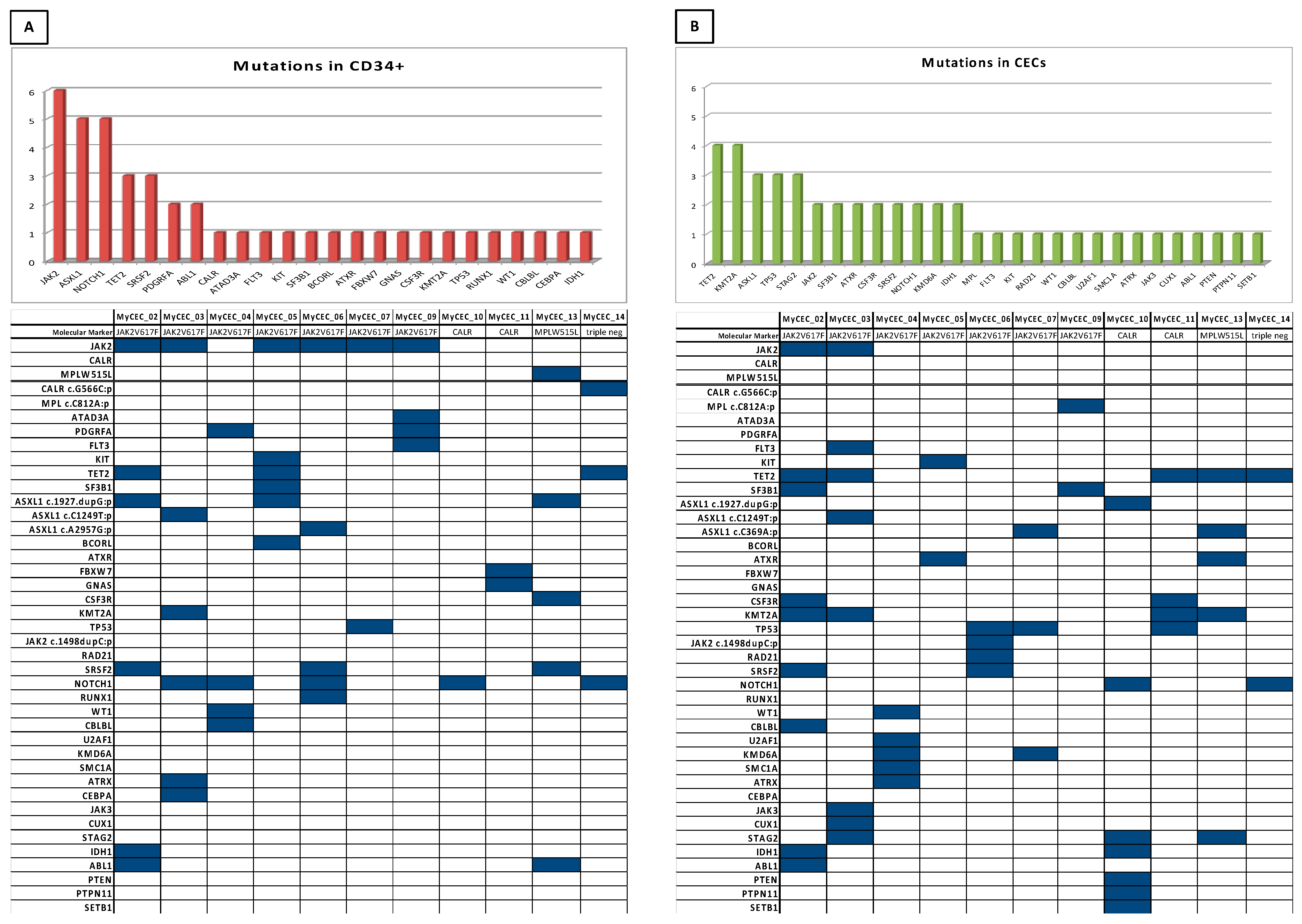
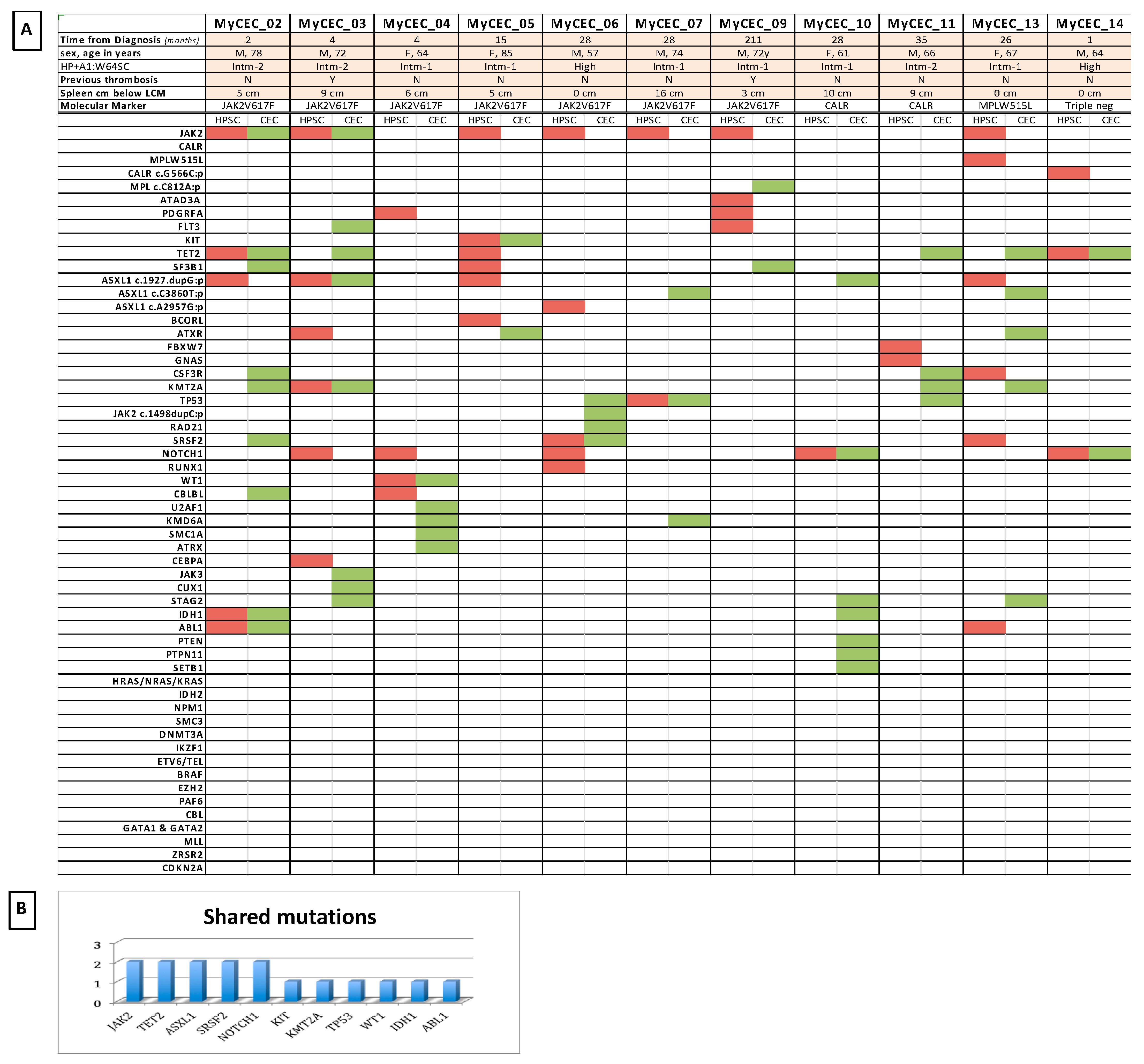
3.3. Comparative NGS Analysis on PMF Patients’ HSPCs and CECs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Circulating Endothelial Cell Identification by CellSearch Protocol
Appendix B. CECs Collection with DEPArray System Protocol
Appendix C. Protocol for DNA Extraction, Amplification and NGS Analysis
References
- Tefferi, A. Myeloproliferative neoplasms: A decade of discoveries and treatment advances. Am. J. Hematol. 2016, 91, 50–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, C.; McLornan, D. Myelofibrosis. Hematology 2014, 19, 120–121. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, A.M.; Lasho, T.L.; Guglielmelli, P.; Biamonte, F.; Pardanani, A.; Pereira, A.; Finke, C.; Score, J.; Gangat, N.; Mannarelli, C.; et al. Mutations and prognosis in primary myelofibrosis. Leukemia 2013, 27, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Rumi, E.; Pietra, D.; Pascutto, C.; Guglielmelli, P.; Martinez-Trillos, A.; Casetti, I.; Colomer, D.; Pieri, L.; Pratcorona, M.; Rotunno, G.; et al. Clinical effect of driver mutations of JAK2, CALR, or MPL in primary myelofibrosis. Blood 2014, 124, 1062–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tefferi, A.; Nicolosi, M.; Mudireddy, M.; Szuber, N.; Finke, C.M.; Lasho, T.L.; Hanson, C.A.; Ketterling, R.P.; Pardanani, A.; Gangat, N.; et al. Driver mutations and prognosis in primary myelofibrosis: Mayo-Careggi MPN alliance study of 1095 patients. Am. J. Hematol. 2018, 93, 348–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pikman, Y.; Lee, B.H.; Mercher, T.; McDowell, E.; Ebert, B.L.; Gozo, M.; Cuker, A.; Wernig, G.; Moore, S.; Galinsky, I.; et al. MPLW515L Is a Novel Somatic Activating Mutation in Myelofibrosis with Myeloid Metaplasia. PLoS Med. 2006, 3, e270. [Google Scholar] [CrossRef] [Green Version]
- Tefferi, A.; Vannucchi, A.M. Genetic Risk Assessment in Myeloproliferative Neoplasms. Mayo Clin. Proc. 2017, 92, 1283–1290. [Google Scholar] [CrossRef]
- Barbui, T.; Finazzi, G.; Falanga, A. Myeloproliferative neoplasms and thrombosis. Blood 2013, 122, 2176–2184. [Google Scholar] [CrossRef]
- Rungjirajittranon, T.; Owattanapanich, W.; Ungprasert, P.; Siritanaratkul, N.; Ruchutrakool, T. A systematic review and meta-analysis of the prevalence of thrombosis and bleeding at diagnosis of Philadelphia-negative myeloproliferative neoplasms. BMC Cancer 2019, 19, 184. [Google Scholar] [CrossRef]
- Farina, M.; D’Adda, M.; Daffini, R.; Polverelli, N.; Ferrari, S.; Bottelli, C.; Gramegna, D.; Cerqui, E.; Micheletti, M.; Bernardi, S.; et al. Jak2 Allelic Ratio Impacts on Vascular Event in Myelofibrosis by Increasing the Risk of Thrombosis. A Single Center Experience on 150 Patients. HemaSphere 2019, 3, 298. [Google Scholar] [CrossRef]
