Abstract
In animals, muscle growth is a quantitative trait controlled by multiple genes. Previously, we showed that the transient receptor potential channel 1 (TRPC1) gene was differentially expressed in muscle tissues between pig breeds with divergent growth traits base on RNA-seq. Here, we characterized TRPC1 expression profiles in different tissues and pig breeds and showed that TRPC1 was highly expressed in the muscle. We found two single nucleotide polymorphisms (SNPs) (C-1763T and C-1604T) in TRPC1 that could affect the promoter region activity and regulate pig growth rate. Functionally, we used RNAi and overexpression to illustrate that TRPC1 promotes myoblast proliferation, migration, differentiation, fusion, and muscle hypertrophy while inhibiting muscle degradation. These processes may be mediated by the activation of Wnt signaling pathways. Altogether, our results revealed that TRPC1 might promote muscle growth and development and plays a key role in Wnt-mediated myogenesis.
1. Introduction
Muscle development and growth determines the quality of meat and production yield, which are important economic traits in pig farming. Myogenesis, which mainly consists of myofiber formation and hypertrophy, is a complex process that includes myocyte proliferation, migration, differentiation, and fusion to form myotubes, followed by maturation of myofibers [,,]. Although some genes reportedly play important roles in muscle development, the specific regulatory mechanisms remain unclear [,,,,].
Transient receptor potential channel 1 (TRPC1), the first of the seven mammalian TRP channels to be cloned [,], is widely distributed in animal tissues []. TRPC1 is a signaling platform located in the cell and organelle membranes that primarily functions as a nonspecific cation channel within pathways controlling Ca2+ entry in response to cell surface receptor activation, thereby affecting cell proliferation and survival [], differentiation [], secretion [,], and migration [,,]. The TRPC1-mediated Ca2+ channel is associated with the regulation of physiological functions in several organs and tissues, including the cardiovascular system [,,], central nervous system [], skeletal and muscular tissues, as well as the immune system [].
TRPC1-siRNA treatment reduced store-operated Ca2+ entry and the expression of myogenic differentiation markers, subsequently suppressing skeletal myogenesis []. TRPC1 overexpression could accelerate myoblast fusion and produce hypertrophic myotubes, while TRPC1 knockdown delayed the onset of Ca2+ entry, reducing the speed of cell migration, and affecting the differentiation and fusion of myoblasts []. Although mice lacking TRPC1 could survive until adulthood, they showed defects in skeletal muscle function characterized by a smaller fiber cross-sectional area, less force per cross-sectional area, and less myofibrillar proteins than the controls []. In agreement with in vitro experiments, TRPC1 knockdown affected myoblast differentiation, resulting in smaller myotubes []. Furthermore, TRPC1 may be related to muscle regeneration and Duchenne muscular dystrophy [,]. Inhibiting TRPC1 expression during reloading resists the recovery of muscle mass and slow myosin heavy chain profiles []. Calcineurin is involved in the regulation of TRPC1 expression during muscle regrowth []. In summary, TRPC1 appears to play an important role in muscle development.
Although full-length or partial mRNA sequences have been reported for the seven TRPC genes (TRPC1-TRPC7) in various mammalian species, including humans, mice, rats, rabbits, and bovines [], most studies on TRPC1 have mainly focused on humans and mice. The pig TRPC1 gene is located on chromosome 13 and consists of 12 exons. In pigs, the TRPC1 gene has been linked with metabolic syndrome and atherosclerosis [,]. However, the properties of TRPC1 and its physiological functions in porcine muscle development are still unknown. Our previous study showed that TRPC1 was a key gene differentially expressed between pigs with divergent growth phenotypes based on transcriptome and proteome profiles []. Therefore, we hypothesized that the pig TRPC1, as a candidate gene for growth traits, may be differentially expressed in vivo and may be involved in muscle growth regulation.
Growth rate and body weight are highly variable among pig breeds. Tibetan pig (TP) is a mini-type native Chinese breed that has a slow growth rate and low adult weight []. Wujin pig (WJ) is another native Chinese breed that has a slow growth rate and normal adult weight, while the Yorkshire (YY) is a popular commercial breed that has a fast growth rate and normal adult weight []. This study investigated the differences in TRPC1 expression and SNPs among several breeds of pigs (YY; TP; WJ; New Huai population, NHP) and analyzed the association of TRPC1 with growth traits in these pigs. In addition, we assessed the functions of TRPC1 on porcine growth traits and explored the possible regulatory pathways. Our findings may provide a theoretical basis for investigating muscle development mechanisms in pigs and other agricultural animals and suggests a novel strategy for agricultural breeding.
2. Materials and Methods
2.1. Experimental Materials
Ear tissue samples from pigs (WJ, n = 29; TP, n = 34; YY, n = 36; NPH, n = 102) were collected for DNA extraction. The TP and WJ are slow-growth indigenous Chinese breeds with small and medium sizes, respectively. YY is the main commercial breed that has characteristics of fast growth and large size. WJ, TP, and YY pigs were raised in Tibet Agriculture and Animal Husbandry College, and the newly bred NHP population was raised in the Anhui Kexin Pig Breeding Farm in Hefei, China. For extraction of total RNA, nine embryos from each group were collected from two pregnant sows 60 days after insemination. The longissimus dorsi (LD) muscle tissues from the 12th rib were sampled and snap-frozen in liquid nitrogen and then stored at −80 °C. Animal care was conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals in China. All experiments were approved by the Committee on the Ethics of Animal Experiments of China Agricultural University (permit number: SKLAB-2012-04-07).
2.2. DNA, RNA, and cDNA Preparation
Genomic DNA was isolated from ear tissue using the standard phenol/chloroform extraction method, dissolved in a TE solution, and stored at −20 °C. Total RNA was extracted from tissues and cells using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and its quality and integrity were verified using a NanoDrop 2000 Biophotometer (Thermo Fisher Scientific Inc, West Palm Beach, FL,USA) and via electrophoresis. RNA samples (2 μg) in a 20 μL reaction volume were reverse transcribed to cDNA using the FastQuant Reverse Transcriptase Kit (TIANGEN, Beijing, China).
2.3. Measurement of Gene Expression
Semi-quantitative real-time polymerase chain reaction (SqRT-PCR) was performed as previously described [], and qRT-PCR was performed using the SYBR Green Master Mix (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Experiments were performed in duplicate, and gene expression levels were calculated using the 2−ΔΔCt method as previously described []. We selected a stable housekeeping gene, β-actin, as the internal reference. All primers used for this part are presented in Table S1.
