NUAK Kinases: Brain–Ovary Axis
Abstract
1. Introduction
1.1. Liver Kinase B (LKB) 1, Adenosine Monophosphate (AMP)-Activated Protein Kinase (AMPK) and AMPK-Related Kinases
1.2. LKB1, AMPK, and AMPK-Related Proteins in Cancer
1.3. The NUAK1 and NUAK2 Protein Kinases
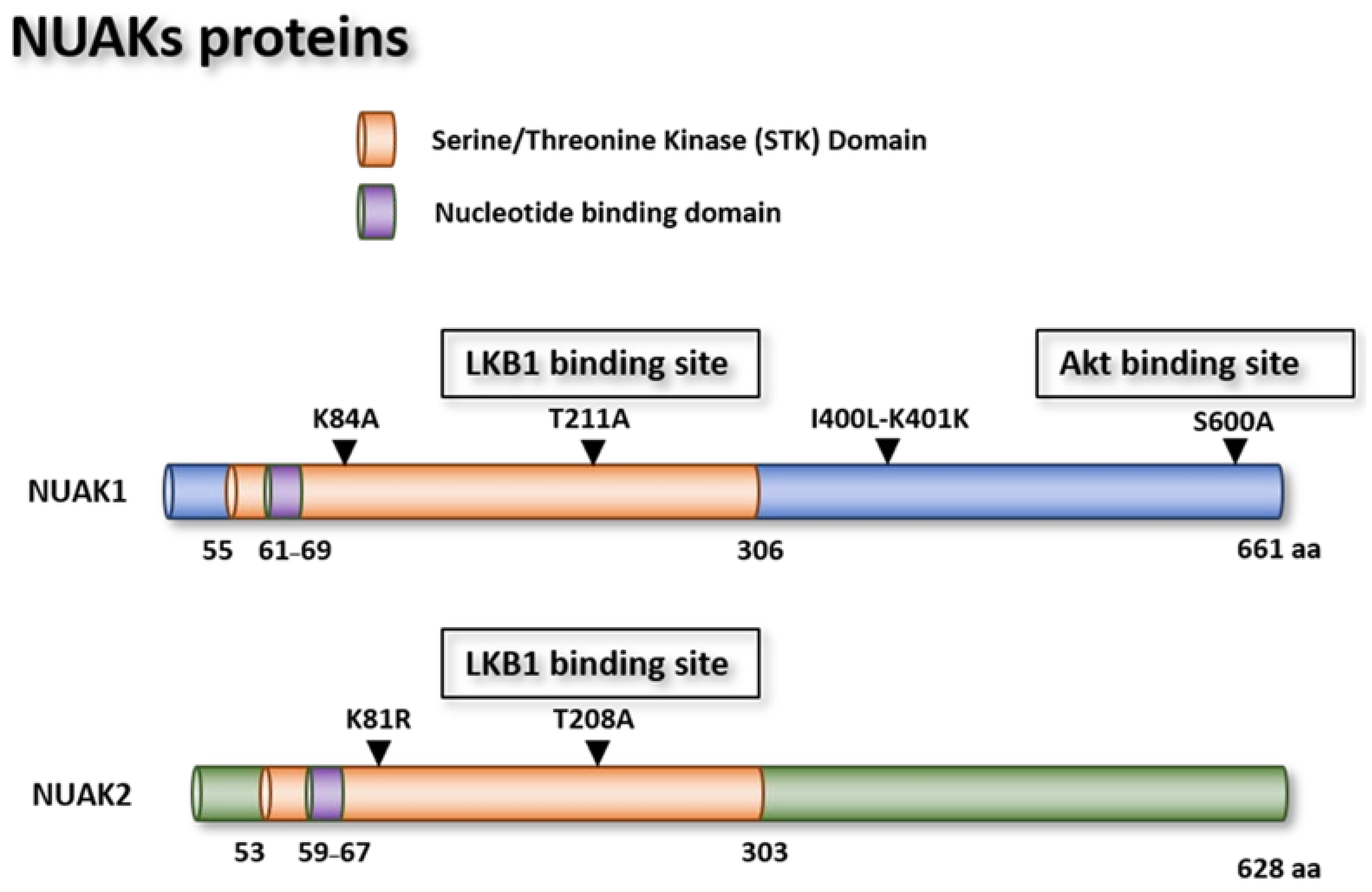
1.3.1. NUAK Genes
NUAK1 Mutants
NUAK1 Tissue Expression
NUAK2 Gene
NUAK2 Mutants
NUAK2 Tissue Expression
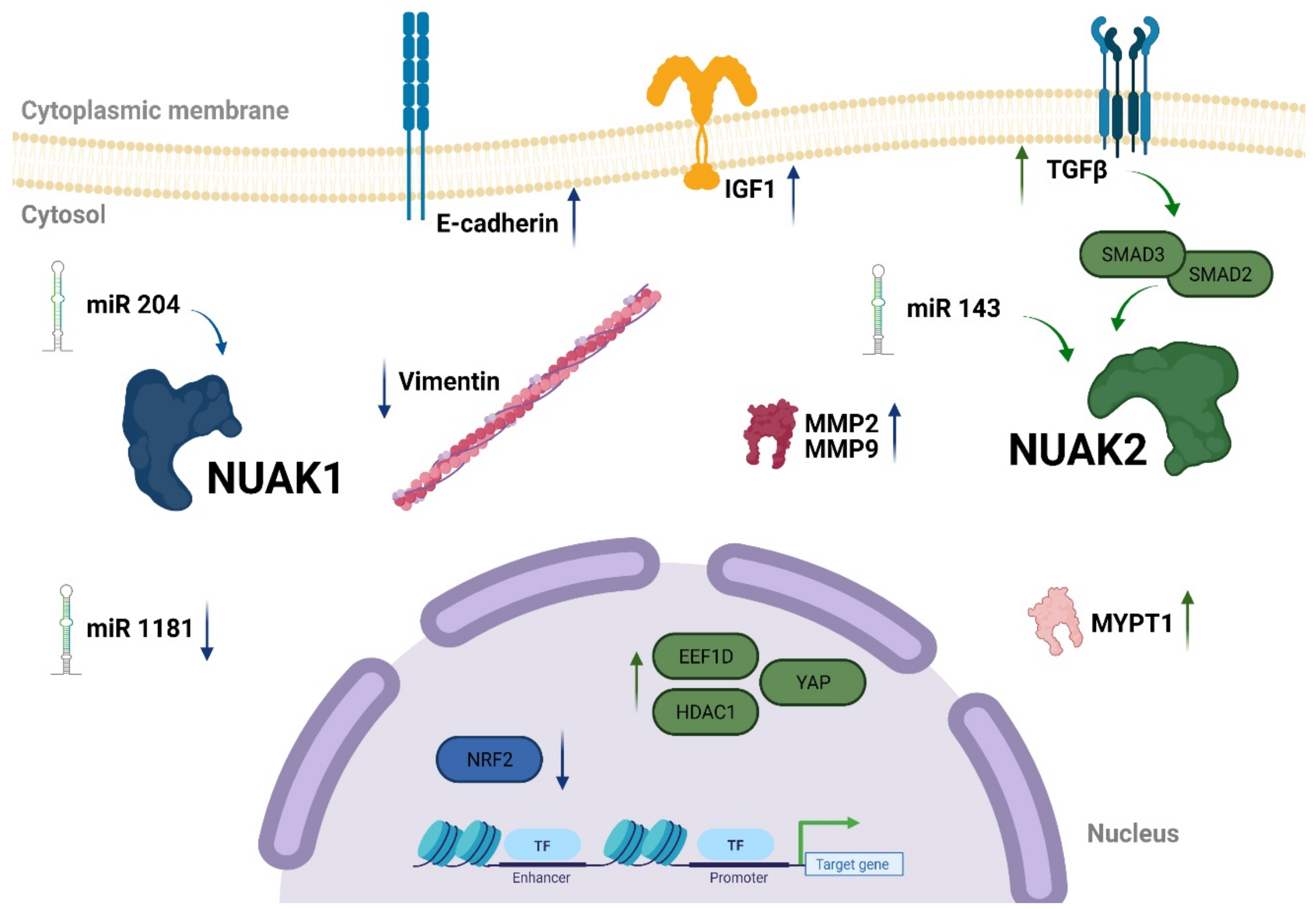
1.3.2. NUAK1 and NUAK2 in Cancer
Apoptosis and Senescence
Migration and Invasiveness
1.3.3. The NUAK1 and NUAK2 Regulatory Network
1.3.4. NUAK1 and NUAK2 in Ovary and Brain
2. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Curi, R.; Newsholme, P.; Newsholme, E.A. Metabolism of pyruvate by isolated rat mesenteric lymphocytes, lymphocyte mitochondria and isolated mouse macrophages. Biochem. J. 1988, 250, 383–388. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Li, L.; Tan, J.; Miao, Y.; Lei, P.; Zhang, Q. ROS and Autophagy: Interactions and Molecular Regulatory Mechanisms. Cell. Mol. Neurobiol. 2015, 35, 615–621. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Elazar, Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007, 17, 422–427. [Google Scholar] [CrossRef]
- Resta, N.; Simone, C.; Mareni, C.; Montera, M.; Gentile, M.; Susca, F.; Gristina, R.; Pozzi, S.; Bertario, L.; Bufo, P.; et al. STK11 mutations in Peutz-Jeghers syndrome and sporadic colon cancer. Cancer Res. 1998, 58, 4799–4801. [Google Scholar]
- Shackelford, D.B.; Shaw, R.J. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer 2009, 9, 563–575. [Google Scholar] [CrossRef]
- Müller, M.; Lutter, D.; Püschel, A.W.; Hardie, D.G.; Monteverde, T.; Muthalagu, N.; Port, J.; Murphy, D.J.; Hardie, D.G.; Ross, F.A.; et al. Europe PMC Funders Group Molecular Pathways: Is AMPK a Friend or a Foe in Cancer? Orphanet J. Rare Dis. 2012, 13, 3–8. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Ross, F.A.; Hawley, S.A.; Auciello, F.R.; Gowans, G.J.; Atrih, A.; Lamont, D.J.; Hardie, D.G. Mechanisms of Paradoxical Activation of AMPK by the Kinase Inhibitors SU6656 and Sorafenib. Cell Chem. Biol. 2017, 24, 813–824.e4. [Google Scholar] [CrossRef]
- Lizcano, J.M.; Göransson, O.; Toth, R.; Deak, M.; Morrice, N.A.; Boudeau, J.; Hawley, S.A.; Udd, L.; Mäkelä, T.P.; Hardie, D.G.; et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004, 23, 833–843. [Google Scholar] [CrossRef]