- Boveri, E.; Passamonti, F.; Rumi, E.; Pietra, D.; Elena, C.; Arcaini, L.; Pascutto, C.; Castello, A.; Cazzola, M.; Magrini, U.; et al. Bone marrow microvessel density in chronic myeloproliferative disorders: A study of 115 patients with clinicopathological and molecular correlations. Br. J. Haematol. 2008, 140, 162–168. [Google Scholar] [CrossRef]
- Grote, K.; Luchtefeld, M.; Schieffer, B. JANUS under stress—Role of JAK/STAT signaling pathway in vascular diseases. Vascul. Pharmacol. 2005, 43, 357–363. [Google Scholar] [CrossRef]
- Bar-Natan, M.; Hoffman, R. New insights into the causes of thrombotic events in patients with myeloproliferative neoplasms raise the possibility of novel therapeutic approaches. Haematologica 2019, 104, 3–6. [Google Scholar] [CrossRef]
- Teofili, L.; Larocca, L.M. Blood and endothelial cells: Together through thick and thin. Blood 2013, 121, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Farina, M.; Russo, D.; Hoffman, R. The possible role of mutated endothelial cells in myeloproliferative neoplasms. Haematologica 2020. [Google Scholar] [CrossRef]
- Oppliger Leibundgut, E.; Horn, M.P.; Brunold, C.; Pfanner-Meyer, B.; Marti, D.; Hirsiger, H.; Tobler, A.; Zwicky, C. Hematopoietic and endothelial progenitor cell trafficking in patients with myeloproliferative diseases. Haematologica 2006, 91, 1465–1472. [Google Scholar]
- Hill, J.M.; Zalos, G.; Halcox, J.P.J.; Schenke, W.H.; Waclawiw, M.A.; Quyyumi, A.A.; Finkel, T. Circulating Endothelial Progenitor Cells, Vascular Function, and Cardiovascular Risk. N. Engl. J. Med. 2003, 348, 593–600. [Google Scholar] [CrossRef]
- Ingram, D.A.; Mead, L.E.; Tanaka, H.; Meade, V.; Fenoglio, A.; Mortell, K.; Pollok, K.; Ferkowicz, M.J.; Gilley, D.; Yoder, M.C. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood 2004, 104, 2752–2760. [Google Scholar] [CrossRef]
- Yoder, M.C.; Mead, L.E.; Prater, D.; Krier, T.R.; Mroueh, K.N.; Li, F.; Krasich, R.; Temm, C.J.; Prchal, J.T.; Ingram, D.A. Redefining endothelial progenitor cells via clonal analysis and hematopoietic stem/progenitor cell principals. Blood 2007, 109, 1801–1809. [Google Scholar] [CrossRef] [Green Version]
- Rosti, V.; Bonetti, E.; Bergamaschi, G.; Campanelli, R.; Guglielmelli, P.; Maestri, M.; Magrini, U.; Massa, M.; Tinelli, C.; Viarengo, G.; et al. High Frequency of Endothelial Colony Forming Cells Marks a Non-Active Myeloproliferative Neoplasm with High Risk of Splanchnic Vein Thrombosis. PLoS ONE 2010, 5, e15277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosti, V.; Villani, L.; Riboni, R.; Poletto, V.; Bonetti, E.; Tozzi, L.; Bergamaschi, G.; Catarsi, P.; Dallera, E.; Novara, F.; et al. Spleen endothelial cells from patients with myelofibrosis harbor the JAK2V617F mutation. Blood 2013, 121, 360–368. [Google Scholar] [CrossRef]
