Targeting Asparagine and Serine Metabolism in Germinal Centre-Derived B Cells Non-Hodgkin Lymphomas (B-NHL)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Preparation of Customised Complete RPMI-1640 Medium
2.3. Isotope Labelling
2.4. Cell Viability Assay
2.5. Western Blotting
2.6. NMR Sample Preparation
2.7. NMR Data Acquisition
2.8. Analysis of NMR Spectra
2.9. Transcriptomic Data and RNA-Seq Data Analysis
3. Results
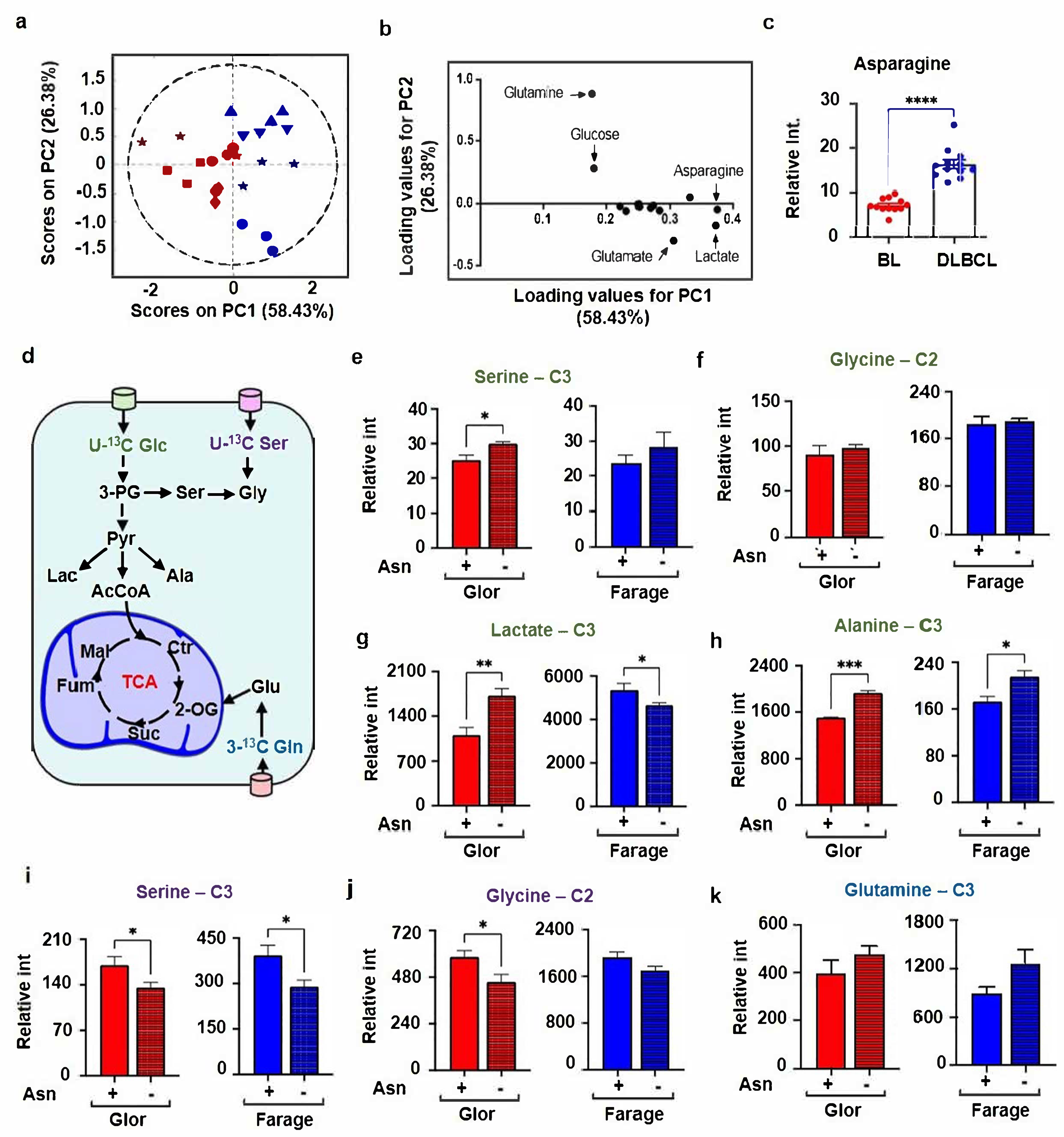
3.1. BL Cells Consumed Dramatically More Extracellular Asparagine Than DLBCL and Regulates Serine Metabolism
3.2. BL Tumours Differentially Express the Genes Involved in Serine Metabolism Compared to DLBCL
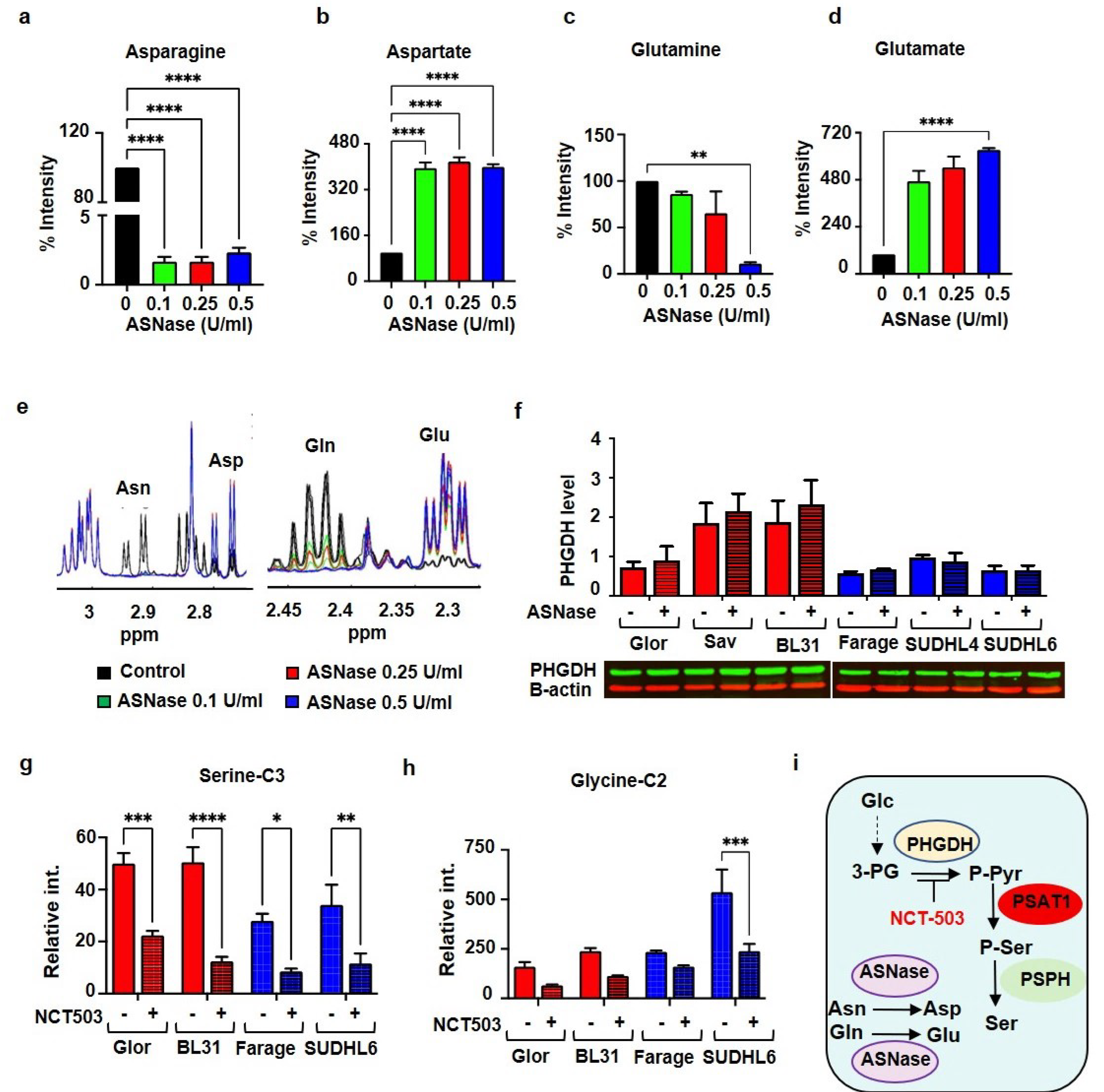
3.3. Asparaginase and NCT503 Reduce Extracellular Asparagine and Serine Production from Glucose Respectively
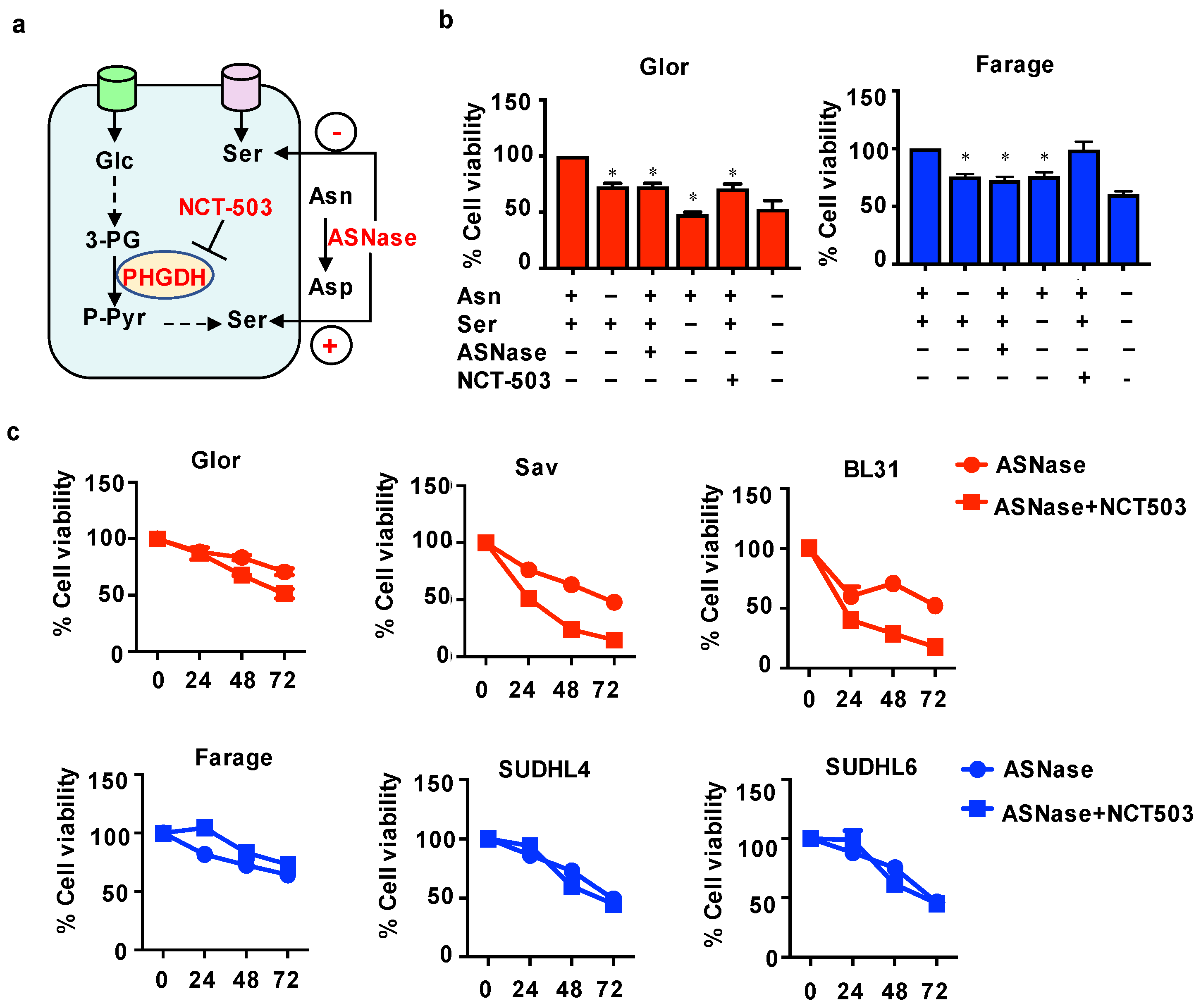
3.4. Combination of ASNase with NCT Increases the Sensitivity of BL Cells to ASNase
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bouska, A.; Bi, C.; Lone, W.; Zhang, W.; Kedwaii, A.; Heavican, T.; Lachel, C.M.; Yu, J.; Ferro, R.; Eldorghamy, N.; et al. Adult high-grade B-cell lymphoma with burkitt lymphoma signature: Genomic features and potential therapeutic targets. Blood 2017, 130, 1819–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monti, S. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005, 105, 1851–1861. [Google Scholar] [CrossRef] [Green Version]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct Types of Diffuse Large B-Cell Lymphoma Identied by Gene Expression Proling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the world health organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [Green Version]
- Flowers, C.; Sinha, R. Nastoupil treatment strategies for patients with diffuse large B-cell lymphoma: Past, present, and future. BLCTT 2012, 87. [Google Scholar] [CrossRef] [Green Version]
- Barnes, J.A.; LaCasce, A.S.; Feng, Y.; Toomey, C.E.; Neuberg, D.; Michaelson, J.S.; Hochberg, E.P.; Abramson, J.S. Evaluation of the addition of rituximab to CODOX-M/IVAC for burkitt’s lymphoma: A retrospective analysis. Ann. Oncol. 2011, 22, 1859–1864. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Salzer, W.; Bostrom, B.; Messinger, Y.; Perissinotti, A.J.; Marini, B. Asparaginase activity levels and monitoring in patients with acute lymphoblastic leukemia. Leuk. Lymphoma 2018, 59, 1797–1806. [Google Scholar] [CrossRef]
- Tabe, Y.; Lorenzi, P.L.; Konopleva, M. Amino acid metabolism in hematologic malignancies and the era of targeted therapy. Blood 2019, 134, 1014–1023. [Google Scholar] [CrossRef]
- Krall, A.S.; Xu, S.; Graeber, T.G.; Braas, D.; Christofk, H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 2016, 7, 11457. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Reid, M.A.; Allen, A.E.; Liu, S.; Liberti, M.V.; Liu, P.; Liu, X.; Dai, Z.; Gao, X.; Wang, Q.; Liu, Y.; et al. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nat. Commun. 2018, 9, 5442. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.; Taurino, G.; Bianchi, M.G.; Kilberg, M.S.; Bussolati, O. Asparagine synthetase in cancer: Beyond acute lymphoblastic leukemia. Front. Oncol. 2020, 9, 1480. [Google Scholar] [CrossRef] [PubMed]
- Tsurusawa, M.; Chin, M.; Iwai, A.; Nomura, K.; Maeba, H.; Taga, T.; Higa, T.; Kuno, T.; Hori, T.; Yamagata, M.; et al. L-asparagine depletion levels and l-asparaginase activity in plasma of children with acute lymphoblastic leukemia under asparaginase treatment. Cancer Chemother. Pharmacol. 2004, 53, 204–208. [Google Scholar] [CrossRef]
- Günther, U.L.; Ludwig, C.; Rüterjans, H. NMRLAB—Advanced NMR data processing in matlab. J. Magn. Reson. 2000, 145, 201–208. [Google Scholar] [CrossRef]
- Ludwig, C.; Günther, U.L. MetaboLab-Advanced NMR data processing and analysis for metabolomics. BMC Bioinf. 2011, 12, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saborano, R.; Eraslan, Z.; Roberts, J.; Khanim, F.; Lalor, P.; Reed, M. A framework for tracer-based metabolism in mammalian Cells by NMR. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. Icoshift: A versatile tool for the rapid alignment of 1D NMR Spectra. J. Mag. Reson. 2010, 202, 190–202. [Google Scholar] [CrossRef]