- Jaleel, M.; Villa, F.; Deak, M.; Toth, R.; Prescott, A.R.; Van Aalten, D.M.F.; Alessi, D.R. The ubiquitin-associated domain of AMPK-related kinases regulates conformation and LKB1-mediated phosphorylation and activation. Biochem. J. 2006, 394, 545–555. [Google Scholar] [CrossRef]
- Suzuki, A.; Kusakai, G.I.; Kishimoto, A.; Lu, J.; Ogura, T.; Lavin, M.F.; Esumi, H. Identification of a novel protein kinase mediating Akt survival signaling to the ATM protein. J. Biol. Chem. 2003, 278, 48–53. [Google Scholar] [CrossRef]
- Crump, J.G.; Zhen, M.; Jin, Y.; Bargmann, C.I. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron 2001, 29, 115–129. [Google Scholar] [CrossRef]
- Kishi, M.; Pan, Y.A.; Crump, J.G.; Sanes, J.R. Mammalian SAD kinases are required for neuronal polarization. Science 2005, 307, 929–932. [Google Scholar] [CrossRef]
- Molina, E.; Hong, L.; Chefetz, I.I. AMPKα-like proteins as LKB1 downstream targets in cell physiology and cancer. J. Mol. Med. 2021, 99, 651–662. [Google Scholar] [CrossRef]
- Feldman, J.D.; Vician, L.; Crispino, M.; Hoe, W.; Baudry, M.; Herschman, H.R. The salt-inducible kinase, SIK, is induced by depolarization in brain. J. Neurochem. 2000, 74, 2227–2238. [Google Scholar] [CrossRef]
- Yang, L.; Xie, N.; Huang, J.; Huang, H.; Xu, S.; Wang, Z.; Cai, J. SIK1-LNC represses the proliferative, migrative, and invasive abilities of lung cancer cells. Oncol. Targets. Ther. 2018, 11, 4197–4206. [Google Scholar] [CrossRef]
- Katoh, Y.; Takemori, H.; Horike, N.; Doi, J.; Muraoka, M.; Min, L.; Okamoto, M. Salt-inducible kinase (SIK) isoforms: Their involvement in steroidogenesis and adipogenesis. Mol. Cell. Endocrinol. 2004, 217, 109–112. [Google Scholar] [CrossRef]
- Horike, N.; Takemori, H.; Katoh, Y.; Doi, J.; Min, L.; Asano, T.; Sun, X.J.; Yamamoto, H.; Kasayama, S.; Muraoka, M.; et al. Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J. Biol. Chem. 2003, 278, 18440–18447. [Google Scholar] [CrossRef]
- Timm, T.; Li, X.Y.; Biernat, J.; Jiao, J.; Mandelkow, E.; Vandekerckhove, J.; Mandelkow, E.M. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 2003, 22, 5090–5101. [Google Scholar] [CrossRef]
- Johnson, L.N.; Noble, M.E.M.; Owen, D.J. Active and inactive protein kinases: Structural basis for regulation. Cell 1996, 85, 149–158. [Google Scholar] [CrossRef]
- Heyer, B.S.; Kochanowski, H.; Solter, D. Expression of Melk, a new protein kinase, during early mouse development. Dev. Dyn. 1999, 215, 344–351. [Google Scholar] [CrossRef]