2.4. SNP Screening, Genotyping, and Correlation Analysis
Four pairs of primers for SNP screening of the TRPC1 gene (NC_010455.5) were designed using Primer Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA) and are listed in Table S2. The amplicon sequences covered 2201 bp regions in the 5′-flanking and exon 1 of the gene. The PCR products amplified from ten individuals of each group were pooled and sequenced using Sanger sequencing to identify SNPs using Chromas Pro (Technelysium, South Brisbane, Australia) and DNAMAN6.0 (Lynnon, San Ramon, CA, USA). Genotypes of the identified SNPs were determined by individual PCR and Sanger sequencing in the WJ, TP, YY, and NHP populations. The 102 individuals in the NHP population were used to measure age at 30 kg and 90 kg body weight.
2.5. Dual-Luciferase Reporter Assays
The promoter regions of TRPC1 were cloned into a PGL3 luciferase reporter vector (Addgene, Cambridge, Mass, USA). The vector was transfected into a 24-well plate. After 48 h, the cells were lysed in 100 μL of lysis buffer and then assayed for promoter activity using a dual-luciferase reporter assay system (Promega, Madison, WI, USA). The Renilla luciferase signal was normalized to the firefly luciferase signal. The enzymatic activity of luciferase was measured using a PerkinElmer 2030 Multilabel Reader (PerkinElmer, Waltham, MA, USA).
2.6. Cell Culture
The C2C12 myoblast cell line (Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China) was cultured at subconfluent densities in a growth medium, made up of Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (PS, Gibco). C2C12 cells were differentiated into myocytes or myotubes in a differentiation medium, consisting of DMEM containing 2% horse serum (Gibco) and 1% PS. All cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C.
2.7. Plasmids Construction, siRNA Synthesis, and Transfection
For the TRPC1-overexpression plasmids, the full-length sequence was cloned into the BamHI (cat. no. R136S, NEB, USA) and NotI (cat. no. R0189S, NEB) restriction sites of the expression vector pcDNA3.1. The full-length TRPC1 sequence was amplified with F/R primers (F: 5′-ATGATGGCGGCCCTGTACCC-3′; R: 5′-TTAATTTCTTGGATAAAACA-3′). TRPC1 siRNA and scrambled siRNA were synthesized following the manufacturer’s protocol (GenePharma, Shanghai, China). The sequences of TRPC1 siRNAs are shown in Table S3. For cell transfection, C2C12 cells and primary myoblasts were transfected with 4 μg of the expression vectors or approximately 10 μL of siRNA oligo using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in each well of a 6-well plate.
2.8. CCK8 and EdU Proliferation Assays
Cell proliferation was measured by CCK8 and 5-ethynyl-2-deoxyuridine (EdU) assays. Cells were cultured in DMEM supplemented with 10% FBS for 24 h. CCK8 (10 µL, Beyotime Biotechnology, Shanghai, China) cells were incubated at 37 °C for 1 h in the dark. The absorbance value was measured using a microplate reader (Biotek, Winooski, VT, USA) at 450 nm. The EdU assay was performed using the EdU assay kit (Ribobio, Guangzhou, China) according to the manufacturer’s instructions. Briefly, 24 or 36 h after transfection, cells were exposed to 50 mM EdU for 2 h. Next, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Subsequently, cells were incubated in Apollo reaction solution for 1 h and stained with Hoechst 33342 for 30 min. The cells were further analyzed by calculating the ratio of EdU cells to the total number of cells.
2.9. Cell Wound Healing and Transwell Migration Assay
Cell migration was measured by the cell wound healing assay described in a previous report []. For the transfected cells, a scratch wound was made across the well using a pipette tip. The wounded monolayers were then washed twice with PBS (Gibco) to remove cell debris and incubated with DMEM media with 2% FBS. After 24 h, the wound area and cell migration were photographed under a microscope (ZEISS, Jena, Germany). The transwell migration assay was performed using 24-well transwell chambers (6.5 mm diameter, 8.0 μm pore size; Corning Inc., Corning, NY, USA), in which 10% FBS stimulated cell migration as a chemotactic factor; thus, serum-free cells in the top chamber are more likely to migrate into the bottom chamber with 10% FBS. Briefly, cells were serum-starved with serum-free medium for 6–8 h. Then, cells in 200 µL serum-free medium were placed into the top chamber, and 500 µL complete DMEM was placed into the bottom chamber. After incubation for 12 h, non-migrated cells were removed from the top surface of the transwell with a cotton swab, and the cells that migrated to the lower surfaces of the inserts were stained with 0.1% crystal violet for 0.5–1 h. The migrated cells were photographed under a microscope (ZEISS, Germany). Notably, to avoid the confounding effects of proliferation on cell migration results, the incubation times were markedly less than the doubling times for these cells.
2.10. Immunofluorescent Staining
For immunofluorescence, 4% paraformaldehyde-fixed cells were washed with PBS and permeabilized with 0.5% Triton X-100 for 15 min. Cells and sections were supplemented with a blocking buffer (Beyotime Biotechnology, Shanghai, China) at room temperature for 2 h and then incubated with mouse anti-myosin heavy chain (cat. no. M4276, Sigma-Aldrich, St. Louis, MO, USA, 1:500) at 4 °C overnight. Cells were washed briefly and incubated with the fluorescently labeled secondary antibodies (Alexa Fluor 594) for 1 h at room temperature. After washing with PBS, DAPI (Roche Applied Science, Nutley, NJ, USA) was used to stain the nuclei for 5 min. The immunofluorescence images were visualized with a fluorescence microscope (Leica image analysis system, Model Q500MC). The number of nuclei present in one myosin positive cell indicated myoblast fusion.
2.11. RNA-seq and Functional Annotation
Total RNA was extracted from control and TRPC1-overexpressed C2C12 cells using TRIzol, as mentioned above. cDNA library construction and high-throughput sequencing were performed on the Hiseq-Xten platform by Annoroad Gene Technology Co., Ltd. (Beijing, China). Each group had three biological replicates. The differentially expressed genes (DEGs) were screened using the criterion |log2 (fold change)| > 1 and P-adjusted ≤ 0.01, and their function was classified using the annotation of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with the DAVID online software (http://david.abcc.ncifcrf.gov/home.jsp).