- Hyberts, S.G.; Milbradt, A.G.; Wagner, A.B.; Arthanari, H.; Wagner, G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional poisson gap scheduling. J. Biomol. NMR 2012, 52, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Abate, F.; Ambrosio, M.R.; Mundo, L.; Laginestra, M.A.; Fuligni, F.; Rossi, M.; Zairis, S.; Gazaneo, S.; De Falco, G.; Lazzi, S.; et al. Distinct viral and mutational spectrum of endemic burkitt lymphoma. PLoS Pathog. 2015, 11, e1005158. [Google Scholar] [CrossRef] [Green Version]
- Teater, M.; Dominguez, P.M.; Redmond, D.; Chen, Z.; Ennishi, D.; Scott, D.W.; Cimmino, L.; Ghione, P.; Chaudhuri, J.; Gascoyne, R.D.; et al. AICDA drives epigenetic heterogeneity and accelerates germinal center-derived lymphomagenesis. Nat. Commun. 2018, 9, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arita, M.; Karsch-Mizrachi, I.; Cochrane, G. The international nucleotide sequence database collaboration. Nucl. Acids Res. 2021, 49, D121–D124. [Google Scholar] [CrossRef] [PubMed]
- Yalamanchili, H.K.; Wan, Y.; Liu, Z. Data analysis pipeline for RNA-seq experiments: From differential expression to cryptic splicing. Curr. Protoc. Bioinf. 2017, 59. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; on behalf of the International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucl. Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the r/bioconductor package BiomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucl. Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein–Protein interaction networks, integrated over the tree of life. Nucl. Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Reina-Campos, M.; Diaz-Meco, M.T.; Moscat, J. The complexity of the serine glycine one-carbon pathway in cancer. J. Cell Biol. 2020, 219, e201907022. [Google Scholar] [CrossRef]