- Jeon, S.M.; Chandel, N.S.; Hay, N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 2012, 485, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.-M.; Hay, N. The dark face of AMPK as an essential tumor promoter. Cell. Logist. 2012, 2, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Ogura, T.; Kishimoto, A.; Minegishi, Y.; Nakajima, N.; Miyazaki, M.; Esumi, H. Critical roles of AMP-activated protein kinase in constitutive tolerance of cancer cells to nutrient deprivation and tumor formation. Oncogene 2002, 21, 6082–6090. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Legembre, P.; Schickel, R.; Barnhart, B.C.; Peter, M.E. Identification of SNF1/AMP kinase-related kinase as an NF-κB- regulated anti-apoptotic kinase involved in CD95-induced motility and invasiveness. J. Biol. Chem. 2004, 279, 46742–46747. [Google Scholar] [CrossRef]
- Suzuki, A.; Lu, J.; Kusakai, G.; Kishimoto, A.; Ogura, T.; Esumi, H. ARK5 Is a Tumor Invasion-Associated Factor Downstream of Akt Signaling. Mol. Cell. Biol. 2004, 24, 3526–3535. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Kusakai, G.I.; Kishimoto, A.; Minegichi, Y.; Ogura, T.; Esumi, H. Induction of cell-cell detachment during glucose starvation through F-actin conversion by SNARK, the fourth member of the AMP-activated protein kinase catalytic subunit family. Biochem. Biophys. Res. Commun. 2003, 311, 156–161. [Google Scholar] [CrossRef]
- Port, J.; Muthalagu, N.; Raja, M.; Ceteci, F.; Monteverde, T.; Kruspig, B.; Hedley, A.; Kalna, G.; Lilla, S.; Neilson, L.; et al. Colorectal tumors require NUAK1 for protection from oxidative stress. Cancer Discov. 2018, 8, 632–647. [Google Scholar] [CrossRef]
- Lefebvre, D.L.; Bai, Y.; Shahmolky, N.; Sharma, M.; Poon, R.; Drucker, D.J.; Rosen, C.F. Identification and characterization of a novel sucrose-non-fermenting protein kinase/AMP-activated protein kinase-related protein kinase, SNARK. Biochem. J. 2001, 355, 297–305. [Google Scholar] [CrossRef]
- Humbert, N.; Navaratnam, N.; Augert, A.; Da Costa, M.; Martien, S.; Wang, J.; Martinez, D.; Abbadie, C.; Carling, D.; De Launoit, Y.; et al. Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. EMBO J. 2010, 29, 376–386. [Google Scholar] [CrossRef]
- Hou, X.; Liu, J.E.; Liu, W.; Liu, C.Y.; Liu, Z.Y.; Sun, Z.Y. A new role of NUAK1: Directly phosphorylating p53 and regulating cell proliferation. Oncogene 2011, 30, 2933–2942. [Google Scholar] [CrossRef] [PubMed]
