Abstract
Hull opening is a key physiological process during reproductive development, strongly affecting the subsequent fertilization and seed development in rice. In this study, we characterized a rice mutant, non-open hull 1 (noh1), which was derived from ethylmethane-sulfonate (EMS)-treated Xinong 1B (Oryza sativa L.). All the spikelets of noh1 developed elongated and thin lodicules, which caused the failure of hull opening and the cleistogamy. In some spikelets of the noh1, sterile lemmas transformed into hull-like organs. qPCR analysis indicated that the expression of A- and E-function genes was significantly upregulated, while the expression of some B-function genes was downregulated in the lodicules of noh1. In addition, the expression of A-function genes was significantly upregulated, while the expression of some sterile-lemma maker genes was downregulated in the sterile lemma of noh1. These data suggested that the lodicule and sterile lemma in noh1 mutant gained glume-like and lemma-like identity, respectively. Genetic analysis showed that the noh1 trait was controlled by a single recessive gene. The NOH1 gene was mapped between the molecular markers ZJ-9 and ZJ-25 on chromosome 1 with a physical region of 60 kb, which contained nine annotated genes. These results provide a foundation for the cloning and functional research of NOH1 gene.
1. Introduction
Rice floret contains four whorls of floral organs, which are made up of one lemma, one palea, two lodicules, six stamens, and one carpel from outside to inside, respectively [1]. After fertilization, the lodicules and stamens gradually degenerate, while the carpel develops into seed. The lemma and palea hook together most of the time to protect the internal organs and seeds. Only during the flowering date, the lemma and palea in each floret open once from 09:00 to 11:00 under normal circumstances. This process generally lasts 40 to 90 min and then the lemma and palea close and never reopen. These two actions are called hull opening (the lemma and the palea together are called a hull) and hull closing, respectively. Hull opening is a necessary condition for plant fertilization, and hull closing protects the development of seed from outside interference after fertilization.
Lodicules, two small, fleshy, and lung-like floral organs, asymmetrically formed inside the lemma, play a crucial role in promoting hull opening/closing by their expansion and shrinkage. During flowering, the water potential of lodicule cells decreases, and then lodicules absorb water and cause the lodicules to swell. The swollen lodicules then push lemma outwards and simultaneously squeeze the palea inwards, which causes the hook of the lemma and palea to release, and thus the lemma and the palea are separated from each other. At this time, the filaments are rapidly elongated, the anthers stick out of the hulls and crack, and then the pollination begins [2]. It is also believed that the excessive water absorption in lodicule cells finally leads the cells to rupture and shrink [2]. With the lodicules losing the supports to the lemma, the lemma and palea lock together again and the floret gets closed [3].
The identity of lodicules is mainly determined by B-class genes OsMADS2, OsMADS4, and OsMADS16 in the ABCDE model, all of which encode transcription factors containing MADS-box domain [4,5,6]. Another two MADS-box genes, OsMADS6 and OsMADS32, are also responsible for regulating lodicule identity [7,8]. In addition to the MADS-box genes, the STAMENLESS1 (SL1) gene encoding a C2H2 zinc finger protein is also involved in the regulation of lodicule development [9]. These genes all regulate the development of the morphological characteristics of the lodicules. Their mutations usually cause the transformation of lodicules into other floral organs, which generally lose their normal function and fail to mediate hull opening/closing. In fact, cleistogamy may be a favorable trait for self-pollination crops (such as rice, wheat) to avoid failure of pollinating caused by adverse weather conditions, while it may prevent transgenes from spreading into the environment by reducing outcrossing rates.
In this study, a rice mutant with failure of hull opening, named non-open hull 1 (noh1), is reported. The lodicules in noh1 mutant transformed into glume-like organs, and lost the water swelling function, which caused the rice floret to remain in a closed state and complete cleistogamy during the whole development process. The NOH1 gene has been located on chromosome 1, which provides a foundation for the cloning and further function research of NOH1.
2. Materials and Methods
2.1. Plant Materials
The noh1 mutant derived from an EMS mutagenesis population which used xian-type (indica) maintainer line Xinong 1B cultivated by Rice Research Institute of Southwest University as donor. The noh1 mutation was stably inherited through successive generations of self-crossing. F1 population was generated by the cross of xian-type (indica) sterile line 56S with noh1. The parent plants, F1 population and F2 population, were all grown in Chongqing for genetic analysis, and then plants with mutant phenotype in F2 population were used to map the NOH1 gene.
