Machine Learning Assisted Classification of Cell Lines and Cell States on Quantitative Phase Images
Abstract
:1. Introduction
2. Digital Holographic Microscopy and Calculation of Cellular Parameters
2.1. Phase Image Reconstruction and Data Processing
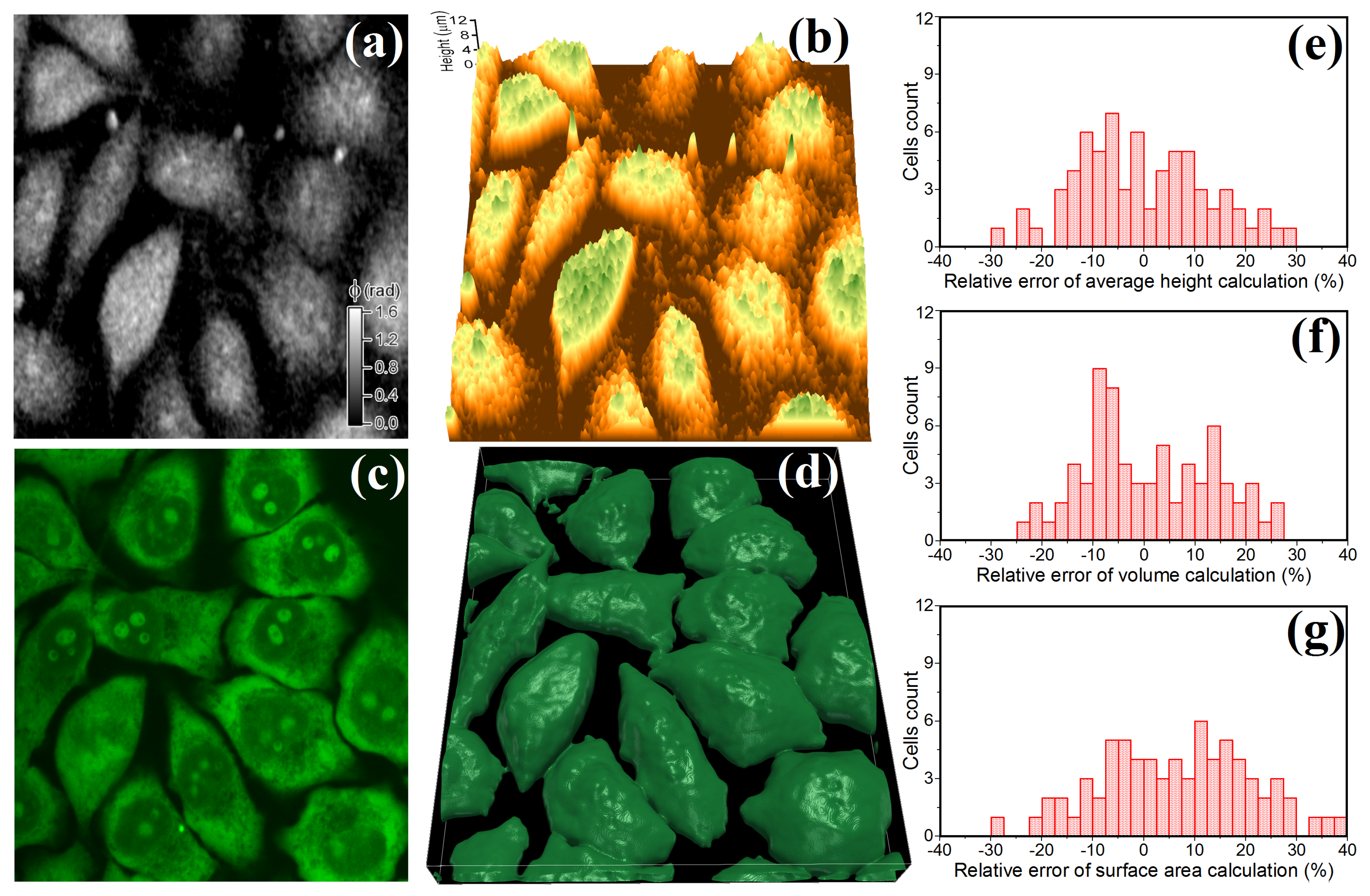
2.2. Evaluation of Cellular Morphology
3. Cell Samples and Photodynamic Treatment
3.1. Preparation of Cell Samples
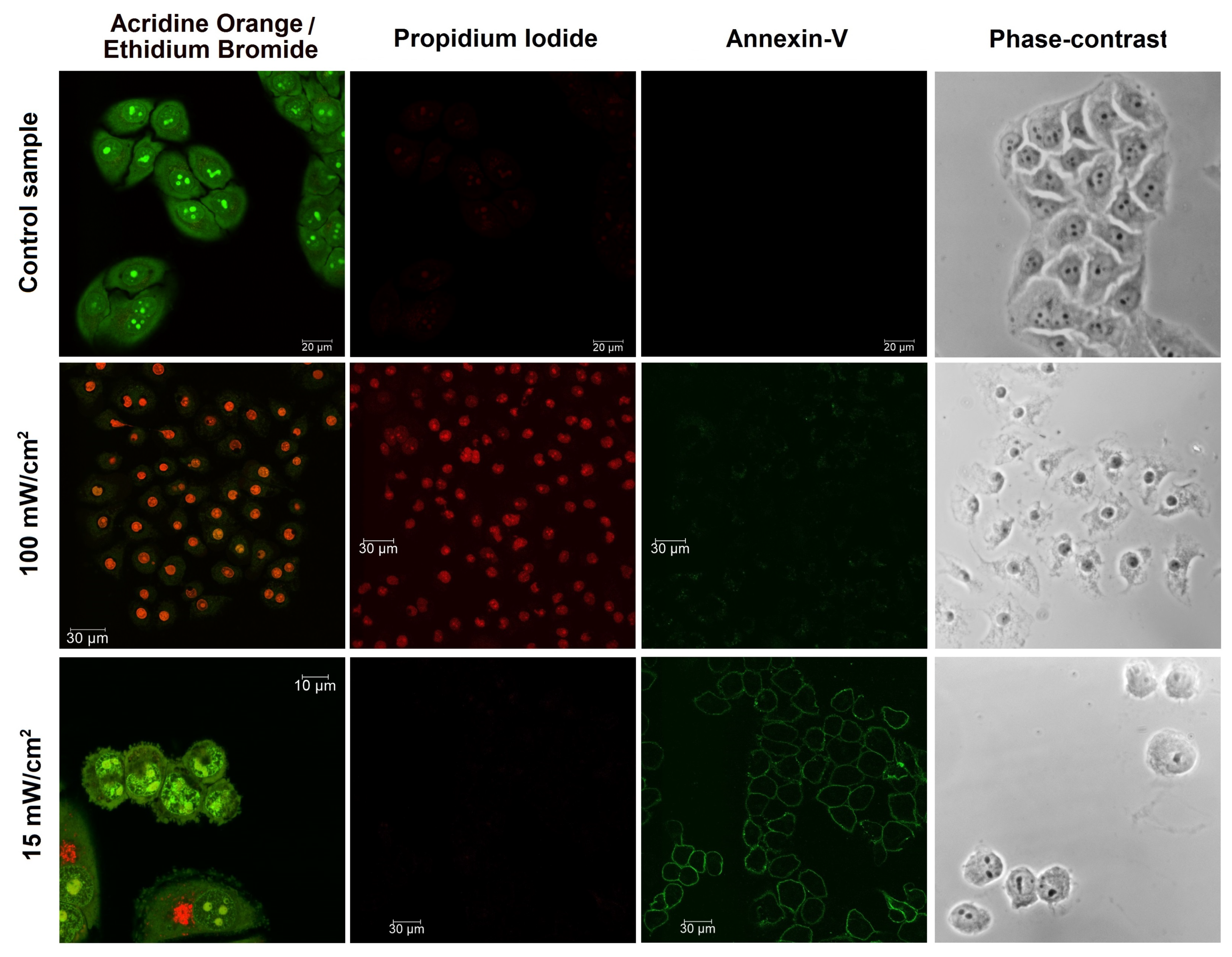
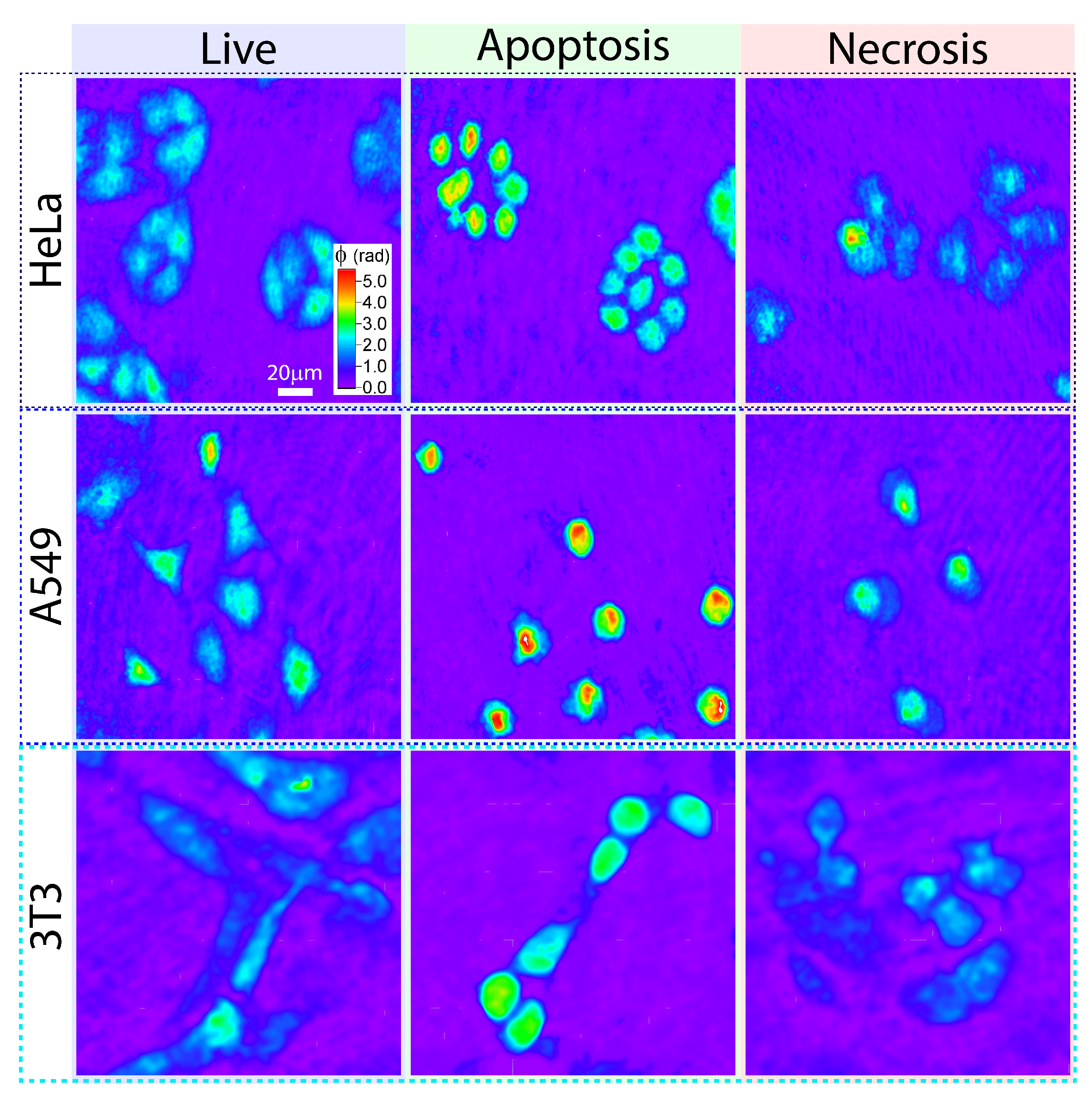
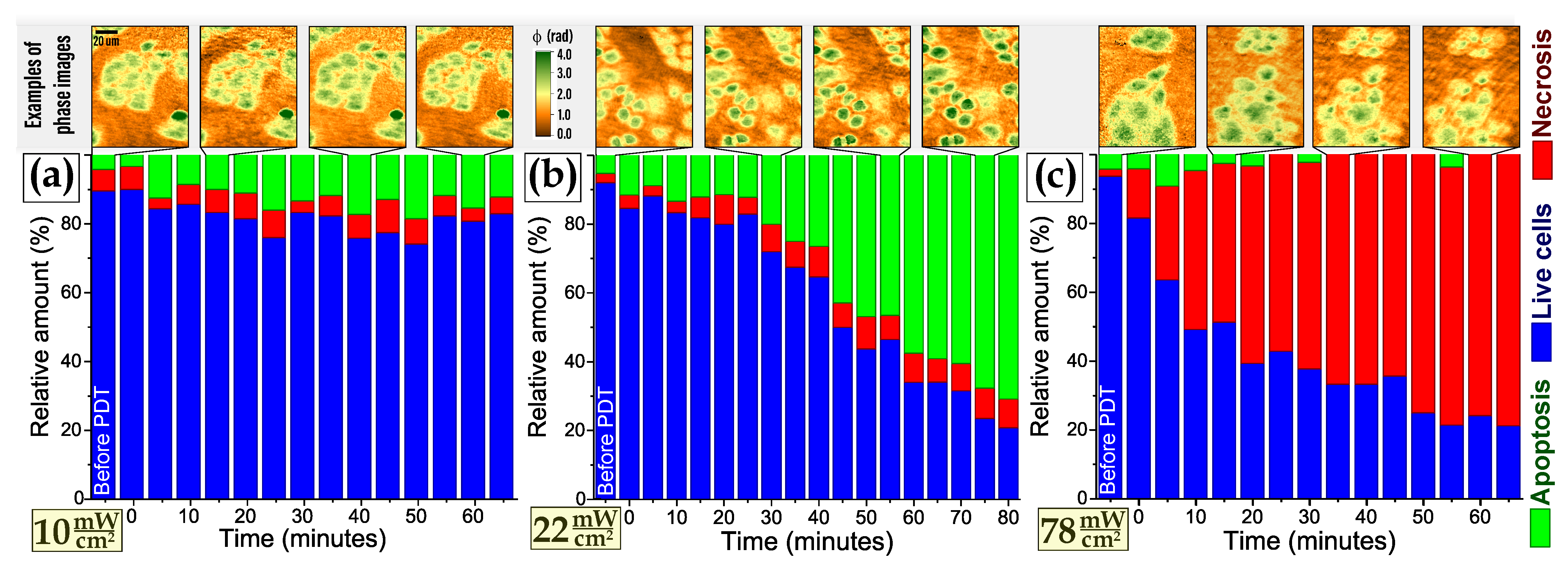
3.2. Cell Death Induced by PDTr
4. Machine-Learning Based Cell Classification Algorithm
4.1. Development of Machine-Learning Cell Classification Algorithm
4.2. Field Test of the Developed Algorithm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheremkhin, P.; Evtikhiev, N.; Krasnov, V.; Rodin, V.; Rymov, D.; Starikov, R. Machine learning methods for digital holography and diffractive optics. Procedia Comput. Sci. 2020, 169, 440–444. [Google Scholar] [CrossRef]
- Wu, Y.; Rivenson, Y.; Zhang, Y.; Wei, Z.; Günaydin, H.; Lin, X.; Ozcan, A. Extended depth-of-field in holographic imaging using deep-learning-based autofocusing and phase recovery. Optica 2018, 5, 704–710. [Google Scholar] [CrossRef] [Green Version]
- Yi, F.; Park, S.; Moon, I. High-throughput label-free cell detection and counting from diffraction patterns with deep fully convolutional neural networks. J. Biomed. Opt. 2021, 26, 036001. [Google Scholar] [CrossRef]
- Nguyen, T.; Bui, V.; Lam, V.; Raub, C.B.; Chang, L.C.; Nehmetallah, G. Automatic phase aberration compensation for digital holographic microscopy based on deep learning background detection. Opt. Express 2017, 25, 15043–15057. [Google Scholar] [CrossRef]
- Lyu, M.; Yuan, C.; Li, D.; Situ, G. Fast autofocusing in digital holography using the magnitude differential. Appl. Opt. 2017, 56, F152–F157. [Google Scholar] [CrossRef] [PubMed]