- Zagórska, A.; Deak, M.; Campbell, D.G.; Banerjee, S.; Hirano, M.; Aizawa, S.; Prescott, A.R.; Alessi, D.R. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Sci. Signal. 2010, 3, ra25. [Google Scholar] [CrossRef]
- Legembre, P.; Barnhart, B.C.; Zheng, L.; Vijayan, S.; Straus, S.E.; Puck, J.; Dale, J.K.; Lenardo, M.; Peter, M.E. Induction of apoptosis and activation of NF-κB by CD95 require different signalling thresholds. EMBO Rep. 2004, 5, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Lin, W.; Zhang, L.; Jilg, N.; Shao, R.X.; Schaefer, E.A.K.; Zhao, H.; Fusco, D.N.; Peng, L.F.; Kato, N.; et al. The AMPK-related kinase SNARK regulates hepatitis C virus replication and pathogenesis through enhancement of TGF-β signaling. J. Hepatol. 2013, 59, 942–948. [Google Scholar] [CrossRef]
- NCBI National Center for Biotechnology Information (Internet). National Library of Medicine: Bethesda, MD, USA, National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 20 August 2021).
- Mo, G.; Zhang, B.; Jiang, Q. Role of ARK5 in cancer and other diseases (Review). Exp. Ther. Med. 2021, 22, 697. [Google Scholar] [CrossRef] [PubMed]
- Hay, N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell 2005, 8, 179–183. [Google Scholar] [CrossRef]
- Fouqué, A.; Legembre, P. Study of the CD95-mediated non-apoptotic signaling pathway: PI3K. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; pp. 103–110. [Google Scholar] [CrossRef]
- Suzuki, A.; Iida, S.; Kato-Uranishi, M.; Tajima, E.; Zhan, F.; Hanamura, I.; Huang, Y.; Ogura, T.; Takahashi, S.; Ueda, R.; et al. ARK5 is transcriptionally regulated by the Large-MAF family and mediates IGF-1-induced cell invasion in multiple myeloma: ARK5 as a new molecular determinant of malignant multiple myeloma. Oncogene 2005, 24, 6936–6944. [Google Scholar] [CrossRef]
- Lundberg, A.S.; Hahn, W.C.; Gupta, P.; Weinberg, R.A. Genes involved in senescence and immortalization. Curr. Opin. Cell Biol. 2000, 12, 705–709. [Google Scholar] [CrossRef]
- Takahashi, A.; Ohtani, N.; Yamakoshi, K.; Iida, S.; Tahara, H.; Nakayama, K.; Nakayama, K.I.; Ide, T.; Saya, H.; Hara, E. Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat. Cell Biol. 2006, 8, 1291–1297. [Google Scholar] [CrossRef]