2.12. Statistical Analyses
Differences in gene expression, absorbance, and the correlation between the genotypes and traits were analyzed using the one-way analysis of variance (ANOVA) and Student’s t-test. Results are expressed as mean ± standard deviation (SD). P < 0.05 was considered significant. A χ2 test was used to analyze the distribution of genotypes and to compare differences in genotype distribution.
3. Results
3.1. TRPC1 is Involved in Growth of Pigs
Expression of the TRPC1 was detected in the heart, LD, liver, back fat (BF), intestine, and leg tissues of the TP pigs (Figure 1A–D). Among these tissues, both the mRNA and protein levels of TRPC1 were higher in the LD, liver, and BF than in other organs, indicating that TRPC1 is predominantly expressed in tissues involved in growth. The YY pigs had higher TRPC1 expression in the LD than the TP and WJ pigs (Figure 1E,F), consistent with our previous transcriptomic results []. Therefore, we inferred that higher TRPC1 expression might promote muscle development and growth.
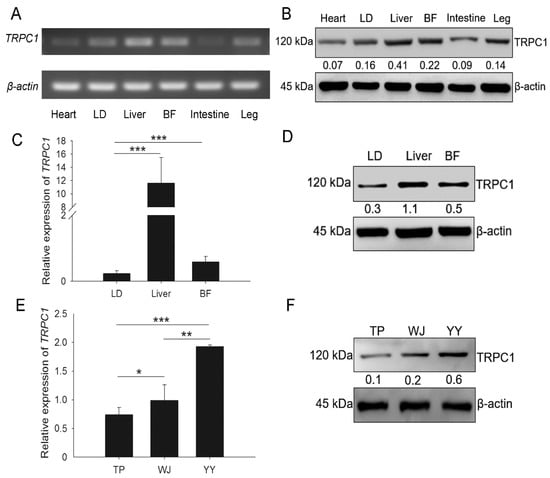
Figure 1.
TRPC1 is involved in the growth of pigs. TRPC1 expression in different tissues of TP pigs at the embryonic stage by SqRT-PCR (A), WB (B,D), and qRT-PCR (C). The TRPC1 mRNA (E) and protein (F) expression levels in the LD of three pig breeds. LD, longissimus dorsi; BF, back fat; TP, Tibetan pig (n = 6), WJ, Wujin pig (n = 6), YY, Yorkshire (n = 6). Each bar represents the means ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001.
3.2. The Genes Flanking TRPC1 Have Two SNPs, C-1763T, and C-1604T
In the 5′-flanking regions of the TRPC1 gene, two SNPs, C-1763T, and C-1604T, were found (Figure 2A). The two SNPs were linked completely and formed two haplotypes, CC and TT. Three genotypes (CC/CC, CT/CT, and TT/TT) were observed in the WJ and TP and one genotype (CC/CC) in the YY populations, and the distributions of the genotypes conformed to the Hardy–Weinberg equilibrium (P > 0.05) (Table 1). YY had higher frequencies of the genotype CC/CC and haplotype CC than the TP and WJ populations (P < 0.05). The promoter with the haplotype CC showed higher transcription activity than TT, as evidenced by the dual-luciferase reporter assays (Figure 2B).
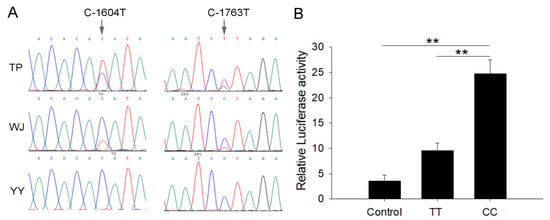
Figure 2.
SNP sites and promoter activity analysis. (A) The sequencing chromatogram at the sites of two SNPs, C-1604T and C-1763T. The arrows indicate the SNP site. (B) Dual-luciferase analysis for promoter activity. Control refers to C2C12 cells co-transfected with PGL3-basic and PRL-TK. CC and TT represent CC and TT homozygous individuals whose promoters are connected to PGL3-basic and co-transfected with PRL-TK cells, respectively. Each bar represents the mean ± SD. ** P < 0.01.

Table 1.
Genotype and haplotype frequencies in the C-1763T and C-1604T in pig populations.
The NHP population had three genotypes at the two SNP sites (Table 2). Because only one individual was TT/TT, we compared the age at 30 kg and 90 kg body weights between the CC/CC and CT/CT genotypes. The results showed that the CC/CC genotype exhibited significantly faster growth than the CT/CT genotype (P < 0.01) (Table 2).

Table 2.
Analysis of the effects of CC, CT, and TT genotypes of the TRPC1 gene in New Huai pigs.
3.3. TRPC1 Promotes Myoblast Proliferation and Migration
One overexpression and four siRNA fragments were constructed, and their detection efficiency was determined. The results showed that siRNA-886 had the highest interference efficiency among the four siRNAs (Figure 3A,B). Thus, TRPC1 knockdown was treated by siRNA-886 transfection. For proliferation, a lower absorbance value was observed in cells following siRNA-886 transfection relative to the control (Figure 3C), whereas TRPC1 overexpression markedly improved the absorbance (Figure 3D). TRPC1 knockdown significantly decreased EdU incorporation (Figure 3E), and TRPC1 overexpression resulted in increased EdU positivity compared with that of the control (Figure 3F). Consistent with the staining results, TRPC1 knockdown significantly downregulated the expression of apoptosis markers (p27, BAD), and TRPC1 overexpression showed the same expression pattern (Figure 3G,H). Indeed, the microscopic views showed that the doubling time of cells was ~24 h, and a dramatic reduction in the number of proliferated cells after TRPC1 knockdown was observed. The opposite effects were observed after TRPC1 overexpression (Figure S1), highlighting the possibility that TRPC1 accelerates myoblast proliferation. Besides, the migration capacity was concomitantly attenuated and promoted when TRPC1 was respectively knocked down and overexpressed (Figure 3I,J).
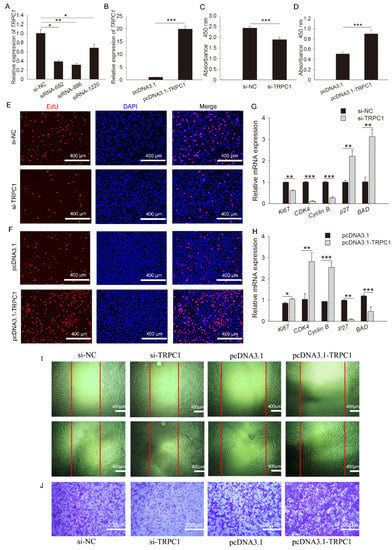
Figure 3.