- Pitkäaho, T.; Manninen, A.; Naughton, T.J. Focus prediction in digital holographic microscopy using deep convolutional neural networks. Appl. Opt. 2019, 58, A202–A208. [Google Scholar] [CrossRef] [PubMed]
- Pitkäaho, T.; Manninen, A.; Naughton, T.J. Performance of autofocus capability of deep convolutional neural networks in digital holographic microscopy. In Digital Holography and Three-Dimensional Imaging; Optical Society of America: Washington, DC, USA, 2017; p. W2A5. [Google Scholar]
- Shao, S.; Mallery, K.; Kumar, S.S.; Hong, J. Machine learning holography for 3D particle field imaging. Opt. Express 2020, 28, 2987–2999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Taft, D.A.; Chen, Y.J.; Zhang, J.; Wallace, C.T.; Xu, M.; Watkins, S.C.; Xing, J. Learn to segment single cells with deep distance estimator and deep cell detector. Comput. Biol. Med. 2019, 108, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, T.; Xu, S.; Sang, C.; Jin, Y.; Qin, Z. Accurate segmentation of overlapping cells in cervical cytology with deep convolutional neural networks. Neurocomputing 2019, 365, 157–170. [Google Scholar] [CrossRef]
- Ma, S.; Fang, R.; Luo, Y.; Liu, Q.; Wang, S.; Zhou, X. Phase aberration compensation via deep learning in digital holographic microscopy. Meas. Sci. Technol. 2021, 32, 105203. [Google Scholar] [CrossRef]
- Pan, F.; Dong, B.; Xiao, W.; Ferraro, P. Stitching sub-aperture in digital holography based on machine learning. Opt. Express 2020, 28, 6537–6551. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Ayoub, A.B.; Psaltis, D. Three-dimensional tomography of red blood cells using deep learning. Adv. Photonics 2020, 2, 026001. [Google Scholar] [CrossRef]
- Jeon, W.; Jeong, W.; Son, K.; Yang, H. Speckle noise reduction for digital holographic images using multi-scale convolutional neural networks. Opt. Lett. 2018, 43, 4240–4243. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Moon, I.; Javidi, B. Cell morphology-based classification of red blood cells using holographic imaging informatics. Biomed. Opt. Express 2016, 7, 2385–2399. [Google Scholar] [CrossRef] [Green Version]