- Hao, Y.; Chun, A.; Cheung, K.; Rashidi, B.; Yang, X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008, 283, 5496–5509. [Google Scholar] [CrossRef]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.C.; Pepe-Mooney, B.; Galli, G.G.; Dill, M.T.; Huang, H.T.; Hao, M.; Wang, Y.; Liang, H.; Calogero, R.A.; Camargo, F.D. NUAK2 is a critical YAP target in liver cancer. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Harris, A.L. Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Namiki, T.; Tanemura, A.; Valencia, J.C.; Coelho, S.G.; Passeron, T.; Kawaguchi, M.; Vieira, W.D.; Ishikawa, M.; Nishijima, W.; Izumo, T.; et al. AMP kinase-related kinase NUAK2 affects tumor growth, migration, and clinical outcome of human melanoma. Proc. Natl. Acad. Sci. USA 2011, 108, 6597–6602. [Google Scholar] [CrossRef] [PubMed]
- Bambang, A.; Tanadi, C.; Sumarpo, A. Deciphering the role of AMPK-related kinase 5 in human cancer progression and metastasis. Biomed. Res. Ther. 2019, 6, 3396–3404. [Google Scholar] [CrossRef]
- Liu, J.; Tang, G.; Huang, H.; Li, H.; Zhang, P.; Xu, L. Expression level of NUAK1 in human nasopharyngeal carcinoma and its prognostic significance. Eur. Arch. Oto-Rhino-Laryngol. 2018, 275, 2563–2573. [Google Scholar] [CrossRef]
- Ye, X.T.; Guo, A.J.; Yin, P.F.; Cao, X.D.; Chang, J.C. Overexpression of NUAK1 is associated with disease-free survival and overall survival in patients with gastric cancer. Med. Oncol. 2014, 31, 61. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, B.; Sun, X.; Lu, S.; Liu, Z.; Liu, Y.; Li, H.; Wang, L.; Wang, X.; Zhao, C. MiR-204 inhibits human NSCLC metastasis through suppression of NUAK1. Br. J. Cancer 2014, 111, 2316–2327. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Zhang, J.; Chen, W.; Pan, S.; Zhi, X.; Wen, L.; Zhou, Y.; Chen, B.W.; Qiu, J.; Zhang, Y.; et al. ARK5 promotes doxorubicin resistance in hepatocellular carcinoma via epithelial-mesenchymal transition. Cancer Lett. 2016, 377, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.Z.; Yu, J.; Liu, H.Y.; Dong, R.H.; Cao, X.C. ARK5 is associated with the invasive and metastatic potential of human breast cancer cells. J. Cancer Res. Clin. Oncol. 2012, 138, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Liu, G.; Xu, N.; You, X.; Zhou, H.; Zhao, X.; Liu, Q. Knockdown of ARK5 Expression Suppresses Invasion and Metastasis of Gastric Cancer. Cell. Physiol. Biochem. 2017, 42, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, J.; Jäck, H.M. Serum microRNAs as powerful cancer biomarkers. Biochim. Biophys. Acta Rev. Cancer 2010, 18, 200–207. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460469. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Li, J.H.; Li, G.; Wang, S.R. Activation of ARK5/miR-1181/HOXA10 axis promotes epithelial-mesenchymal transition in ovarian cancer. Oncol. Rep. 2015, 34, 1193–1202. [Google Scholar] [CrossRef]