TRPC1 promoted myoblast proliferation and migration. (A) Interference efficiency detection of synthetic siRNA fragments. (B) Efficiency detection of plasmid overexpression. (C,D) CCK8 assay of proliferated myoblasts transfected with RNAi or overexpression fragments. (E,F) EdU staining for proliferated cells following si-TRPC1 or pcDNA3.1-TRPC1 transfection. Nuclei were stained with DAPI, n = 3 in each group, scale bar = 400 μm. (G,H) The mRNA expression levels of proliferation-related genes. (I) Wound healing migration assay of C2C12 myoblasts. Some cells were scraped off with a pipette tip to obtain an acellular area. Twenty-four hours later, cells migrated into the acellular area were stained and counted. Cells were migrated in control conditions, Scale bar = 400 μm. (J) Transwell test for myoblasts. Purple represents migrated cells, n = 3 in each group, scale bar = 200 μm. NC: negative control. The data represent the means ± SD of three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001.
3.4. TRPC1 Promotes Cell Fusion, Differentiation, and Muscle Hypertrophy but Inhibits Muscle Degradation
TRPC1 expression increased with myoblast differentiation and reached a peak at D4 (Figure 4A); thus, the subsequent differentiation was treated for 4 days (Figure 4B). TRPC1 overexpression remarkably facilitated myogenic differentiation, as demonstrated by the increased protein expression of myogenic marker MyoG (Figure 4C) and the mRNA level of MyHC, MyoG, and MyoD (Figure 4D), and induced myotube formation (Figure 4F). Meanwhile, the TRPC1 silenced cells exhibited decreased myogenic marker expression (Figure 4C,E) and formed fewer myotubes (Figure 4G). These results suggest that TRPC1 positively regulates myoblast differentiation and myotube formation.
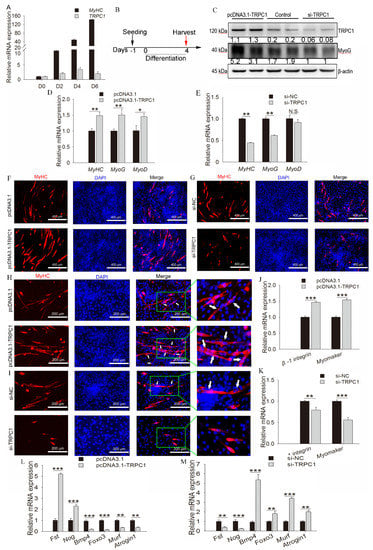
Figure 4.
TRPC1 promoted cell fusion, differentiation, and muscle hypertrophy but inhibited muscle degradation. (A) qRT-PCR results showed the expression profiles of the TRPC1 gene during differentiation. MyHC is the myogenic differentiation marker gene. (B) Schematic of cell differentiation time. (C) The protein level of TRPC1 and differentiation marker MyoG. (D,E) The mRNA expression of differentiation marker genes MyoD, MyoG, and MyHC quantified by qRT-PCR. (F,G) Immunofluorescence staining for MyHC protein in control or pcDNA3.1-TRPC1/si-TRPC1-treated myoblasts cultured for 4 days in differentiation medium. MyHC protein expression is shown in red, and nuclei in blue (DAPI). White scale bar = 400 μm. (H,I) Myoblast fusion analysis by immunofluorescence staining for MyHC, and the arrows represent the multinucleated myotubes, white scale bar = 200 μm. (J,K) The mRNA expression of fusion marker genes quantified by qRT-PCR. (L,M) The mRNA expression of muscle degradation markers (Atrogin1, Bmp4, Murf, and Foxo3) and muscle hypertrophy genes (Fst and Nog) quantified by qRT-PCR (n = 3, respectively). NC: negative control. The data represent the means ± SD of three independent experiments. N.S.: not significant, * P < 0.05, ** P < 0.01, *** P < 0.001.
Upon overexpression of TRPC1 mRNA, myoblasts fused to form long, multinucleated MyHC-positive myotubes accompanied by the upregulation of the fusion marker genes Myomaker and β-1integrin (Figure 4H,J). In contrast, most C2C12 silencing TRPC1 remained as short, rounded, and mononucleated myoblasts and downregulated the mRNA levels of fusion markers (Figure 4I,K), suggesting that TRPC1 facilitates myoblast fusion into one myotube.
Moreover, TRPC1 overexpression significantly promoted the expression of muscle hypertrophy genes (Fst and Nog) while inhibiting the expression of muscle degradation markers, including Atrogin1, Bmp4, Murf, and Foxo3 (Figure 4L). In TRPC1 knockdown cells, the expression of muscle degradation genes was increased while hypertrophy-related genes were downregulated (Figure 4M). These results indicated that TRPC1 promoted muscle hypertrophy and suppressed muscle degradation.
3.5. Regulative Pathway of TRPC1 on Myogenesis
To identify the regulatory pathway of TRPC1 in myogenesis, the TRPC1-overexpressed and control C2C12 cells after 2 days of culture were subjected to RNA-sequencing. Including TRPC1, 1392 DEGs were screened (Figure S2), and enriched GO terms were mainly associated with myotubes or myoblast growth, fusion, and differentiation (Figure S3). The representative KEGG pathways enriched by DEGs mainly contained the Wnt, PI3K-Akt, calcium, and MAPK signaling pathways (Figure S4). Considering key roles in cell proliferation and differentiation, Wnt signaling might be the pivotal pathway underlying the role of TRPC1 in myogenesis. Several DEGs involved in the Wnt signaling pathway were selected for validation by quantitative real-time polymerase chain reaction (qRT-PCR) (Figure 5A,B). TRPC1 overexpression upregulated the expression of the Wnt receptor LRP6 mRNA, downstream TCF/LEF, and target genes CCND, and downregulated the expression of negative regulators of Wnt (Axin2, GSK3β, and SPFR2/4) (Figure 5A). TRPC1 knockdown significantly reduced the expression of LRP6, TCF/LEF, and CCND and increased SPFR2/4 expression (Figure 5B). These results suggested that the promoted-growth phenotype of C2C12 cells induced by TRPC1 might be through the Wnt signaling pathway.