- Rivenson, Y.; Liu, T.; Wei, Z.; Zhang, Y.; de Haan, K.; Ozcan, A. PhaseStain: The digital staining of label-free quantitative phase microscopy images using deep learning. Light. Sci. Appl. 2019, 8, 23. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar]
- Mir, M.; Bhaduri, B.; Wang, R.; Zhu, R.; Popescu, G. Quantitative phase imaging. Prog. Opt. 2012, 57, 217. [Google Scholar]
- Park, Y.; Depeursinge, C.; Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photonics 2018, 12, 578–589. [Google Scholar] [CrossRef]
- Li, Y.; Di, J.; Wang, K.; Wang, S.; Zhao, J. Classification of cell morphology with quantitative phase microscopy and machine learning. Opt. Express 2020, 28, 23916–23927. [Google Scholar] [CrossRef] [PubMed]
- Vicar, T.; Raudenska, M.; Gumulec, J.; Balvan, J. The quantitative-phase dynamics of apoptosis and lytic cell death. Sci. Rep. 2020, 10, 1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roitshtain, D.; Wolbromsky, L.; Bal, E.; Greenspan, H.; Satterwhite, L.L.; Shaked, N.T. Quantitative phase microscopy spatial signatures of cancer cells. Cytom. Part A 2017, 91, 482–493. [Google Scholar] [CrossRef] [Green Version]
- Nissim, N.; Dudaie, M.; Barnea, I.; Shaked, N.T. Real-Time Stain-Free Classification of Cancer Cells and Blood Cells Using Interferometric Phase Microscopy and Machine Learning. Cytom. Part A 2021, 99, 511–523. [Google Scholar] [CrossRef]
- Ozaki, Y.; Yamada, H.; Kikuchi, H.; Hirotsu, A.; Murakami, T.; Matsumoto, T.; Kawabata, T.; Hiramatsu, Y.; Kamiya, K.; Yamauchi, T.; et al. Label-free classification of cells based on supervised machine learning of subcellular structures. PLoS ONE 2019, 14, e0211347. [Google Scholar] [CrossRef]
- Dubey, V.; Popova, D.; Ahmad, A.; Acharya, G.; Basnet, P.; Mehta, D.S.; Ahluwalia, B.S. Partially spatially coherent digital holographic microscopy and machine learning for quantitative analysis of human spermatozoa under oxidative stress condition. Sci. Rep. 2019, 9, 3564. [Google Scholar] [CrossRef]