- Fu, T.G.; Wang, L.; Li, W.; Li, J.Z.; Li, J. MiR-143 inhibits oncogenic traits by degrading NUAK2 in glioblastoma. Int. J. Mol. Med. 2016, 37, 1627–1635. [Google Scholar] [CrossRef]
- Li, J.; Feng, B.; Nie, Y.; Jiao, P.; Lin, X.; Huang, M.; An, R.; He, Q.; Zhou, H.E.; Salomon, A.; et al. Sucrose nonfermenting-related kinase regulates both adipose inflammation and energy homeostasis in mice and humans. Diabetes 2018, 67, 400–411. [Google Scholar] [CrossRef]
- Bonnard, C.; Navaratnam, N.; Ghosh, K.; Chan, P.W.; Tan, T.T.; Pomp, O.; Yu Jin Ng, A.; Tohari, S.; Changede, R.; Carling, D.; et al. A loss-of-function NUAK2 mutation in humans causes anencephaly due to impaired Hippo-YAP signaling. J. Exp. Med. 2020, 217, e20191561. [Google Scholar] [CrossRef] [PubMed]
- Kolliopoulos, C.; Raja, E.; Razmara, M.; Heldin, P.; Heldin, C.H.; Moustakas, A.; Van Der Heide, L.P. Transforming growth factor (TGF) induces NUAK kinase expression to fine-tune its signaling output. J. Biol. Chem. 2019, 294, 4119–4136. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Black, B.L.; Derynck, R. TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001, 15, 2950–2966. [Google Scholar] [CrossRef] [PubMed]
- Van de Vis, R.A.J.; Moustakas, A.; van der Heide, L.P. NUAK1 and NUAK2 Fine-Tune TGF-β Signaling. Cancers 2021, 13, 3377. [Google Scholar] [CrossRef]
- Pakneshan, S.; Safarpour, D.; Tavassoli, F.; Jabbari, B. Brain metastasis from ovarian cancer: A systematic review. J. Neurooncol. 2014, 119, 1–6. [Google Scholar] [CrossRef]
- Franklin, T.B.; Perrot-Sinal, T.S. Sex and ovarian steroids modulate brain-derived neurotrophic factor (BDNF) protein levels in rat hippocampus under stressful and non-stressful conditions. Psychoneuroendocrinology 2006, 31, 38–48. [Google Scholar] [CrossRef]
- Kuznetsov, V.A.; Tang, Z.; Ivshina, A.V. Identification of common oncogenic and early developmental pathways in the ovarian carcinomas controlling by distinct prognostically significant microRNA subsets. BMC Genomics 2017, 18, 692. [Google Scholar] [CrossRef]
- Tessarollo, L. Pleiotropic functions of neurotrophins in development. Cytokine Growth Factor Rev. 1998, 9, 125–137. [Google Scholar] [CrossRef]
- Dissen, G.A.; Romero, C.; Hirshfield, A.N.; Ojeda, S.R. Nerve growth factor is required for early follicular development in the mammalian ovary. Endocrinology 2001, 142, 2078–2086. [Google Scholar] [CrossRef]