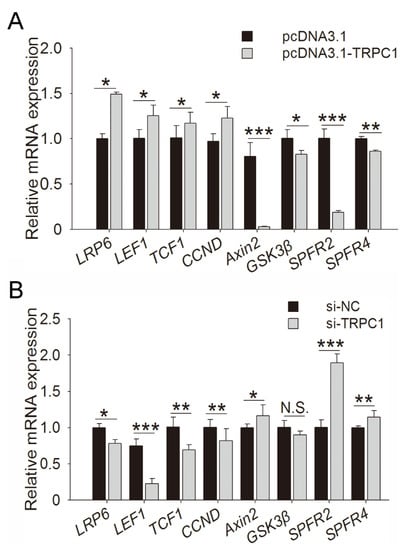
Figure 5.
Regulation of TRPC1 in myogenesis. The mRNA expression of Wnt-related genes in C2C12 cells in which TRPC1 was overexpressed (A) or knocked down (B), respectively. NC: negative control. The data represent the means ± SD of three independent experiments. N.S.: not significant, * P < 0.05, ** P < 0.01, *** P < 0.001.
4. Discussion
The embryonic development stage is important for postnatal muscle growth. Most commonly, myogenesis and myofibers are formed during the embryonic stage. Approximately 60 days post-insemination, the myofiber formation of porcine embryos is at a critical stage [,]. The two Chinese indigenous breeds (TP and WJ) showed slow body growth and small size, whereas an introduced YY breed showed faster growth. This might indicate that YY formed more myofibers than TP and WJ during embryonic development. The higher expression level of TRPC1 might lead to greater myofiber formation in YY than in TP and WJ, which is consistent with the results of our previous study []. Although TRPC1 exists in multiple tissues, it is mainly expressed in the LD tissue, which indicates that TRPC1 plays important regulatory roles in skeletal muscle development and facilitates postnatal muscle growth in pigs. High expression in BF implied its fat deposition regulation, which is consistent with previous research [].
Previous studies have reported that the TRPC1 gene polymorphisms are associated with type 2 diabetes and diabetic nephropathy in humans [,,,]. In the present study, we found two SNPs (C-1763T and C-1604T) that formed two haplotypes (CC and TT) in the 5′-flanking region of the TRPC1 gene in pigs. The genotypes CC/CC and haplotype CC were associated with fast body growth among the breeds (TP, WJ, and YY) and within a population (NHP). The haplotype CC in the promoter region caused a new transcription factor binding site for Myod1 compared with the TT, as evidenced by an analysis performed with the online software CONSITE (http://consite.genereg.net). The dual-luciferase reporter assays also demonstrated that the haplotype CC would increase transcription activity. Therefore, our results suggest that these two SNPs might regulate the expression of TRPC1 and postnatal body growth in pigs, and they are potential molecular markers that could be used in pig breeding. Certainly, the regulations of the SNPs on gene expression and muscle development need further to be tested in porcine primary myoblasts.
During myogenesis and muscle regeneration, myoblasts proliferate, migrate, differentiate, and fuse to form multinucleated myotubes that eventually develop into mature muscle fibers []. Some studies have demonstrated an essential role of TRPC1 in myogenesis by interference or knockout methods [,,,,]. Our gain and loss of function experiments further confirmed that TRPC1 induced more proliferated and migrated cells and increased the expression of cell survival markers. TRPC1−/− mice developed smaller fibers accompanied by decreased expression of myogenic factors []. Our results showed that TRPC1 was more highly expressed in differentiated C2C12 cells, indicating a potential role for myogenic differentiation. Supporting this result, we found that TRPC1 overexpression enhanced myoblast differentiation and fusion characterized by more polynuclear myotubes and upregulated expression of myogenic and fusion genes. In addition, TRP channels have been implicated in muscle hypertrophy and regeneration []. We also observed that TRPC1 promoted muscle hypertrophy but inhibited muscle degradation. Our data collectively suggest that porcine TRPC1 is a potent regulator of myogenesis and provides a cytological basis for explaining the TRPC1-mediated positive regulation on growth rate.
The Wnt/β-catenin signaling plays a crucial role in muscle development in African Cladopus [], avian [], and mammals [], and also affects adult muscle regeneration as well as the proliferation and differentiation of myogenic cell lines in vitro [,,]. Activated Wnt/β-catenin signaling increases muscle mass [,] and promotes myoblast proliferation and differentiation [], while reducing Wnt/β-catenin inhibited myogenic differentiation and the expression of Myomaker, and Myomixer []. We observed that TRPC1 increased transcription of receptors, key factors, and target genes of the canonical Wnt pathway, suggesting that it positively regulates via activation of Wnt signals. The canonical Wnt pathway is only a part of the complex regulatory network for muscle formation. Whether TRPC1 interacts with noncanonical Wnt signaling or is associated with other signaling pathways obtained from KEGG analysis warrants further study.
In conclusion, our study revealed that TRPC1 is highly expressed in the muscle of fast-growing pigs and can positively regulate myoblast proliferation, migration, differentiation, fusion, and hypertrophy by activating the Wnt signaling pathway, ensuring myoblasts differentiate into mature muscle fibers, thereby accelerating the growth rate of pigs. We explored the biological function and preliminary regulatory mechanism of TRPC1 and obtained molecular markers that could be used for genetic improvement. These results provide insights into the molecular mechanism of muscle development, which can be implemented in future breeding programs for pigs.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/10/1/147/s1, Figure S1: The microscopic images of proliferated cells, Figure S2: Differentially expressed genes identification. Volcano plot displaying DEGs between TRPC1-overexpression group and control, Figure S3: Significantly enriched GO terms of differentially expressed genes. Figure S4: KEGG-enriched scatter plot of DEGs between control and TRPC1-overexpressed C2C12, Table S1: The primer sequences for SqRT-PCR and qRT-PCR, Table S2: Sequence, targets, and amplication size of the primer used for SNP identification of the pig TRPC1 gene, Table S3: The sequences of TRPC1 siRNA fragment.