- Singla, N.; Srivastava, V.; Mehta, D.S. Development of full-field optical spatial coherence tomography system for automated identification of malaria using the multilevel ensemble classifier. J. Biophotonics 2018, 11, e201700279. [Google Scholar] [CrossRef]
- Wang, Y.; Ju, P.; Wang, S.; Su, J.; Zhai, W.; Wu, C. Identification of living and dead microalgae cells with digital holography and verified in the East China Sea. Mar. Pollut. Bull. 2021, 163, 111927. [Google Scholar] [CrossRef]
- Chen, X.; Kandel, M.E.; Popescu, G. Spatial light interference microscopy: Principle and applications to biomedicine. Adv. Opt. Photonics 2021, 13, 353–425. [Google Scholar] [CrossRef]
- Bhaduri, B.; Pham, H.; Mir, M.; Popescu, G. Diffraction phase microscopy with white light. Opt. Lett. 2012, 37, 1094–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girshovitz, P.; Shaked, N.T. Generalized cell morphological parameters based on interferometric phase microscopy and their application to cell life cycle characterization. Biomed. Opt. Express 2012, 3, 1757–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mir, M.; Wang, Z.; Shen, Z.; Bednarz, M.; Bashir, R.; Golding, I.; Prasanth, S.G.; Popescu, G. Optical measurement of cycle-dependent cell growth. Proc. Natl. Acad. Sci. USA 2011, 108, 13124–13129. [Google Scholar] [CrossRef] [Green Version]
- Popescu, G. Quantitative phase imaging of nanoscale cell structure and dynamics. Methods Cell Biol. 2008, 90, 87–115. [Google Scholar] [PubMed]
- El-Schich, Z.; Leida Mölder, A.; Gjörloff Wingren, A. Quantitative phase imaging for label-free analysis of cancer cells—focus on digital holographic microscopy. Appl. Sci. 2018, 8, 1027. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.W.; Belashov, A.V.; Zhikhoreva, A.A.; Semenova, I.V.; Cheng, C.J.; Su, L.Y.; Wu, C.H. Application of digital holographic tomography in antitumor effect of cantharides complex on 4T1 breast cancer cells. Appl. Opt. 2021, 60, 3365–3373. [Google Scholar] [CrossRef]
