Abstract
Soybean (Glycine max (L.) Merr.) flowering time and plant height are critical agronomic traits that significantly influence yield and environmental adaptability. To clarify the regulatory mechanisms of flowering-related genes and their associations with plant height, a genome-wide identification of such genes in soybean were performed. This analysis used Arabidopsis thaliana flowering genes as references, employing BLASTP searches and pathway classification. All of the identified flowering-related genes were classified into eight regulatory pathways, with the photoperiod pathway (Ph) being the most prominent. Evolutionary and expression analyses revealed that core regulators (e.g., GmFTs, GmSOC1s) are conserved across pathways and are preferentially expressed in shoot apical meristems (SAMs). Additionally, both flowering-related genes and key hormones (e.g., IAA, GA, ABA) exhibited rhythmic responses to light signals. CRISPR-Cas9-mediated validation confirmed that genes GmSAUR46b regulates both flowering time and plant height, as mutants of this gene showed early flowering and reduced height. Notably, a large proportion of previously mapped flowering genes overlapped with our identified ones, while some remained undetected, likely due to whole-genome duplication and adaptive evolution, which generate new regulatory networks. Most of the identified flowering-related genes, however, have not been mapped, which highlights substantial uncharacterized potential in soybean flowering and plant height regulation. This provides a valuable molecular framework to guide soybean molecular breeding for enhanced yield and environmental adaptability.
1. Introduction
Soybean (Glycine max (L.) Merr.) is a cornerstone of global agriculture and one of the most economically valuable crops, serving as a primary source of vegetable oil and plant-based protein [1,2]. Modern cultivated soybeans originated from the domestication of wild soybean (G. soja Sieb. & Zucc.) in China approximately 5000 years ago, after which they gradually spread worldwide [1,2,3,4]. A critical factor influencing soybean growth performance and yield potential is the tightly regulated transition from vegetative to reproductive growth, with flowering marking the initiation of this shift [2].
Flowering time in soybean is more than just a developmental milestone [5]; it is a highly plastic trait intricately linked to the plant’s ability to synchronize its life cycle with environmental cues, including photoperiod [2,5,6,7,8,9,10,11,12,13,14,15,16,17,18], temperature [19,20,21,22,23,24], and hormonal signals [25,26]. Both delayed and early flowering can disrupt the balance of growth, resulting in reduced pod formation, suboptimal seed development, and ultimately significant yield losses [27,28,29]. Given the growing global demand for soybean products and the challenges posed by climate change [30,31,32], a deeper understanding of the genetic mechanisms controlling flowering time has become urgently necessary for sustainable agriculture.
As a model plant with well-characterized genetic systems [33,34,35,36,37], Arabidopsis thaliana has provided a theoretical framework for flowering regulation research in other species [38]. Based on environmental influencing factors, flowering regulatory pathways can be categorized into eight types: the photoperiod pathway (Ph); the flower development/apical meristem response pathway (Fd) [34,35,36,39,40,41]; the hormone pathway (Ho) [42,43,44,45,46,47]; the vernalization pathway (Ve) [48]; the ambient temperature pathway (At) [49,50,51]; the aging pathway (Ag); the autonomous pathway (Au) [52,53]; and the sugar pathway (Su) [54]. These pathways integrate internal and external signals through coordinated crosstalk to precisely control the floral transition [33,55,56,57,58,59]. For instance, the photoperiod pathway detects day-length variations via photoreceptors and activates floral integrator genes, such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) [39,54,60,61]. In the hormone pathway, gibberellin (GA) facilitates flowering by alleviating the DELLA-mediated repression of genes like LEAFY (LFY) and SOC1 [46,62]. The vernalization pathway centers on FLOWERING LOCUS C (FLC), a major repressor; cold exposure inhibits gene FLC expression through epigenetic modifications, thereby activating flowering-promoting genes [49,50,51,63,64]. Additionally, critical crosstalk exists between these pathways—for example, the photoreceptor Cryptochrome 2 (CRY2) and the splicing factor CRY2 INTERACTING SPLICING FACTOR 1 (CIS1) coordinately regulate flowering under low-temperature conditions [65]—and root-specific genes (e.g., FRIGIDA (FRI)) also contribute to flowering regulation [66,67]. These discoveries have not only enhanced our understanding of plant development, but also provided a theoretical framework for studying flowering regulation in other plant species.
As a typical short-day crop, soybean’s precise regulation of flowering time depends on a complex gene interaction network [5,6,68,69]. This network begins with the perception of photoperiod signals, proceeds through a cascade of core regulatory factors, and ultimately controls the expression of flowering-inducing genes, enabling adaptation to varying light environments. Within this regulatory system, E1 family genes (E1s) [17,70,71,72,73] act as a central hub linking light signals to downstream flowering pathways. The regulation of their expression and function relies on the synergistic action of upstream photoreceptor—E3 [74,75] and E4 [74,76,77], which are homologous to phytochrome A (phyA), specifically phyA3 and phyA2—and transcriptional regulators such as Tof11 (homologous to A. thaliana PSEUDO-RESPONSE-REGULATOR3a, PRR3a), Tof12 (homologous to A. thaliana PRR3b) [78,79], Tof5 (homologous to A. thaliana FRUITFULL2a, FUL2a) [80], and Tof16 (homologous to A. thaliana LATE ELONGATED HYPOCOTYL1a, LHY1a) [78,81,82], among others. Together, these components form a multi-level regulatory network (Table 1).