Author Contributions
Conceptualization, H.Z. and B.Z.; methodology, Y.F.; software, R.N.; validation, H.Z.; formal analysis, Y.F., X.T. and R.N.; investigation, P.S., R.Z. and X.T.; resources, R.Z. and P.S.; data curation, Y.F. and X.T.; writing—original draft preparation, Y.F.; writing—review and editing, H.Z.; visualization, Y.F., X.T. and R.N.; supervision, H.Z. and B.Z.; project administration, H.Z.; funding acquisition, R.Z. and P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Major Special Project on New Varieties Cultivation for Transgenic Organisms (No. 2016ZX08009-003-006), the Central Government guides Local Science and Technology (No. YDZX20195400004426), and the China Agricultural University partner assistance on research cooperation (2020SF001).
Institutional Review Board Statement
All experiments were approved by the Committee on the Ethics of Animal Experiments of China Agricultural University (permit number: SKLAB-2012-04-07).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original data in this study are openly available in Dataverse Project, reference number [GSE160972].
Acknowledgments
We are grateful to Qingyong Meng for providing antibodies. We would like to thank Xiaoxiang Hu and Zhengxing Lian for providing the fluorescence microscope and microplate reader.
Conflicts of Interest
The authors declare that they have no conflict of interest with the contents of this article.
References
- Abmayr, S.M.; Pavlath, G.K. Myoblast fusion: Lessons from flies and mice. Development 2012, 139, 641–656. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.E.; Merkel, R.A.; Young, R.B. Cellular aspect of muscle growth: Myogenic cell proliferation. J. Anim. Sci. 1979, 49, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Wigmore, P.M.; Stickland, N.C. Muscle development in large and small pig fetuses. J. Anat. 1983, 137, 235–245. [Google Scholar] [PubMed]
- Buckingham, M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev. 2006, 16, 525–532. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Liu, X. Characterization of transcriptional complexity during longissimus muscle development in bovines using high-throughput sequencing. PLoS ONE 2013, 8, e64356. [Google Scholar] [CrossRef] [PubMed]
- Hindi, S.M.; Tajrishi, M.M.; Kumar, A. Signaling mechanisms in mammalian myoblast fusion. Sci. Signal. 2013, 6, re2. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jin, P.; Duan, R.; Chen, E.H. Mechanisms of myoblast fusion during muscle development. Curr. Opin. Genet. Dev. 2015, 32, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Te Pas, M.F.; Keuning, E.; Hulsegge, B.; Hoving-Bolink, A.H.; Evans, G.; Mulder, H.A. Longissimus muscle transcriptome profiles related to carcass and meat quality traits in fresh meat Pietrain carcasses. J. Anim. Sci. 2010, 88, 4044–4055. [Google Scholar] [CrossRef] [PubMed]
- Wes, P.D.; Chevesich, J.; Jeromin, A.; Rosenberg, C.; Stetten, G.; Montell, C. TRPC1, a human homolog of a Drosophila store-operated channel. Proc. Natl. Acad. Sci. USA 1995, 92, 9652–9656. [Google Scholar] [CrossRef]
- Zhu, X.; Chu, P.B.; Peyton, M.; Birnbaumer, L. Molecular cloning of a widely expressed human homologue for the Drosophila trp gene. FEBS Lett. 1995, 373, 193–198. [Google Scholar] [CrossRef]
- Sours, S.; Du, J.; Chu, S.; Ding, M.; Zhou, X.J.; Ma, R. Expression of canonical transient receptor potential (TRPC) proteins in human glomerular mesangial cells. Am. J. Physiol. Renal. Physiol. 2006, 290, F1507–F1515. [Google Scholar] [CrossRef] [PubMed]
- Fiorio Pla, A.; Maric, D.; Brazer, S.C.; Giacobini, P.; Liu, X.; Chang, Y.H.; Ambudkar, I.S.; Barker, J.L. Canonical transient receptor potential 1 plays a role in basic fibroblast growth factor (bFGF)/FGF receptor-1-induced Ca2+ entry and embryonic rat neural stem cell proliferation. J. Neurosci. 2005, 25, 2687–2701. [Google Scholar] [PubMed]
- Louis, M.; Zanou, N.; Van Schoor, M.; Gailly, P. TRPC1 regulates skeletal myoblast migration and differentiation. J. Cell Sci. 2008, 121, 3951–3959. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, K.T.; Bandyopadhyay, B.C.; Pani, B.; Dietrich, A.; Paria, B.C.; Swaim, W.D.; Beech, D.; Yildrim, E.; Singh, B.B.; et al. Attenuation of store-operated Ca2+ current impairs salivary gland fluid secretion in TRPC1−/− mice. Proc. Natl. Acad. Sci. USA 2007, 104, 17542–17547. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Birnbaumer, L.; Singh, B.B. TRPC1 regulates calcium-activated chloride channels in salivary gland cells. J. Cell. Physiol. 2015, 230, 2848–2856. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.N.; Platoshyn, O.; Golovina, V.A.; Liu, L.; Zou, T.; Marasa, B.S.; Turner, D.J.; Yuan, J.X.-J.; Wang, J.-Y. TRPC1 functions as a store-operated Ca2+ channel in intestinal epithelial cells and regulates early mucosal restitution after wounding. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G782–G792. [Google Scholar] [CrossRef] [PubMed]
- Bomben, V.C.; Turner, K.L.; Barclay, T.-T.C.; Sontheimer, H. Transient receptor potential canonical channels are essential for chemotactic migration of human malignant gliomas. J. Cell. Physiol. 2011, 226, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Dreja, K.; Shah, S.S.; Cheong, A.; Xu, S.Z.; Sukumar, P.; Naylor, J.; Forte, A.; Cipollaro, M.; McHugh, D.; et al. Upregulated TRPC1 channel in vascular injury in vivo and its role in human neointimal hyperplasia. Circ. Res. 2006, 98, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Seth, M.; Zhang, Z.S.; Mao, L.; Graham, V.; Burch, J.; Stiber, J.; Tsiokas, L.; Winn, M.; Abramowitz, J.; Rockman, H.A.; et al. TRPC1 channels are critical for hypertrophic signaling in the heart. Circ. Res. 2009, 105, 1023–1030. [Google Scholar] [CrossRef] [PubMed]