- Zhikhoreva, A.A.; Belashov, A.V.; Danilova, A.B.; Avdonkina, N.A.; Baldueva, I.A.; Gelfond, M.; Nekhaeva, T.L.; Semenova, I.V.; Vasyutinskii, O.S. Significant difference in response of malignant tumor cells of individual patients to photodynamic treatment as revealed by digital holographic microscopy. J. Photochem. Photobiol. Biol. 2021, 32, 112235. [Google Scholar] [CrossRef] [PubMed]
- Belashov, A.V.; Zhikhoreva, A.A.; Belyaeva, T.N.; Kornilova, E.S.; Salova, A.V.; Semenova, I.V.; Vasyutinskii, O.S. In vitro monitoring of photoinduced necrosis in HeLa cells using digital holographic microscopy and machine learning. JOSA A 2020, 37, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Liebling, M.; Blu, T.; Unser, M. Complex-wave retrieval from a single off-axis hologram. J. Opt. Soc. Am. A 2004, 21, 367–377. [Google Scholar] [CrossRef] [Green Version]
- Katkovnik, V.; Shevkunov, I.A.; Petrov, N.V.; Egiazarian, K. Wavefront reconstruction in digital off-axis holography via sparse coding of amplitude and absolute phase. Opt. Lett. 2015, 40, 2417–2420. [Google Scholar] [CrossRef]
- Goldstein, R.M.; Zebker, H.A.; Werner, C.L. Satellite radar interferometry: Two-dimensional phase unwrapping. Radio Sci. 1988, 23, 713–720. [Google Scholar] [CrossRef] [Green Version]
- Zhikhoreva, A.A.; Belashov, A.V.; Gorbenko, D.A.; Avdonkina, N.A.; Baldueva, I.A.; Danilova, A.B.; Gelfond, M.L.; Nekhaeva, T.L.; Semenova, I.V.; Vasyutinskii, O.S. Morphological Changes in Malignant Tumor Cells at Photodynamic Treatment Assessed by Digital Holographic Microscopy. Russ. J. Phys. Chem. B 2019, 13, 394–400. [Google Scholar] [CrossRef]
- Belashov, A.V.; Zhikhoreva, A.A.; Belyaeva, T.N.; Nikolsky, N.N.; Semenova, I.V.; Kornilova, E.S.; Vasyutinskii, O.S. Quantitative assessment of changes in cellular morphology at photodynamic treatment in vitro by means of digital holographic microscopy. Biomed. Opt. Express 2019, 10, 4975–4986. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Choi, Y.; Fang-Yen, C.; Sung, Y.; Dasari, R.R.; Feld, M.S.; Choi, W. High-speed synthetic aperture microscopy for live cell imaging. Opt. Lett. 2011, 36, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Weiner, A.M. Optical phase imaging using a synthetic aperture phase retrieval technique. Opt. Express 2014, 22, 9380–9394. [Google Scholar] [CrossRef] [PubMed]
- Bianco, V.; Memmolo, P.; Paturzo, M.; Finizio, A.; Javidi, B.; Ferraro, P. Quasi noise-free digital holography. Light. Sci. Appl. 2016, 5, e16142. [Google Scholar] [CrossRef]
- Baczewska, M.; Eder, K.; Ketelhut, S.; Kemper, B.; Kujawińska, M. Refractive index changes of cells and cellular compartments upon paraformaldehyde fixation acquired by tomographic phase microscopy. Cytom. Part A 2021, 99, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.E.; Dierker, C.; Vollmer, A.; Ketelhut, S.; Kemper, B.; Schnekenburger, J. Multimodal label-free in vitro toxicity testing with digital holographic microscopy. In Biophotonics: Photonic Solutions for Better Health Care IV; International Society for Optics and Photonics: Bellingham, DC, USA, 2014; Volume 9129, p. 912940. [Google Scholar]
- Mann, C.J.; Yu, L.; Kim, M.K. Movies of cellular and sub-cellular motion by digital holographic microscopy. BioMed. Eng. OnLine 2006, 5, 21. [Google Scholar] [CrossRef] [Green Version]
- Simon, B.; Debailleul, M.; Beghin, A.; Tourneur, Y.; Haeberlé, O. High-resolution tomographic diffractive microscopy of biological samples. J. Biophotonics 2010, 3, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Shen, Y.; Huang, L.; Lv, G.; Lei, C.; Fan, X.; Lin, F.; Zhang, Y.; Wu, L.; Yang, Y. In vitro cytotoxicity of gold nanorods in A549 cells. Environ. Toxicol. Pharmacol. 2015, 39, 871–878. [Google Scholar] [CrossRef]
- Patel, R.M.; Patel, S.K. Cytotoxic activity of methanolic extract of Artocarpus heterophyllus against A549, Hela and MCF-7 cell lines. J. Appl. Pharm. Sci. 2011, 1, 167–171. [Google Scholar]