2.2. Morphological and Histological Analysis of noh1
During flowering stage, the phenotypic characteristics of the mutants and wild-type spikelets were investigated using a Nikon SMZ1500 stereoscope (Nikon Instruments Shanghai Inc., China) and a Hitachi SU3500 (Hitachi High-Technologies Corporation, Tokyo, Japan) scanning electron microscope with a −20 °C cooling stage under a low-vacuum environment.
For paraffin section, the mutant and wild-type spikelets at heading stage were fixed in FAA (50% (v/v) ethanol, 0.9 N glacial acetic, and 3.7% (v/v) formaldehyde) and placed at 4 °C for at least 16 h after pumping the air in the tissue. Next, those spikelets were dehydrated by a gradient ethanol series, infiltrated by xylene, embedded into paraffin (Sigma-Aldrich Inc., Shanghai, China), cut into 8-μm-thick slices, and then pasted on the microscope slides (RM2245; Leica, Hamburg, Germany). These slices were dyed sequentially with 1% (w/v) safranin (Amresco Inc., Framingham, MS, USA) and 1% (w/v) Fast Green (Amresco Inc., Framingham, MA, USA) and then dehydrated through an ethanol series, infiltrated with xylene, and finally mounted beneath a coverslip. Light microscopy was performed using a Nikon E600 microscope.
2.3. Molecular Mapping of NOH1
Locating the target gene was performed according to the BSA (bulked segregant analysis) method [10]. DNA of the parents, F2 population, wild-type, and mutant gene pools were extracted from similar sized fresh leaves following the previously reported CTAB (hexadecyl trimethyl ammonium bromide) method [11]. The quality and quantity of DNA were estimated using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA) and 1% (w/v) agarose gel electrophoresis. SSR (simple sequence repeats) markers that were distributed evenly on the 12 chromosomes were employed for gene mapping from the Web (http://www.gramene.org/microsat/) (see Table S1 in the Supplementary Materials). All the primers were synthesized by the Shanghai Invitrogen Company. The total volume of the PCR amplifications was 15 µL, which contained 1.5 µL 10× PCR buffer, 1 µL 50 ng µL−1 DNA, 0.75 µL 2.5 mmol L−1 dNTPs, 9.5 µL ddH2O, 1 µL 10 mmol L−1 forward and reverse primer, and 0.25 µL 5 U µL−1 rTaq DNA polymerase (Takara Bio Inc., Dalian, China). Amplification was performed with a MyCycler Thermal Cycler (Bio-Rad, Foster City, CA, USA) under the following conditions: 5 min at 94 °C for DNA strand separation, followed by 35 cycles of denaturing at 94 °C for 30 s, annealing at 56 °C for 30 s, extension at 72 °C for 30s, and finally an extension at 72 °C for 7 min. Amplified products were separated by electrophoresis on 10.0% polyacrylamide gels, and then silver staining was used to observe the color of the band patterns [12].
2.4. Linkage Map Construction
Bands for molecular markers were identified in both parents, 56S and noh1, and labeled as A and B, respectively, and the heterozygote that contains two parent bands was labeled as H. The linkage relationship was analyzed by MAPMAKER3.0 [13], and the recombination rate was represented as the number of recombinants. Meanwhile, we constructed the physical map based on the rice genome sequence information offered by the Gramene website (http://www.gramene.org/).
2.5. qPCR Analysis
After appearance of the non-open hull phenotype at the flowering stage, we collected about 60 pairs of lodicules from several plants with the corresponding phenotype using a Nikon SMA1500 stereoscope and extracted total RNA from the lodicules of wild-type and noh1 florets using the RNAprep Pure Plant Kit (Tiangen Biochemical Technology Co., Ltd., Beijing, China). The first-strand cDNA was synthesized from 1 µg total RNA with oligo(dT)18 primers in a 20 µL reaction volume using the PrimeScript® Reagent Kit With gDNA Eraser (Takara Bio Inc., Dalian, China). Several representative floral specific genes were selected by consulting the literature and their expression levels (OsMADS1, OsMADS2, OsMADS4, OsMADS6, OsMADS16, OsMADS14, OsMADS15, OsMADS34, G1, ASP1 and DL) were detected by qPCR. The qPCR analysis was performed with three replicates using SYBR premix Ex Taq II Kit (Takara Bio Inc., Dalian, China) in an ABI 7500 Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA). ACTIN was used as an endogenous control (see Table S4 in the Supplementary Materials).