- Guy, A.; Danaee, A.; Paschalaki, K.; Boureau, L.; Rivière, E.; Etienne, G.; Mansier, O.; Laffan, M.; Sekhar, M.; James, C. Absence of JAK2V617F Mutated Endothelial Colony-Forming Cells in Patients with JAK2V617F Myeloproliferative Neoplasms and Splanchnic Vein Thrombosis. HemaSphere 2020, 4, e364. [Google Scholar] [CrossRef] [PubMed]
- Teofili, L.; Martini, M.; Iachininoto, M.G.; Capodimonti, S.; Nuzzolo, E.R.; Torti, L.; Cenci, T.; Larocca, L.M.; Leone, G. Endothelial progenitor cells are clonal and exhibit the JAK2V617F mutation in a subset of thrombotic patients with Ph-negative myeloproliferative neoplasms. Blood 2011, 117, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Helman, R.; Pereira, W.d.O.; Marti, L.C.; Campregher, P.V.; Puga, R.D.; Hamerschlak, N.; Chiattone, C.S.; Santos, F.P.d.S. Granulocyte whole exome sequencing and endothelial JAK2V617F in patients with JAK2V617F positive Budd-Chiari Syndrome without myeloproliferative neoplasm. Br. J. Haematol. 2018, 180, 443–445. [Google Scholar] [CrossRef]
- Sozer, S.; Fiel, M.I.; Schiano, T.; Xu, M.; Mascarenhas, J.; Hoffman, R. The presence of JAK2V617F mutation in the liver endothelial cells of patients with Budd-Chiari syndrome. Blood 2009, 113, 5246–5249. [Google Scholar] [CrossRef] [PubMed]
- Widemann, A.; Sabatier, F.; Arnaud, L.; Bonello, L.; Al-Massarani, G.; Paganelli, F.; Poncelet, P.; Dignat-George, F. CD146-based immunomagnetic enrichment followed by multiparameter flow cytometry: A new approach to counting circulating endothelial cells. J. Thromb. Haemost. 2008, 6, 869–876. [Google Scholar] [CrossRef]
- Almici, C.; Skert, C.; Bruno, B.; Bianchetti, A.; Verardi, R.; Di Palma, A.; Neva, A.; Braga, S.; Piccinelli, G.; Piovani, G.; et al. Circulating endothelial cell count: A reliable marker of endothelial damage in patients undergoing hematopoietic stem cell transplantation. Bone Marrow Transplant. 2017, 52, 1637–1642. [Google Scholar] [CrossRef] [Green Version]
- Torres, C.; Fonseca, A.M.; Leander, M.; Matos, R.; Morais, S.; Campos, M.; Lima, M. Circulating endothelial cells in patients with venous thromboembolism and myeloproliferative neoplasms. PLoS ONE 2013, 8, e81574. [Google Scholar] [CrossRef] [Green Version]
- Passamonti, F.; Cervantes, F.; Vannucchi, A.M.; Morra, E.; Rumi, E.; Pereira, A.; Guglielmelli, P.; Pungolino, E.; Caramella, M.; Maffioli, M.; et al. A dynamic prognostic model to predict survival in primary myelofibrosis: A study by the IWG-MRT (International Working Group for Myeloproliferative Neoplasms Research and Treatment). Blood 2010, 115, 1703–1708. [Google Scholar] [CrossRef]
- Tefferi, A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia 2010, 24, 1128–1138. [Google Scholar] [CrossRef]