- Danihelová, M.; Veverka, M.; Šturdík, E.; Jantová, S. Antioxidant action and cytotoxicity on HeLa and NIH-3T3 cells of new quercetin derivatives. Interdiscip. Toxicol. 2013, 6, 209. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Chang, J.E.; Jheon, S.; Han, S.J.; Kim, J.K. Enhanced production of reactive oxygen species in HeLa cells under concurrent low-dose carboplatin and Photofrin® photodynamic therapy. Oncol. Rep. 2018, 40, 339–345. [Google Scholar]
- Lee, Y.H.; Cheng, F.Y.; Chiu, H.W.; Tsai, J.C.; Fang, C.Y.; Chen, C.W.; Wang, Y.J. Cytotoxicity, oxidative stress, apoptosis and the autophagic effects of silver nanoparticles in mouse embryonic fibroblasts. Biomaterials 2014, 35, 4706–4715. [Google Scholar] [CrossRef]
- Liang, P.; Kolodieznyi, D.; Creeger, Y.; Ballou, B.; Bruchez, M.P. Subcellular Singlet Oxygen and Cell Death: Location Matters. Front. Chem. 2020, 8, 1045. [Google Scholar] [CrossRef] [PubMed]
- Castano, A.P.; Demidova, T.N.; Hamblin, M.R. Mechanisms in photodynamic therapy: Part two—Cellular signaling, cell metabolism and modes of cell death. Photodiagnosis Photodyn. Ther. 2005, 2, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Plaetzer, K.; Kiesslich, T.; Verwanger, T.; Krammer, B. The modes of cell death induced by PDT: An overview. Med. Laser Appl. 2003, 18, 7–19. [Google Scholar] [CrossRef]
- Bianco, V.; Memmolo, P.; Merola, F.; Carcagni, P.; Distante, C.; Ferraro, P. High-accuracy identification of micro-plastics by holographic microscopy enabled support vector machine. In Quantitative Phase Imaging V; International Society for Optics and Photonics: Bellingham, DC, USA, 2019; Volume 10887, p. 108870F. [Google Scholar]
- Strbkova, L.; Zicha, D.; Vesely, P.; Chmelik, R. Automated classification of cell morphology by coherence-controlled holographic microscopy. J. Biomed. Opt. 2017, 22, 086008. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Rocca, A.; Marino, A.; Rocca, V.; Moscato, S.; de Vito, G.; Piazza, V.; Mazzolai, B.; Mattoli, V.; Ngo-Anh, T.J.; Ciofani, G. Barium titanate nanoparticles and hypergravity stimulation improve differentiation of mesenchymal stem cells into osteoblasts. Int. J. Nanomed. 2015, 10, 433. [Google Scholar]