3. Results
3.1. Morphological Analysis of noh1 Mutant
While no significant defect was observed during the vegetative stage, some significant abnormalities were exhibited in the noh1 spikelets at the reproductive stage in comparison with the wild-type spikelets. Generally, the wild-type spikelet has two pairs of glumes (rudimentary glume and sterile lemma) and one fertile top floret, consisting of a lemma and a palea in whorl 1, two lodicules in whorl 2, six stamens in whorl 3, and one carpel in whorl 4 (Figure 1a,b). The lodicules of the wild-type look fleshy and semitransparent with smooth surfaces (Figure 1c–e). However, in the noh1 floret, the lodicules were elongated and became much thinner and narrower than those in the wild-type, while the identities of other floral organs were normal (Figure 1h–l). Furthermore, it was observed that these slim lodicules developed linear cells in parallel arrays in their smooth upper epidermis, which was similar to the epidermis of wild-type sterile lemma (Figure 1e,g,l). In addition, most of the sterile lemmas in noh1 mutants were also obviously elongated and showed abundant trichomes and protrusions in their upper epidermis, which was almost the same as that in the hull (lemma and palea) (Figure 1f,m).
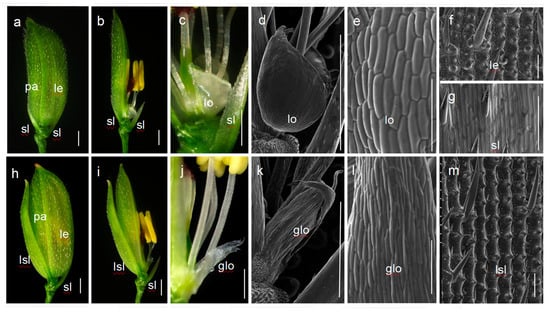
Figure 1.
Morphological analysis of spikelet of the wild-type (Xinong 1B) and noh1 mutant. (a–c) the wild-type spikelet; (d–e) the wild-type lodicule; (f) the wild-type lemma; (g) the wild-type sterile lemma; (h–j) the noh1 spikelet; (k–l) the glume-like lodicule in noh1; (m) the lemma-like sterile lemma of noh1. glo: glume-like lodicule; lsl: lemma-like sterile lemma; le: lemma; lo: lodicule; pa: palea; sl: sterile lemma; bars represent 1000 μm (a–d,h–k) and 100 μm (e–g,l,m).
3.2. Histological Analysis of noh1 Mutant
To further clarify the identities of floral organs in noh1 mutants, a histological analysis was performed. In the wild-type florets, the lodicules were composed of lots of large parenchymatous cells and radial small vascular bundles (Figure 2a,b), while the sterile lemma consisted of three layers including upper and lower epidermal cells and middle parenchymatous cells (Figure 2a,c). However, in the thinner lodicules of noh1 florets, the number of parenchymatous cell layers decreased significantly, and even the number of vascular bundles was reduced to only one (Figure 2d,e), which caused it to look like the cell structure of the wild-type sterile lemma. Furthermore, the wild-type lemma consisted of four cell layers including the silicified upper epidermis, fibrous sclerenchyma, spongy parenchyma cells, and lower epidermis, with five vascular bundles. The wild-type glume contained three kinds of cell types, smooth upper epidermal cells, parenchymatous cells, and lower epidermal cells, with only one vascular bundle. It was found that the cell structure of the elongated sterile lemma in noh1 mutants was not similar to the wild-type glume but to the wild-type lemma (Figure 2c,f).
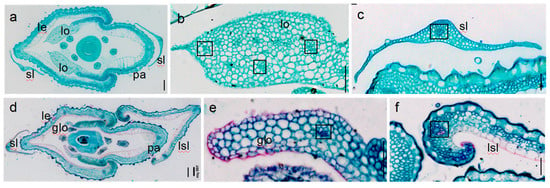
Figure 2.
Histological analysis of spikelet at flowering stage in the wild-type and noh1 plants; (a–c) the cross-sections of the wild-type spikelet; (d–f) the cross-sections of the noh1 spikelet. glo: glume-like lodicule; lsl: lemma-like sterile lemma; le: lemma; lo: lodicule; pa: palea; sl: sterile lemma; □: vascular bundle; bars represent 100 μm (a,d) and 50 μm (b,c,e,f).