- Dave, S.S.; Fu, K.; Wright, G.W.; Lam, L.T.; Kluin, P.; Boerma, E.-J.; Greiner, T.C.; Weisenburger, D.D.; Rosenwald, A.; Ott, G.; et al. Molecular diagnosis of burkitt’s lymphoma. N. Engl. J. Med. 2006, 354, 2431–2442. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, R.; Young, R.M.; Ceribelli, M.; Jhavar, S.; Xiao, W.; Zhang, M.; Wright, G.; Shaffer, A.L.; Hodson, D.J.; Buras, E.; et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 2012, 490, 116–120. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eraslan, Z.; Papatzikas, G.; Cazier, J.-B.; Khanim, F.L.; Günther, U.L. Targeting Asparagine and Serine Metabolism in Germinal Centre-Derived B Cells Non-Hodgkin Lymphomas (B-NHL). Cells 2021, 10, 2589. https://doi.org/10.3390/cells10102589
Eraslan Z, Papatzikas G, Cazier J-B, Khanim FL, Günther UL. Targeting Asparagine and Serine Metabolism in Germinal Centre-Derived B Cells Non-Hodgkin Lymphomas (B-NHL). Cells. 2021; 10(10):2589. https://doi.org/10.3390/cells10102589
Chicago/Turabian StyleEraslan, Zuhal, Grigorios Papatzikas, Jean-Baptiste Cazier, Farhat L. Khanim, and Ulrich L. Günther. 2021. "Targeting Asparagine and Serine Metabolism in Germinal Centre-Derived B Cells Non-Hodgkin Lymphomas (B-NHL)" Cells 10, no. 10: 2589. https://doi.org/10.3390/cells10102589