- Guglielmelli, P.; Lasho, T.L.; Rotunno, G.; Score, J.; Mannarelli, C.; Pancrazzi, A.; Biamonte, F.; Pardanani, A.; Zoi, K.; Reiter, A.; et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: An international study of 797 patients. Leukemia 2014, 28, 1804–1810. [Google Scholar] [CrossRef] [PubMed]
- Avecilla, S.T.; Goss, C.; Bleau, S.; Tonon, J.A.; Meagher, R.C. How do i perform hematopoietic progenitor cell selection? Transfusion 2016, 56, 1008–1012. [Google Scholar] [CrossRef] [Green Version]
- Vembadi, A.; Menachery, A.; Qasaimeh, M.A. Cell Cytometry: Review and Perspective on Biotechnological Advances. Front. Bioeng. Biotechnol. 2019, 7, 147. [Google Scholar] [CrossRef]
- Rowand, J.L.; Martin, G.; Doyle, G.V.; Miller, M.C.; Pierce, M.S.; Connelly, M.C.; Rao, C.; Terstappen, L.W.M.M. Endothelial cells in peripheral blood of healthy subjects and patients with metastatic carcinomas. Cytom. Part A 2007, 71A, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Di Trapani, M.; Manaresi, N.; Medoro, G. DEPArrayTM system: An automatic image-based sorter for isolation of pure circulating tumor cells. Cytom. Part A 2018, 93, 1260–1266. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, A.B.; Romani, A.; Freida, D.; Medoro, G.; Abonnenc, M.; Altomare, L.; Chartier, I.; Guergour, D.; Villiers, C.; Marche, P.N.; et al. Electronic sorting and recovery of single live cells from microlitre sized samples. Lab Chip 2006, 6, 121–126. [Google Scholar] [CrossRef]
- Bernardi, S.; Farina, M.; Zanaglio, C.; Cattina, F.; Polverelli, N.; Schieppati, F.; Re, F.; Foroni, C.; Malagola, M.; Dunbar, A.J.; et al. ETV6: A Candidate Gene for Predisposition to “Blend Pedigrees”? A Case Report from the NEXT-Famly Clinical Trial. Case Rep. Hematol. 2020, 2020, 2795656. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.G.; Maconachie, G.D.E.; Sheth, V.; McLean, R.J.; Gottlob, I. Development and clinical utility of a novel diagnostic nystagmus gene panel using targeted next-generation sequencing. Eur. J. Hum. Genet. 2017, 25, 725–734. [Google Scholar] [CrossRef] [Green Version]
- Cottrell, C.E.; Al-Kateb, H.; Bredemeyer, A.J.; Duncavage, E.J.; Spencer, D.H.; Abel, H.J.; Lockwood, C.M.; Hagemann, I.S.; O’Guin, S.M.; Burcea, L.C.; et al. Validation of a Next-Generation Sequencing Assay for Clinical Molecular Oncology. J. Mol. Diagn. 2014, 16, 89–105. [Google Scholar] [CrossRef]
- Chang, X.; Wang, K. Wannovar: Annotating genetic variants for personal genomes via the web. J. Med. Genet. 2012, 49, 433–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Kanda, Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Millner, L.M.; Linder, M.W.; Valdes, R., Jr. Circulating tumor cells: A review of present methods and the need to identify heterogeneous phenotypes. Ann. Clin. Lab. Sci. 2013, 43, 295–304. [Google Scholar]