3.3. Morphological Analysis of noh1 Lodicules during Flowering
In the wild-type floret of rice, about one hour before flowering, the lemma and the palea were still interlocked tightly and the lodicules were not plump enough (Figure 3a,e,i). At about 20 min after floret opening, the lodicules absorbed water and notably expanded, while the filaments were elongated and the anthers were cracked, which allowed pollination to occur (Figure 3b,f,j). About 1.5 h after blossoming, the lodicules gradually shrank and the hull began to close (Figure 3c,g,k). About 48 h after flowering, the hull had closed completely with full shrinkage of the lodicules, and the ovary began to develop (Figure 3d,h,l). Therefore, the process from hull opening to closing is highly relevant to the expansion and atrophy of the lodicules.
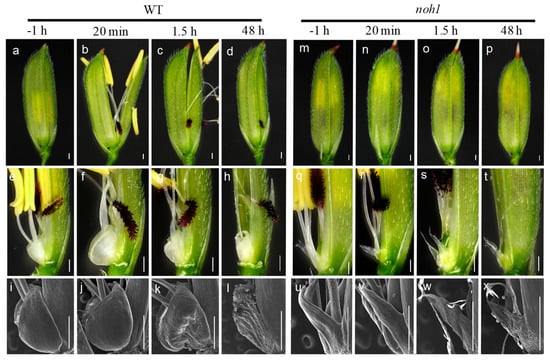
Figure 3.
Morphological observation of wild-type (WT) and noh1 mutant florets during flowering; (a–d) show successively the WT florets at one hour before opening, 20 m after opening, 1.5 h after opening. and 48 h after opening; (m–p) show successively the noh1 florets at one hour before opening, 20 min after opening, 1.5 h after opening, and 48 h after opening; (e–h) show successively the morphological features of the WT lodicule at one hour before opening, 20 min after opening, 1.5 h after opening, and 48 h after opening; (q–t) show successively the morphological features of the noh1 lodicule at one hour before opening, 20 min after opening, 1.5 h after opening, and 48 h after opening; (i–l) show successively the microscopic structure of the WT lodicule at one hour before opening, 20 min after opening, 1.5 h after opening, and 48 h after opening; (u–x) show successively the microscopic structure of the noh1 lodicule at one hour before opening, 20 min after opening, 1.5 h after opening, and 48 h after opening; bars represent 500 μm.
Compared with the wild-type, obvious differences were observed during the noh1 floret opening. Firstly, the hull in noh1 florets never opened during the whole flowering stage (Figure 3m–p). Secondly, comparing the florets with elongated filaments (which means that the florets have been opened or pollinated) with the florets with non-elongated filaments (which means that the florets have not been opened or pollinated), no obvious swelling was observed in the lodicules during the noh1 floret opening or pollination (Figure 3u,v). Fortunately, the other processes related to floret opening, such as filament elongation, anther dehiscence, and pollination, were almost normal so that the noh1 florets were still fertile.
3.4. Expression Analysis of Floral Organ Identity Genes in Lodicule of noh1 and Wild-Type
Given that the noh1 lodicules and sterile lemma showed glume/hull-like identity in morphological features, the expression of some spikelet/floral organ identity genes were detected, which includes OsMADS6 (which was expressed in the lodicule and palea) [7], DL (which was expressed in the lemma and pistil) [14,15], G1 (which was expressed in the sterile lemma and palea) [16], ASP1 (which was mainly expressed in the sterile lemma) [17], A class genes OsMADS14 and OsMADS15 (which were expressed in the lemma and palea) [18,19], B class genes OsMADS2, OsMADS4, and OsMADS16/SPW1 (which were expressed in the lodicule and stamen) [4,5,6], E class gene OsMADS1 (which was mainly expressed in the lemma and palea), and OsMADS34 (which was mainly expressed in the sterile lemma, lemma, and palea) [20,21]. Compared with that of the wild-type plants, it was not surprising that the relative expression level of three B class genes OsMADS2, OsMADS4, and OsMADS16, which were all involved in the development of lodicules, were downregulated in the lodicules of noh1 mutants. On the contrary, the genes related to the identities of the sterile lemma and hull were significantly upregulated, such as OsMADS1, OsMADS14 and DL, or even extremely upregulated, such as OsMADS15 and G1 (Figure 4a,b). In the elongated sterile lemma of noh1, the expression of OsMADS34, G1, and ASP1 was significantly downregulated, while the expression levels of OsMADS1, OsMADS14, OsMADS15, and DL were upregulated with varying degrees (Figure 4c,d). In addition, the expression of OsMADS6, which was related to palea but not lemma, was also significantly downregulated in the elongated sterile lemma of noh1 mutants.