- Ghensi, P.; Bressan, E.; Gardin, C.; Ferroni, L.; Soldini, M.C.; Mandelli, F.; Soldini, C.; Zavan, B. The biological properties of OGI surfaces positively act on osteogenic and angiogenic commitment of mesenchymal stem cells. Materials 2017, 10, 1321. [Google Scholar] [CrossRef] [Green Version]
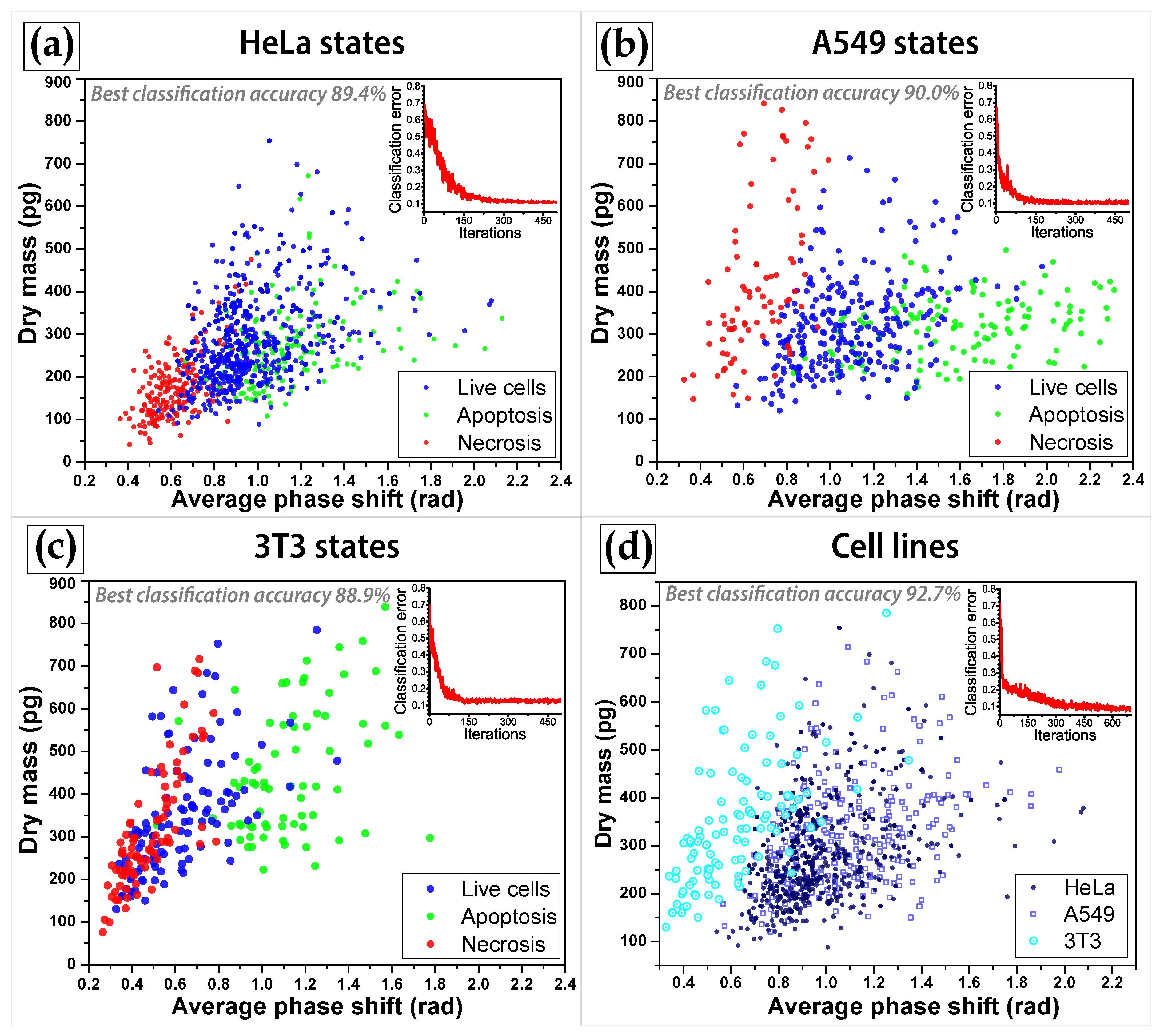
| Classification between | SVM | k-NN | EC |
|---|---|---|---|
| Hela states | 89.4% | 84.4% | 85.0% |
| A549 states | 90.0% | 89.0% | 82.0% |
| 3T3 states | 88.9% | 82.7% | 83.9% |
| cell lines | 92.7% | 78.1% | 81.7% |
| cell states in three cell lines | 77.8% | 66.5% | 66.9% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belashov, A.V.; Zhikhoreva, A.A.; Belyaeva, T.N.; Salova, A.V.; Kornilova, E.S.; Semenova, I.V.; Vasyutinskii, O.S. Machine Learning Assisted Classification of Cell Lines and Cell States on Quantitative Phase Images. Cells 2021, 10, 2587. https://doi.org/10.3390/cells10102587
Belashov AV, Zhikhoreva AA, Belyaeva TN, Salova AV, Kornilova ES, Semenova IV, Vasyutinskii OS. Machine Learning Assisted Classification of Cell Lines and Cell States on Quantitative Phase Images. Cells. 2021; 10(10):2587. https://doi.org/10.3390/cells10102587
Chicago/Turabian StyleBelashov, Andrey V., Anna A. Zhikhoreva, Tatiana N. Belyaeva, Anna V. Salova, Elena S. Kornilova, Irina V. Semenova, and Oleg S. Vasyutinskii. 2021. "Machine Learning Assisted Classification of Cell Lines and Cell States on Quantitative Phase Images" Cells 10, no. 10: 2587. https://doi.org/10.3390/cells10102587
APA StyleBelashov, A. V., Zhikhoreva, A. A., Belyaeva, T. N., Salova, A. V., Kornilova, E. S., Semenova, I. V., & Vasyutinskii, O. S. (2021). Machine Learning Assisted Classification of Cell Lines and Cell States on Quantitative Phase Images. Cells, 10(10), 2587. https://doi.org/10.3390/cells10102587