- Smirnov, D.A.; Foulk, B.W.; Doyle, G.V.; Connelly, M.C.; Terstappen, L.W.M.M.; O’Hara, S.M. Global gene expression profiling of circulating endothelial cells in patients with metastatic carcinomas. Cancer Res. 2006, 66, 2918–2922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yildiz, A.; Güryildirim, M.; Pepeler, M.S.; Yazol, M.; Oktar, S.Ö.; Acar, K. Assessment of Endothelial Dysfunction with Flow-Mediated Dilatation in Myeloproliferative Disorders. Clin. Appl. Thromb. 2018, 24, 1102–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farinacci, M.; Krahn, T.; Dinh, W.; Volk, H.-D.; Düngen, H.-D.; Wagner, J.; Konen, T.; von Ahsen, O. Circulating endothelial cells as biomarker for cardiovascular diseases. Res. Pract. Thromb. Haemost. 2019, 3, 49–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilera-Diaz, A.; Vazquez, I.; Ariceta, B.; Mañú, A.; Blasco-Iturri, Z.; Palomino-Echeverría, S.; Larrayoz, M.J.; García-Sanz, R.; Prieto-Conde, M.I.; Chillón, M.D.C.; et al. Assessment of the clinical utility of four NGS panels in myeloid malignancies. Suggestions for NGS panel choice or design. PLoS ONE 2020, 15, e0227986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundberg, P.; Karow, A.; Nienhold, R.; Looser, R.; Hao-Shen, H.; Nissen, I.; Girsberger, S.; Lehmann, T.; Passweg, J.; Stern, M.; et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 2014, 123, 2220–2228. [Google Scholar] [CrossRef] [Green Version]
- Murray, P.D.F. The Development in vitro of the Blood of the Early Chick Embryo. Proc. R. Soc. B Biol. Sci. 1932, 111, 497–521. [Google Scholar] [CrossRef] [Green Version]
- Ueno, H.; Weissman, I.L. Clonal Analysis of Mouse Development Reveals a Polyclonal Origin for Yolk Sac Blood Islands. Dev. Cell 2006, 11, 519–533. [Google Scholar] [CrossRef] [Green Version]
- Vogeli, K.M.; Jin, S.-W.; Martin, G.R.; Stainier, D.Y.R. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature 2006, 443, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Van Egeren, D.; Escabi, J.; Nguyen, M.; Liu, S.; Reilly, C.R.; Patel, S.; Kamaz, B.; Kalyva, M.; DeAngelo, D.J.; Galinsky, I.; et al. Reconstructing the Lineage Histories and Differentiation Trajectories of Individual Cancer Cells in Myeloproliferative Neoplasms. Cell Stem Cell 2021, 28, 514–523.e9. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.; Lee, J.; Moore, L.; Baxter, J.E.; Hewinson, J.; Dawson, K.J.; Menzies, A.; Godfrey, A.L.; Green, A.R.; Campbell, P.J.; et al. Driver Mutation Acquisition in Utero and Childhood Followed By Lifelong Clonal Evolution Underlie Myeloproliferative Neoplasms. Blood 2020, 136, LBA-1. [Google Scholar] [CrossRef]
- Loomans, C.J.M.; Wan, H.; de Crom, R.; van Haperen, R.; de Boer, H.C.; Leenen, P.J.M.; Drexhage, H.A.; Rabelink, T.J.; van Zonneveld, A.J.; Staal, F.J.T. Angiogenic murine endothelial progenitor cells are derived from a myeloid bone marrow fraction and can be identified by endothelial NO synthase expression. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1760–1767. [Google Scholar] [CrossRef] [Green Version]