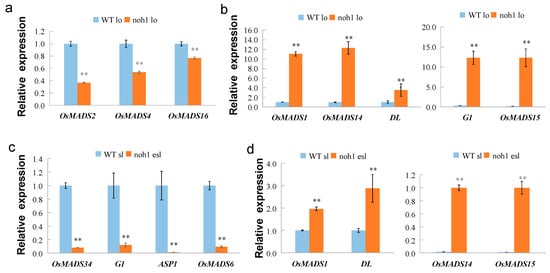
Figure 4.
Relative expression levels of floral organ identity genes in the wild type (WT) and noh1 floral organs; (a) B class genes downregulated in the noh1 lodicule; (b) genes partially upregulated in the noh1 lodicule; (c) genes partially downregulated in the noh1 elongated sterile lemma (about 15 pairs of sterile lemmas with corresponding phenotype were collected by using a pointy tweezer to extract the total RNA); (d) genes partially upregulated in the noh1 elongated sterile lemma. lo: lodicule; sl: sterile lemma; esl: elongated sterile lemma; error bars indicate SD. ** indicates p < 0.01.
3.5. Genetic Analysis
The noh1 mutant was crossed with the sterile line 56S. All F1 plants exhibited normal phenotype, while F2 plants showed the normal phenotype or noh1 mutant phenotype. Genetic analysis of the F2 progeny showed that the segregation ratio of WT plants and mutant individuals was a good fit to 3:1 (536 of 2242 were mutant individuals; χ2 = 1.37 < χ20.05 = 3.84), indicating that the noh1 trait was controlled by a single recessive gene.
3.6. Gene Mapping of NOH1
Recessive individuals in the F2 population were used as a mapping population to localize the NOH1 gene. Ten wild-type plants and ten mutant plants in the F2 segregating population were randomly selected to construct wild-type and mutant DNA pools, respectively. About 430 pairs of primers for SSR and IN/DEL (insertion-deletion) markers, which were uniformly distributed on the 12 chromosomes of the parents 56S and noh1 (see Table S1 in the Supplementary Materials), were used. A total of 112 pairs of primers for molecular markers showed polymorphism between the parents and were employed to screen the wild-type gene pool and the mutant gene pool (see Table S2 in the Supplementary Materials). By the linkage analysis, the target gene was linked to the polymorphic markers M33, M69, M76, and M86 on chromosome 1 (Table 1). Therefore, we used the four markers to survey the 300 mutant individuals, and then the NOH1 gene was localized between SSR markers M33 and M76 on the long arm of chromosome 1 with recombinants of 47 and 3, respectively (Figure 5a).

Table 1.
Polymorphic markers for gene mapping.
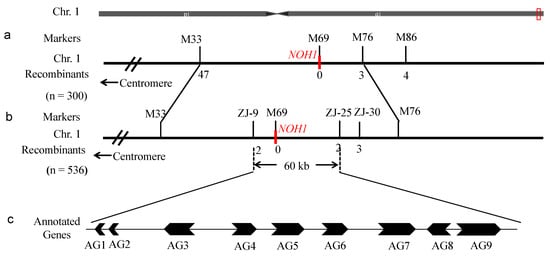
Figure 5.
Localization of the target gene NOH1 on Chromosome 1 (Chr.1) of rice. (a) Primary mapping of NOH1 on chromosome 1 based on 300 individuals; (b) NOH1 was fine-mapped to an interval of 60 kb by 536 individuals; (c) Nine genes were annotated on the 60 kb region. AG1: miR172b; AG2: miR806a; AG3: LOC_Os01g74130; AG4: LOC_Os01g74140; AG5: LOC_Os01g74146; AG6: LOC_Os01g74152; AG7: LOC_Os01g74160; AG8: LOC_Os01g74170; AG9: LOC_Os01g74180.
In order to further locate NOH1, 36 pairs of primers for new markers between M33 and M76 were synthesized (see Table S3 in the Supplementary Materials). Of those primers, ZJ-9, ZJ-25, and ZJ-30 exhibited polymorphism (Table 1). Then we used these primers to further analyze all of the 536 mutant individuals. Ultimately, the NOH1 gene was localized between SSR markers ZJ-9 and ZJ-25 with recombinants of 2 and 2, respectively (Figure 5b). The physical distance was approximate 60 kb with nine annotated genes (seven Open Reading Frames and two non-coding RNA genes), referring to the BAC (Bacterial Artificial Chromosome) (AP003277) of the sequencing variety Nipponbare (Figure 5c).