- Dissen, G.A.; Hill, D.F.; Costa, M.E.; Ma, Y.J.; Ojeda, S.R. Nerve growth factor receptors in the peripubertal rat ovary. Mol. Endocrinol. 1991, 5, 1642–1650. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Borella, F.; Bertero, L.; Morrone, A.; Gambella, A.; Bovetti, M.; Cosma, S.; Carosso, A.; Katsaros, D.; Gemmiti, S.; Preti, M.; et al. Brain metastases from ovarian cancer: Current evidence in diagnosis, treatment, and prognosis. Cancers 2020, 12, 2156. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Akesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Barnes, A.P.; Lilley, B.N.; Pan, Y.A.; Plummer, L.J.; Powell, A.W.; Raines, A.N.; Sanes, J.R.; Polleux, F. LKB1 and SAD Kinases Define a Pathway Required for the Polarization of Cortical Neurons. Cell 2007, 129, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Courchet, J.; Lewis, T.L.; Lee, S.; Courchet, V.; Liou, D.Y.; Aizawa, S.; Polleux, F. Terminal axon branching is regulated by the LKB1-NUAK1 kinase pathway via presynaptic mitochondrial capture. Cell 2013, 153, 1510–1525. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, Y.; Hsu, C.W.; Martinez-Traverso, I.M.; Zhang, M.; Bai, Y.; Ishii, M.; Maxson, R.E.; Olson, E.N.; Dickinson, M.E.; et al. Yap and taz play a crucial role in neural crest-derived craniofacial development. Development 2016, 143, 504–515. [Google Scholar] [CrossRef]
- Lasagna-Reeves, C.A.; de Haro, M.; Hao, S.; Park, J.; Rousseaux, M.W.C.; Al-Ramahi, I.; Jafar-Nejad, P.; Vilanova-Velez, L.; See, L.; De Maio, A.; et al. Reduction of Nuak1 Decreases Tau and Reverses Phenotypes in a Tauopathy Mouse Model. Neuron 2016, 92, 407–418. [Google Scholar] [CrossRef]
- Phippen, N.T.; Bateman, N.W.; Wang, G.; Conrads, K.A.; Ao, W.; Teng, P.N.; Litzi, T.A.; Oliver, J.; Larry Maxwell, G.; Hamilton, C.A.; et al. NUAK1 (ARK5) is associated with poor prognosis in ovarian cancer. Front. Oncol. 2016, 6, 213. [Google Scholar] [CrossRef]
- Wang, S.; Shuwei, L.I.; Wang, H.; Wei, L.I.; Yuxue, G.A.O.; Wang, X.; Fang, C.; Zhang, B.; Xiuning, S.U.N.; Ruifang, L.I.; et al. Knockdown of ARK5 expression suppresses invasion of ovarian cancer cells. Mol. Med. Rep. 2019, 19, 2927–2934. [Google Scholar] [CrossRef]