- Dudley, A.C.; Udagawa, T.; Melero-Martin, J.M.; Shih, S.-C.; Curatolo, A.; Moses, M.A.; Klagsbrun, M. Bone marrow is a reservoir for proangiogenic myelomonocytic cells but not endothelial cells in spontaneous tumors. Blood 2010, 116, 3367–3371. [Google Scholar] [CrossRef]
- Bernardi, S.; Farina, M. Exosomes and extracellular vesicles in myeloid Neoplasia: The multiple and complex roles played by these “magic bullets”. Biology 2021, 10, 105. [Google Scholar] [CrossRef]
- Bernardi, S.; Balbi, C. Extracellular vesicles: From biomarkers to therapeutic tools. Biology 2020, 9, 258. [Google Scholar] [CrossRef]
- Zhan, H.; Lin, C.H.S.; Segal, Y.; Kaushansky, K. The JAK2V617F-bearing vascular niche promotes clonal expansion in myeloproliferative neoplasms. Leukemia 2018, 32, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Guy, A.; Gourdou-Latyszenok, V.; Le Lay, N.; Peghaire, C.; Kilani, B.; Dias, J.V.; Duplaa, C.; Renault, M.-A.; Denis, C.; Villeval, J.L.; et al. Vascular endothelial cell expression of JAK2V617F is sufficient to promote a pro-thrombotic state due to increased P-selectin expression. Haematologica 2019, 104, 70–81. [Google Scholar] [CrossRef] [Green Version]
- Etheridge, S.L.; Roh, M.E.; Cosgrove, M.E.; Sangkhae, V.; Fox, N.E.; Chen, J.; López, J.A.; Kaushansky, K.; Hitchcock, I.S. JAK2V 617 F-positive endothelial cells contribute to clotting abnormalities in myeloproliferative neoplasms. Proc. Natl. Acad. Sci. USA 2014, 111, 2295–2300. [Google Scholar] [CrossRef] [Green Version]
- Guadall, A.; Lesteven, E.; Letort, G.; Awan Toor, S.; Delord, M.; Pognant, D.; Brusson, M.; Verger, E.; Maslah, N.; Giraudier, S.; et al. Endothelial Cells Harbouring the JAK2V617F Mutation Display Pro-Adherent and Pro-Thrombotic Features. Thromb. Haemost. 2018, 118, 1586–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castiglione, M.; Jiang, Y.P.; Mazzeo, C.; Lee, S.; Chen, J.S.; Kaushansky, K.; Yin, W.; Lin, R.Z.; Zheng, H.; Zhan, H. Endothelial JAK2V617F mutation leads to thrombosis, vasculopathy, and cardiomyopathy in a murine model of myeloproliferative neoplasm. J. Thromb. Haemost. 2020. [Google Scholar] [CrossRef] [PubMed]
- Polzer, B.; Medoro, G.; Pasch, S.; Fontana, F.; Zorzino, L.; Pestka, A.; Andergassen, U.; Meier-Stiegen, F.; Czyz, Z.T.; Alberter, B.; et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 2014, 6, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Rapone, C.; Bregola, G.; Aversa, R.; de Meo, A.; Signorini, G.; Sergio, M.; Ferrarini, A.; Lanzellotto, R.; Medoro, G.; et al. Isolation and genetic analysis of pure cells from forensic biological mixtures: The precision of a digital approach. Forensic Sci. Int. Genet. 2017, 29, 225–241. [Google Scholar] [CrossRef] [Green Version]