- Kunichika, N.; Yu, Y.; Remillard, C.V.; Platoshyn, O.; Zhang, S.; Yuan, J.X.-J. 2004 Overexpression of TRPC1 enhances pulmonary vasoconstriction induced by capacitative Ca2+ entry. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2004, 287, L962–L969. [Google Scholar] [CrossRef]
- Shim, S.; Goh, E.L.; Ge, S.; Sailor, K.; Yuan, J.P.; Roderick, H.L.; Bootman, M.D.; Worley, P.F.; Song, H.; Ming, G. XTRPC1-Dependent chemotropic guidance of neuronal growth cones. Nat. Neurosci. 2005, 8, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Nesin, V.; Tsiokas, L. TRPC1. Handb. Exp. Pharmacol. 2014, 222, 15–51. [Google Scholar] [PubMed]
- Formigli, L.; Sassoli, C.; Squecco, R.; Bini, F.; Martinesi, M.; Chellini, F.; Luciani, G.; Sbrana, F.; Zecchi-Orlandini, S.; Francini, F.; et al. Regulation of transient receptor potential canonical channel 1 (TRPC1) by sphingosine 1-phosphate in C2C12 myoblasts and its relevance for a role of mechanotransduction in skeletal muscle differentiation. J. Cell Sci. 2009, 122, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Antigny, F.; Koenig, S.; Bernheim, L.; Frieden, M. During post-natal human myogenesis, normal myotube size requires TRPC1- and TRPC4-mediated Ca2+ entry. J. Cell Sci. 2013, 126, 2525–2533. [Google Scholar] [CrossRef] [PubMed]
- Zanou, N.; Shapovalov, G.; Louis, M.; Tajeddine, N.; Gallo, C.; Van Schoor, M.; Anguish, I.; Cao, M.L.; Schakman, O.; Dietrich, A.; et al. Role of TRPC1 channel in skeletal muscle function. Am. J. Physiol. Cell Physiol. 2010, 298, C149–C162. [Google Scholar] [CrossRef] [PubMed]
- Antigny, F.; Sabourin, J.; Sauc, S.; Bernheim, L.; Koenig, S.; Frieden, M. TRPC1 and TRPC4 channels functionally interact with STIM1L to promote myogenesis and maintain fast repetitive Ca2+ release in human myotubes. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Vandebrouck, C.; Martin, D.; Colson-Van Schoor, M.; Debaix, H.; Gailly, P. Involvement of TRPC in the abnormal calcium influx observed in dystrophic (mdx) mouse skeletal muscle fibers. J. Cell Biol. 2002, 158, 1089–1096. [Google Scholar] [CrossRef]
- Gervásio, O.L.; Whitehead, N.P.; Yeung, E.W.; Phillips, W.D.; Allen, D.G. TRPC1 binds to caveolin-3 and is regulated by Src kinase—role in Duchenne muscular dystrophy. J. Cell Sci. 2008, 121, 2246–2255. [Google Scholar] [CrossRef]
- Xia, L.; Cheung, K.K.; Yeung, S.S.; Yeung, E.W. The involvement of transient receptor potential canonical type 1 in skeletal muscle regrowth after unloading-induced atrophy. J. Physiol. 2016, 594, 3111–3126. [Google Scholar] [CrossRef]
- Zhang, B.T.; Yeung, S.S.; Cheung, K.K.; Chai, Z.Y.; Yeung, E.W. Adaptive responses of TRPC1 and TRPC3 during skeletal muscle atrophy and regrowth. Muscle Nerve 2014, 49, 691–699. [Google Scholar] [CrossRef]
- Liu, X.; Wang, W.; Singh, B.B.; Lockwich, T.; Jadlowiec, J.; O’Connell, B.; Wellner, R.; Zhu, M.X.; Ambudkar, I.S. Trp1, a candidate protein for the store-operated Ca2+ influx mechanism in salivary gland cells. J. Biol. Chem. 2000, 275, 3403–3411. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Oboukhova, E.A.; Kumar, S.; Sturek, M.; Obukhov, A.G. Canonical transient receptor potential channels expression is elevated in a porcine model of metabolic syndrome. Mol. Endocrinol. 2009, 23, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, X.; Riley, A.M.; Hiett, S.C.; Temm, C.J.; Beli, E.; Long, X.; Chakraborty, S.; Alloosh, M.; White, F.A.; et al. Long-Term spironolactone treatment reduces coronary TRPC expression, vasoconstriction, and atherosclerosis in metabolic syndrome pigs. Basic Res. Cardiol. 2017, 112, 54. [Google Scholar] [CrossRef] [PubMed]
- Shang, P.; Wang, Z.; Chamba, Y.; Zhang, B.; Zhang, H. A comparison of prenatal muscle transcriptome and proteome profiles between pigs with divergent growth phenotypes. J. Cell Biochem. 2019, 120, 5277–5286. [Google Scholar] [CrossRef] [PubMed]
- Yue, M.; Tian, Y.G.; Wang, Y.J.; Gu, Y.; Bayaer, N.; Hu, Q.; Gu, W.W. Associated analysis of single nucleotide polymorphisms found on exon 3 of the IGF-1 gene with Tibetan miniature pig growth traits. Genet. Mol. Res. 2014, 13, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ma, C.; Qiao, F.; Song, Y.; Du, M. Lipolysis in intramuscular lipids during processing of traditional Xuanwei ham. Meat Sci. 2005, 71, 670–675. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Q.; Zhang, B.; Lu, Y.; Yang, Y.; Ban, D.; Zhang, H. Single nucleotide polymorphism scanning and expression of the FRZB gene in pig populations. Gene 2014, 543, 198–203. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jin, J.J.; Lv, W.; Xia, P.; Xu, Z.Y.; Zheng, A.D.; Wang, X.J.; Wang, S.S.; Zeng, R.; Luo, H.M.; Li, G.L.; et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. USA 2018, 115, E9802–E9811. [Google Scholar] [CrossRef]
- Buckingham, M.; Bajard, L.; Chang, T.; Daubas, P.; Hadchouel, J.; Meilhac, S.; Montarras, D.; Rocancourt, D.; Relaix, F. The formation of skeletal muscle: From somite to limb. J. Anat. 2003, 202, 59–68. [Google Scholar] [CrossRef]
- Guo, B.; Greenwood, P.L.; Cafe, L.M.; Zhou, G.; Zhang, W.; Dalrymple, B.P. Transcriptome analysis of cattle muscle identifies potential markers for skeletal muscle growth rate and major cell types. BMC Genomics 2015, 16, 177. [Google Scholar] [CrossRef] [PubMed]