4. Discussion and Conclusions
The lodicule is a grass-specific organ that is generally regarded as a homolog of dicot petal [22,23], which plays a vital role in floret opening in most grass species. The expansion and atrophy of lodicules can promote the opening and closing of the hull, respectively. It was reported earlier that the rice B-class genes OsMADS2, OsMADS4, and OsMADS16 specified the identity of lodicules [4,5,6,15,24]. In the mutants that had non-functional copies of these genes, the lodicules were elongated and/or transformed into glume-like structures. Moreover, recent studies revealed that OsMADS6 and OsMADS32 were also responsible for the regulation of lodicule identity, and that mutations of these genes resulted in the transformation of the lodicule into a glume-like structure [25,26]. In the present study, compared with the WT plants, the lodicule identity genes OsMADS2, OsMADS4, and OsMADS16 were significantly downregulated and the lemma identity-related genes OsMADS1, OsMADS14, DL, OsMADS15, and G1 were largely upregulated in lodicules of noh1 mutants, which indicated that those elongated lodicules in the noh1 mutants lost the identity of the lodicule and might acquire the glume-like (both lemma and sterile lemma) identity to some extent. Considering morphological and histological characteristics, there was strong support for those elongated lodicules being partially transformed into sterile lemma-like organs. Therefore, this change of the noh1 lodicule made it lose the water swelling function, which eventually led to the failure of the hulls to be pushed open, similar to the mutants of B-class genes, OsMADS6, OsMADS16, and OsMADS32. However, most mutants of these genes were either completely or partially sterile because of the defects of other floral organs, although the florets in noh1 and spw1-cls (a allele mutation in the OsMADS16 gene, which only led to transformation of lodicules, but not the stamens [27]) plants were perfectly fertile due to the normal function of the other three floral organs. Therefore, cleistogamy might be an efficient strategy to consider for preventing gene flow from genetically modified crops, and even has positive effect on the seeds setting rate for the self-pollination crops especially during the rainy season; thus, the noh1 mutant could provide a favorable trait for conventional breeding [27].
The sterile lemma is a unique organ structure of Gramineae and is always considered a degenerate lemma [28,29]. In this study, some noh1 sterile lemmas were elongated, and displayed rough upper epidermis with abundant trichomes and protrusions, which were similar to the wild-type lemma. Previous studies revealed that G1, OsMADS34, and ASP1 genes determined the sterile lemma identity, and inhibited the transformation to the lemma [16,17,21,30,31,32,33]. In the elongated sterile lemma of noh1 plants, the expression levels of OsMADS1, OsMADS14, OsMADS15, and DL (the lemma identity-related genes) were upregulated, while OsMADS34, G1, ASP1, and OsMADS6 expressions were downregulated significantly. These results indicated that the elongated sterile lemma had lost the sterile lemma identity and simultaneously gained the identity of the lemma instead of palea in noh1 mutants. Therefore, NOH1 might take effect in the sterile lemma development by regulating one or several of these genes.
In this study, the NOH1 gene was mapped on the long arm of chromosome 1 with a physical distance of 60 kb between the molecular markers ZJ-9 and ZJ-25. According to the gene annotation information provided by the Gramene website (http://www.gramene.org/), seven annotated genes and two non-coding RNA are included within this interval: a retrotransposon protein (LOC_Os01g74130), a transcription factor (LOC_Os01g74140), a cell cycle switch protein (LOC_Os01g74146), two enzymes (LOC_Os01g74152 and LOC_Os01g74160), a expressed protein (LOC_Os01g74170), an adaptin (LOC_Os01g74180), and non-coding RNA genes miR172b and miR806a. Among these genes, LOC_Os01g74140 encodes a WRKY transcription factor, which regulates many metabolic processes and participates in the process of disease resistance, injury, senescence, growth, and gibberellin signal transduction in plants [34]. LOC_Os01g74146 is a B-type cell cycle switch gene in rice that is highly expressed in floral organs and chaff. Particularly, Cly1, one of the miR172 target genes in barley, was reportedly responsible for lodicule development, a single nucleotide substitution of which at the miR172 target sequence led to smaller lodicules and cleistogamy [35]. In this study, we could not detect any sequence differences in all of the seven protein-coding genes between the noh1 mutant and the wild-type. As there are a lot of repetitive regions near the miR172b and miR806a, it still remains unclear whether there is any mutation located there. Next, resequencing and/or epigenetic analysis will be used to determine which kind of mutation results in the noh1 trait.