- Bolognesi, C.; Forcato, C.; Buson, G.; Fontana, F.; Mangano, C.; Doffini, A.; Sero, V.; Lanzellotto, R.; Signorini, G.; Calanca, A.; et al. Digital Sorting of Pure Cell Populations Enables Unambiguous Genetic Analysis of Heterogeneous Formalin-Fixed Paraffin-Embedded Tumors by Next Generation Sequencing. Sci. Rep. 2016, 6, 20944. [Google Scholar] [CrossRef]
- Tenedini, E.; Bernardis, I.; Artusi, V.; Artuso, L.; Roncaglia, E.; Guglielmelli, P.; Pieri, L.; Bogani, C.; Biamonte, F.; Rotunno, G.; et al. Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia 2014, 28, 1052–1059. [Google Scholar] [CrossRef] [Green Version]
- Rotunno, G.; Pacilli, A.; Artusi, V.; Rumi, E.; Maffioli, M.; Delaini, F.; Brogi, G.; Fanelli, T.; Pancrazzi, A.; Pietra, D.; et al. Epidemiology and clinical relevance of mutations in postpolycythemia vera and postessential thrombocythemia myelofibrosis: A study on 359 patients of the AGIMM group. Am. J. Hematol. 2016, 91, 681–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salati, S.; Zini, R.; Nuzzo, S.; Guglielmelli, P.; Pennucci, V.; Prudente, Z.; Ruberti, S.; Rontauroli, S.; Norfo, R.; Bianchi, E.; et al. Integrative analysis of copy number and gene expression data suggests novel pathogenetic mechanisms in primary myelofibrosis. Int. J. Cancer 2016, 138, 1657–1669. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]

| Features | PMF Patients | Healthy Controls | p Value |
|---|---|---|---|
| N or Median (% or Range) | N or Median (% or Range) | ||
| Age (years) | 71.5 (54–85) | 65 (35–84) | 0.22 |
| Male | 9/14 (64%) | 1/5 (20%) | 0.14 |
| PMF | 14/ 14 | 0/5 | |
| Months from Diagnosis | 20.5 (1–211) | NA | |
| WBC (×109/L) | 7.3 (3.8–117) | 5.5 (3.9–9.1) | 0.35 |
| Hb (g/dL) | 10.7 (8–14.8) | 13.6 (12–14.5) | 0.01 |
| PLT (×109/L) | 211 (50–885) | 257 (179–412) | 0.77 |
| Constitutional Symptoms | 4 (29%) | NA | |
| Altered karyotypes | 3 (21%) | NA | |
| Previous Thrombosis | 2 (14%) | 0 (0%) | 0.99 |
| Splenomegaly | |||
| N° patients | 11 (79%) | 0 (0%) | |
| cm below LMC | 5 (0–16) | 0 | |
| Treatment | |||
| Hydroxyurea | 4 (29%) | 0 (0%) | |
| None | 10 (71%) | 5 (100%) | |
| BM fibrosis | |||
| WHO grade 1 | 7 (50%) | NA | |
| WHO grade 2 | 6 (43%) | NA | |
| WHO grade 3 | 1 (7%) | NA | |
| DIPSS (at samples collection) | |||
| Low | 0 (0%) | NA | |
| Intermediate 1 | 7 (50%) | NA | |
| Intermediate 2 | 5 (36%) | NA | |
| High | 2 (14%) | NA | |
| Driver Mutations | |||
| JAK2 | 9 (64%) | NA | |
| CALR | 2 (14%) | NA | |
| MPL | 2 (14%) | NA | |
| Triple negative | 1 (7%) | NA |
| Features | PMF Patients | Healthy Controls | p Value | ||
|---|---|---|---|---|---|
| CEC Median (Range); n pts | p Value | CEC Median (Range); n pts | p Value | ||
| CECs detected | 109 (15–1448); n = 14 | 17 (11–19); n = 5 | 0.001 | ||
| CECs collected | 16.5 (0–118); n = 14 | 8 (2–11); n = 5 | 0.6 | ||