- Krout, D.; Schaar, A.; Sun, Y.; Sukumaran, P.; Roemmich, J.N.; Singh, B.B.; Claycombe-Larson, K.J. The TRPC1 Ca2+-permeable channel inhibits exercise-induced protection against high-fat diet-induced obesity and type II diabetes. J. Biol. Chem. 2017, 292, 20799–20807. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Jin, X.; Li, Q.; Wang, W.; Wang, Y.; Zhang, J. Association of TRPC1 gene polymorphisms with type 2 diabetes and diabetic nephropathy in Han Chinese population. Endocr. Res. 2013, 38, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Freedman, B.I.; Flekac, M.; Santos, E.; Hicks, P.J.; Bowden, D.W.; Efendic, S.; Brismar, K.; Gu, H.F. Evaluation of genetic association and expression reduction of TRPC1 in the development of diabetic nephropathy. Am. J. Nephrol. 2009, 29, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Tabur, S.; Oztuzcu, S.; Duzen, I.V.; Eraydin, A.; Eroglu, S.; Ozkaya, M.; Demiryürek, A.T. Role of the transient receptor potential (TRP) channel gene expressions and TRP melastatin (TRPM) channel gene polymorphisms in obesity-related metabolic syndrome. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 1388–1397. [Google Scholar]
- Everett, K.V.; Chioza, B.A.; Georgoula, C.; Reece, A.; Gardiner, R.M.; Chung, E.M.K. Infantile hypertrophic pyloric stenosis: Evaluation of three positional candidate genes, TRPC1, TRPC5 and TRPC6, by association analysis and re-sequencing. Hum. Genet. 2009, 126, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Vincent, S.D. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr. Opin. Genet. Dev. 2009, 19, 444–453. [Google Scholar] [CrossRef]
- Yap, J.L.Y.; Tai, Y.K.; Frohlich, J.; Fong, C.H.H.; Yin, J.N.; Foo, Z.L.; Ramanan, S.; Beyer, C.; Toh, S.J.; Casarosa, M.; et al. Ambient and supplemental magnetic fields promote myogenesis via a TRPC1-mitochondrial axis: Evidence of a magnetic mitohormetic mechanism. FASEB J. 2019, 33, 12853–12872. [Google Scholar] [CrossRef]
- Meacci, E.; Bini, F.; Sassoli, C.; Martinesi, M.; Squecco, R.; Chellini, F.; Zecchi-Orlandini, S.; Francini, F.; Formigli, L. Functional interaction between TRPC1 channel and connexin-43 protein: A novel pathway underlying S1P action on skeletal myogenesis. Cell. Mol. Life Sci. 2010, 67, 4269–4285. [Google Scholar] [CrossRef]
- Li, M.; Chen, C.; Zhou, Z.; Xu, S.; Yu, Z. A TRPC1-mediated increase in store-operated Ca2+ entry is required for the proliferation of adult hippocampal neural progenitor cells. Cell Calcium 2012, 51, 486–496. [Google Scholar] [CrossRef]
- Cheung, K.K.; Yeung, S.S.; Au, S.W.; Lam, L.S.; Dai, Z.Q.; Li, Y.H.; Yeung, E.W. Expression and association of TRPC1 with TRPC3 during skeletal myogenesis. Muscle Nerve 2011, 44, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Zanou, N.; Schakman, O.; Louis, P.; Ruegg, U.T.; Dietrich, A.; Birnbaumer, L.; Gailly, P. Trpc1 ion channel modulates phosphatidylinositol 3-kinase/Akt pathway during myoblast differentiation and muscle regeneration. J. Biol. Chem. 2012, 287, 14524–14534. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, A.; Chubanov, V.; Kalwa, H.; Rost, B.R.; Gudermann, T. Cation channels of the transient receptor potential superfamily: Their role in physiological and pathophysiological processes of smooth muscle cells. Pharmacol. Therapeut. 2006, 112, 744–760. [Google Scholar] [CrossRef] [PubMed]
- Kazanskaya, O.; Glinka, A.; Barco Barrantes, I.; Stannek, P.; Niehrs, C.; Wu, W. R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 2004, 7, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Tanaka, M.; Münsterberg, A. Expression of (beta)-catenin in the developing chick myotome is regulated by myogenic signals. Development 2000, 127, 4105–4113. [Google Scholar]
- Maretto, S.; Cordenonsi, M.; Dupont, S.; Braghetta, P.; Broccoli, V.; Hassan, A.B.; Volpin, D.; Bressan, G.M.; Piccolo, S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 3299–3304. [Google Scholar] [CrossRef]
- Otto, A.; Schmidt, C.; Luke, G.; Allen, S.; Valasek, P.; Muntoni, F.; Lawrence-Watt, D.; Patel, K. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J. Cell Sci. 2008, 121, 2939–2950. [Google Scholar] [CrossRef]
- Brack, A.S.; Murphy-Seiler, F.; Hanifi, J.; Deka, J.; Eyckerman, S.; Keller, C.; Aguet, M.; Rando, T.A. BCL9 is an essential component of canonical Wnt signaling that mediates the differentiation of myogenic progenitors during muscle regeneration. Dev. Biol. 2009, 335, 93–105. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, Y.; Gu, L.; Lan, M.; Liu, C.; Wang, M.; Su, Y.; Ge, M.; Wang, T.; Yu, Y.; et al. Islr regulates canonical Wnt signaling-mediated skeletal muscle regeneration by stabilizing Dishevelled-2 and preventing autophagy. Nat. Commun. 2018, 9, 5129. [Google Scholar] [CrossRef]
- Le Grand, F.; Jones, A.E.; Seale, V.; Scimè, A.; Rudnicki, M.A. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell 2009, 4, 535–547. [Google Scholar] [CrossRef]
- Von Maltzahn, J.; Bentzinger, C.F.; Rudnicki, M.A. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat. Cell Biol. 2011, 14, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Takata, H.; Terada, K.; Oka, H.; Sunada, Y.; Moriguchi, T.; Nohno, T. Involvement of Wnt4 signaling during myogenic proliferation and differentiation of skeletal muscle. Dev. Dyn. 2007, 236, 2800–2807. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; You, W.; Shan, T. Myomaker, and Myomixer-Myomerger-Minion modulate the efficiency of skeletal muscle development with melatonin supplementation through Wnt/β-catenin pathway. Exp. Cell Res. 2019, 385, 111705. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).