- Fritz, J.L.; Collins, O.; Saxena, P.; Buensuceso, A.; Valdes, Y.R.; Francis, K.E.; Brown, K.R.; Larsen, B.; Colwill, K.; Gingras, A.C.; et al. A novel role for NUAK1 in promoting ovarian cancer metastasis through regulation of fibronectin production in Spheroids. Cancers 2020, 12, 1250. [Google Scholar] [CrossRef]
- Riester, M.; Wei, W.; Waldron, L.; Culhane, A.C.; Trippa, L.; Oliva, E.; Kim, S.H.; Michor, F.; Huttenhower, C.; Parmigiani, G.; et al. Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. J. Natl. Cancer Inst. 2014, 106, dju048. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed]



| Gene | Mutant | Function | Model | Reference |
|---|---|---|---|---|
| NUAK1 | S600A | Abrogates Akt activation | HepG2 (human liver cancer) Wi-38 (human diploid fibroblast, HDF) | [28,31] |
| K84A | Abolishes NUAK1 kinase activity | Wi-38 (human diploid fibroblast, HDF) A549 (human lung cancer) | ||
| T211A | Abrogates LKB1 activation | In vitro kinase assay | [10] | |
| I400K L401K | Abrogates binding with myosin cytoskeleton proteins | HEK 293T (human embryonic kidney) | [34] | |
| NUAK2 | K82R | Dead kinase mutant | ACHN (renal carcinoma cell line) | [35] |
| K81M T208A | Abrogates phosphotransferase activity of NUAK2 and LKB1 activation | Huh 7.5.1 (human hepatocyte derived cellular carcinoma) In vitro kinase assay | [10,36] |
| Gene | Cancer Type | Gender | Number of Samples | Gene Expression | Survival Rate | p-Value |
|---|---|---|---|---|---|---|
| NUAK1 | Ovarian cancer | Total | 373 | High | 24% | 0.0041 |
| Low | 38% | |||||
| Glioma | Total | 153 | High | 8% | 0.051 | |
| Low | 10% | |||||
| Female | 54 | High | 7% | 0.033 | ||
| Low | 13% | |||||
| Male | 99 | High | 8% | 0.28 | ||
| Low | 9% | |||||
| NUAK2 | Ovarian cancer | Total | 373 | High | 37% | 0.092 |
| Low | 28% | |||||
| Glioma | Total | 153 | High | 6% | 0.012 | |
| Low | 13% | |||||
| Female | 54 | High | 5% | 0.12 | ||
| Low | 18% | |||||
| Male | 99 | High | 6% | 0.06 | ||
| Low | 12% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molina, E.; Hong, L.; Chefetz, I. NUAK Kinases: Brain–Ovary Axis. Cells 2021, 10, 2760. https://doi.org/10.3390/cells10102760
Molina E, Hong L, Chefetz I. NUAK Kinases: Brain–Ovary Axis. Cells. 2021; 10(10):2760. https://doi.org/10.3390/cells10102760
Chicago/Turabian StyleMolina, Ester, Linda Hong, and Ilana Chefetz. 2021. "NUAK Kinases: Brain–Ovary Axis" Cells 10, no. 10: 2760. https://doi.org/10.3390/cells10102760
APA StyleMolina, E., Hong, L., & Chefetz, I. (2021). NUAK Kinases: Brain–Ovary Axis. Cells, 10(10), 2760. https://doi.org/10.3390/cells10102760