The noh1 mutant displayed glume-like lodicules and lemma-like sterile lemma. The glume-like lodicules caused the noh1 floret to lose the capability of hull opening and cleistogamy, which might be a favorable trait for conventional breeding. Genetic analysis revealed that the noh1 trait was controlled by a single recessive gene and finally mapped between ZJ-9 and ZJ-25 on chromosome 1 with a 60 kb region. Results of our research lay a foundation for map-based cloning and function analysis of NOH1 gene.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4395/9/2/56/s1, Table S1: Polymorphic markers for polymorphic screening, Table S2: Polymorphic markers for gene pool screening, Table S3: Primers synthesized for fine mapping on chromosome 1, Table S4: Primers used for qPCR analysis.
Author Contributions
Data curation: J.Z. and H.Z.; Formal analysis: X.Z. and H.Z.; Funding acquisition: Y.L. (Yinghua Ling) and Y.L. (Yunfeng Li); Methodology: J.Z.; Software: H.Z.; Validation: H.W., J.T., and H.C.; Writing—original draft: J.Z.; Writing—review and editing: Y.L. (Yunfeng Li).
Funding
This research was funded by the National Natural Science Foundation of China, grant numbers [31730063 and 31271304], National Key Program for Research and Development, grant number [2017YFD0100202], Project of CQ CSTC, grant number [CSTC2017jcyjBX0062], Graduate student scientific research innovation projects in Chongqing, grant number [CYS2015066], and the Fundamental Research Funds for the Central Universities, grant number [XDJK2016A013].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yoshida, H.; Nagato, Y. Flower development in rice. J. Exp. Bot. 2011, 62, 4719–4730. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhou, X.; Wu, X. Advances in study of opening mechanism in rice florets. Sci. Agric. Sin. 2004, 37, 188–195. [Google Scholar] [CrossRef]
- Wang, Z.; Gu, Y.; Gao, Y. Studies on the mechanism of the anthesis of rice Ⅲ. Structure of the lodicule and changes of its contents during flowering. Acta Agron. Sin. 1991, 17, 96–101. (In Chinese) [Google Scholar]
- Xiao, H.; Wang, Y.; Liu, D.; Wang, W.; Li, X.; Zhao, X.; Xu, J.; Zhai, W.; Zhu, L. Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol. Biol. 2003, 52, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.R.; Prasad, K.; Vijayraghavan, U. Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 2007, 176, 283–294. [Google Scholar] [CrossRef]
- Yao, S.G.; Ohmori, S.; Kimizu, M.; Yoshida, H. Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol. 2008, 49, 853–857. [Google Scholar] [CrossRef]
- Li, H.; Liang, W.; Jia, R.; Yin, C.; Zong, J.; Kong, H.; Zhang, D. The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 2010, 20, 299–313. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Cai, Q.; Hu, Y.; Jin, Z.; Zhao, X.; Fan, W.; Huang, Q.; Luo, Z.; Chen, M.; et al. OsMADS32 interacts with PI-like proteins and regulates rice flower development. J. Integr. Plant Biol. 2015, 57, 504–513. [Google Scholar] [CrossRef]
- Xiao, H.; Tang, J.; Li, Y.; Wang, W.; Li, X.; Jin, L.; Xie, R.; Luo, H.; Zhao, X.; Meng, Z. STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J. 2009, 59, 789–801. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Yang, Z.; Zhong, B.; Li, Y.; Xie, R.; Zhao, F.; Ling, Y.; He, G. Genetic analysis and fine mapping of a dynamic rolled leaf gene, RL10(t), in rice (Oryza sativa L.). Genome 2007, 50, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newberg, L.A. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Nagasawa, N.; Kawasaki, S.; Matsuoka, M.; Nagato, Y.; Hirano, H.Y. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 2004, 16, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, N.; Miyoshi, M.; Sano, Y.; Satoh, H.; Hirano, H.; Sakai, H.; Nagato, Y. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 2003, 130, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Suzaki, T.; Tanaka, W.; Hirano, H.Y. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 2009, 106, 20103–20108. [Google Scholar] [CrossRef]