| Sex | 0.53 | NA | |||
| Male | 120 (31–1448); n = 9 | 17; n = 1 | NA | ||
| Female | 116 (54–290); n = 5 | 16 (11–19); n = 4 | 0.02 | ||
| Age | 0.21 | 0.2 | |||
| ≥70 years | 54 (15–399); n = 7 | 12 (11–13); n = 2 | 0.06 | ||
| <70 years | 120 (22–1448); n = 7 | 19 (17–19); n = 3 | 0.02 | ||
| Time from diagnosis | 0.62 | ||||
| <2 years | 67 (21–399); n = 7 | NA | |||
| >2 years | 116 (15–1448); n = 7 | NA | |||
| White blood count | 0.36 | NA | |||
| >10 × 109/L | 67 (11–1448); n = 5 | 0 | NA | ||
| ≤10 × 109/L | 123 (15–1448); n = 9 | 17 (11–19); n = 5 | 0.007 | ||
| Constitutional symptoms | 0.95 | ||||
| Yes | 93.5 (22–399); n = 4 | NA | |||
| No | 109 (15–1448); n = 10 | NA | |||
| History of thrombosis | 0.30 | ||||
| Yes | 217.5 (21–399); n = 4 | 0 | |||
| No | 84.5 (15–1448); n = 10 | 17 (11–19); n = 5 | |||
| Splenomegaly | 0.99 | ||||
| Yes | 116 (15–1448); n = 11 | 0 | |||
| No | 102 (22–290); n = 3 | 17 (11–19); n = 5 | |||
| Treatment | 0.94 | ||||
| Hydroxyurea | 102 (54–290); n = 5 | 0 | |||
| No treatment | 116 (15–1448); n = 9 | 17 (11–19); n = 5 | |||
| DIPSS | 0.90 | ||||
| Interm1 | 116 (25–145); n = 7 | NA | |||
| Interm2-High | 102 (21–1448); n = 7 | NA | |||
| Driver mutations | 0.30 | ||||
| JAK2 | 67 (15–399); n = 9 | NA | |||
| Non JAK2 mutations | 120 (22–1448); n = 5 | NA | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farina, M.; Bernardi, S.; Polverelli, N.; D’Adda, M.; Malagola, M.; Bosio, K.; Re, F.; Almici, C.; Dunbar, A.; Levine, R.L.; et al. Comparative Mutational Profiling of Hematopoietic Progenitor Cells and Circulating Endothelial Cells (CECs) in Patients with Primary Myelofibrosis. Cells 2021, 10, 2764. https://doi.org/10.3390/cells10102764
Farina M, Bernardi S, Polverelli N, D’Adda M, Malagola M, Bosio K, Re F, Almici C, Dunbar A, Levine RL, et al. Comparative Mutational Profiling of Hematopoietic Progenitor Cells and Circulating Endothelial Cells (CECs) in Patients with Primary Myelofibrosis. Cells. 2021; 10(10):2764. https://doi.org/10.3390/cells10102764
Chicago/Turabian StyleFarina, Mirko, Simona Bernardi, Nicola Polverelli, Mariella D’Adda, Michele Malagola, Katia Bosio, Federica Re, Camillo Almici, Andrew Dunbar, Ross L. Levine, and et al. 2021. "Comparative Mutational Profiling of Hematopoietic Progenitor Cells and Circulating Endothelial Cells (CECs) in Patients with Primary Myelofibrosis" Cells 10, no. 10: 2764. https://doi.org/10.3390/cells10102764
APA StyleFarina, M., Bernardi, S., Polverelli, N., D’Adda, M., Malagola, M., Bosio, K., Re, F., Almici, C., Dunbar, A., Levine, R. L., & Russo, D. (2021). Comparative Mutational Profiling of Hematopoietic Progenitor Cells and Circulating Endothelial Cells (CECs) in Patients with Primary Myelofibrosis. Cells, 10(10), 2764. https://doi.org/10.3390/cells10102764