- Yoshida, A.; Ohmori, Y.; Kitano, H.; Taguchi-Shiobara, F.; Hirano, H.Y. ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 2012, 70, 327–339. [Google Scholar] [CrossRef]
- Pelucchi, N.; Fornara, F.; Favalli, C.; Masiero, S.; Lago, C.; Pè, M.E.; Colombo, L.; Kater, M.M. Comparative analysis of rice MADS-box genes expressed during flower development. Sex. Plant Reprod. 2002, 15, 113–122. [Google Scholar] [CrossRef]
- Wang, K.J.; Tang, D.; Hong, L. L.; Xu, W.Y.; Huang, J.; Li, M.; Gu, M.H.; Xue, Y.B.; Cheng, Z.K. DEP and AFO Regulate Reproductive Habit in Rice. PLoS Genet. 2010, 6, e1000818. [Google Scholar] [CrossRef]
- Prasad, K.; Parameswaran, S.; Vijayraghavan, U. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 2005, 43, 915–928. [Google Scholar] [CrossRef]
- Gao, X.; Liang, W.; Yin, C.; Ji, S.; Wang, H.; Su, X.; Guo, C.; Kong, H.; Xue, H.; Zhang, D. The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 2010, 153, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Bommert, P.; Satohnagasawa, N.; Jackson, D.; Hirano, H.Y. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 2005, 46, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Whipple, C.J.; Zanis, M.J.; Kellogg, E.A.; Schmidt, R.J. Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc. Natl. Acad. Sci. USA 2007, 104, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; Vijayraghavan, U. Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 2003, 165, 2301–2305. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, S.; Kimizu, M.; Sugita, M.; Miyao, A.; Hirochika, H.; Uchida, E.; Nagato, Y.; Yoshida, H. MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 2009, 21, 3008–3025. [Google Scholar] [CrossRef] [PubMed]
- Sang, X.; Li, Y.; Luo, Z.; Ren, D.; Fang, L.; Wang, N.; Zhao, F.; Ling, Y.; Yang, Z.; Liu, Y.; He, G. CHIMERIC FLORAL ORGANS 1, encoding a Monocot-specific MADS-box Protein, Regulates Floral Organ Identity in Rice. Plant Physiol. 2012, 160, 788–807. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Itoh, J.I.; Ohmori, S.; Miyoshi, K.; Horigome, A.; Uchida1, E.; Kimizu, M.; Matsumura, Y.; Kusaba, M.; Satoh, H.; Nagato, Y. superwoman1-cleistogamy, a hopeful allele for gene containment in GM rice. Plant Biotechnol J. 2007, 5, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.A. The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza. Rice 2009, 2, 1–14. [Google Scholar] [CrossRef]
- Ren, D.; Li, Y.; Zhao, F.; Sang, X.; Shi, J.; Wang, N.; Guo, S.; Ling, Y.; Zhang, C.; Yang, Z.; He, G. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 2013, 162, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Qian, Q.; Zhu, K.; Tang, D.; Huang, Z.; Gao, L.; Li, M.; Gu, M.; Cheng, Z. ELE restrains empty glumes from developing into lemmas. J. Genet. Genom. 2010, 37, 101–115. [Google Scholar] [CrossRef]
- Kobayashi, K.; Maekawa, M.; Miyao, A.; Hirohiko, H.; Kyozuka, J. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 2010, 51, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wu, F.; Du, X.; Shi, X.; Liu, Y.; Liu, S.; Hu, Y.; Theißen, G.; Meng, Z. The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol. 2014, 202, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yoshida, A.; Takahashi, M.; Maekawa, M.; Kojima, M.; Sakakibara, H.; Kyozuka, J. SAD1, an RNA polymerase I subunit A34.5 of rice, interacts with Mediator and controls various aspects of plant development. Plant J. 2015, 81, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Qiao, M.; Liu, Z.; Xiang, F. Diversification function of WRKY transcription factor. Chin. Bull. Life Sci. 2010, 22, 345–351. [Google Scholar] [CrossRef]
- Nair, S.K.; Wang, N.; Turuspekov, Y.; Pourkheirandish, M.; Sinsuwongwat, S.; Chen, G.; Sameri, M.; Tagiri, A.; Honda, I.; Watanabe, Y.; et al. Cleistogamous Flowering in Barley Arises from the Suppression of MicroRNA-Guided HvAP2 mRNA Cleavage. Proc. Natl. Acad. Sci. USA 2010, 107, 490–495. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).