Under long-day (LD) conditions, the photoreceptor E3 (phyA3) [74,75] and E4 (phyA2) [74,76,77] serve as key components for light signal perception and induce the expression of E1 genes through multiple pathways [2,10]. On one hand, E3/E4 can directly interact with E1 and its homologous proteins to enhance their stability [10]. On the other hand, E3/E4 promote the expression of the transcription factors Tof11 and Tof12 [8]; these two proteins subsequently bind to the promoters of the clock genes LHYs and repress their transcription [13]. Since LHYs can directly bind to the E1 promoter and inhibit its expression, the repression of LHYs by Tof11/Tof12 indirectly alleviates the repression of E1 by LHYs, ultimately upregulating E1 transcription. Additionally, genes such as E2 (a GI homolog) [73,83,84], FLAVIN-BINDING, KELCH REPEAT, F BOX 1s (FKF1s) [85], COL [86,87], and TOE4a/b (AP2/ERF transcription factors) [11] synergistically promote E1 expression through different mechanisms, collectively reinforcing the core inhibitory function of E1 under LD conditions (Table 1).
The downstream targets of E1 primarily include focus on FT family genes (FT2a, FT5a, FT4 (E10) [88,89], FT1a [90,91]), among which FT2a and FT5a are key flowering-inducing factors in soybean [92,93,94,95,96]. Protein E1 downregulates the expression of FT2a and FT5a either directly or indirectly (e.g., by inhibiting Tof5) [72]. As a positive regulator, Tof5 can directly bind to the promoters of FT2a and FT5a to promote their transcription; however, E1’s inhibition of Tof5 indirectly reduces the expression of FT2a and FT5a [80]. This suppression further inhibits the activation of downstream floral identity genes (e.g., AP1 [97], Tof18, a SOC1 homolog [12]), ultimately delaying flowering under LD conditions. Furthermore, under LD environments, GmNF-YC4 acts as another important regulator that directly binds to the promoters of FT2a and FT5a, repressing their transcription and thereby enhancing the flowering-inhibiting effect [98]. The miRNA pathway (miR156b–miR172–TOE4a) is also involved in this process: miR156b indirectly regulates miR172 by inhibiting the expression of SPLs [99,100,101,102], which in turn affects TOE4a levels. TOE4a then contributes to delaying flowering by downregulating FT2a and FT5a expression [103] (Table 1).
In contrast, under short-day (SD) conditions, the function of E3/E4 is significantly diminished, resulting in a reduced inductive effect on E1 expression [71,89,104,105]. On one hand, the decreased activity of E3/E4 enhances the stability of LUX proteins within the circadian clock complex EC (Evening Complex), allowing LUX to strongly repress E1 expression. On the other hand, the repression of J (a gene homologous to A. thaliana ELF3) [5,106,107] and Tof16 by E3/E4 is alleviated; proteins both J and Tof16 can directly bind to the E1 promoter and inhibit its transcription [108,109]. This dual repression causes a sharp decline in E1 expression levels. The reduced abundance of E1 relieves its repression of FT2a and FT5a, while the activity of positive regulators such as Tof5 is restored. Collectively, these factors promote the expression of FT2a and FT5a [80], ultimately activating floral genes such as AP1s and accelerating flowering under SD conditions [104] (Table 1).
Notably, key genes within this regulatory network also exhibit signatures of evolutionary adaptive. For example, different alleles of Tof5 (Tof5H1, Tof5H2) have facilitated the adaptation of cultivated soybean (Glycine max) and wild soybean (Glycine soja) to the light environments of high-latitude regions through artificial selection and natural selection, respectively [80]. This further underscores the central role of this gene network in the ecological adaptation of soybean. In summary, the photoperiodic flowering regulatory network in soybean achieves precise responses to varying photoperiod conditions through the cascade regulation of E3/E4–Tof11/Tof12–E1–Tof5–FT2a/FT5a, complemented by auxiliary mechanisms such as GmNF-YC4 and the miRNA pathway [110] (Table 1).

Table 1.
Information on genes associated with soybean flowering that have been identified.
Table 1.
Information on genes associated with soybean flowering that have been identified.
| Locus | Gene | Accession Number | A. thaliana Homologous Gene Names | Flowering Pathways | Encoded Proteins | Biological or Molecular Functions | Function to Flowering | Reference |
|---|---|---|---|---|---|---|---|---|
| E1 | E1 | Glyma.06G207800 | AP2/B3-like transcriptional factor family protein | Transcriptional factor | Inhibit | [70,71] | ||
| E1La | Glyma.04G156400 | B3-domain protein | Transcriptional factor | Inhibit | [72] | |||
| E1Lb | Glyma.04G143300 | B3-domain protein | Transcriptional factor | Inhibit | [72,73] | |||
| E2 | GmGI | Glyma.10G221500 1 | GI | Ho, Ph, Su | Inhibit | [70,73,84] | ||
| E3 | GmphyA3 | Glyma.19G224200 1 | PHYA | Ph | Phytochrome A | Photoreceptor | Inhibit | [74,75,83] |
| E4 | GmphyA2 | Glyma.20G090000 1 | PHYA | Ph | Phytochrome A | Photoreceptor | Inhibit | [76,77] |
| GmphyA1 | Glyma.10G141400 1 | PHYA | Ph | Phytochrome A | Photoreceptor | Unknown | [76] | |
| E5 | Inexistence | [84,111] | ||||||
| E6 | Unknown | Promote | [107,112,113] | |||||
| E7 | Unknown | Inhibit | [114] | |||||
| E8 | Unknown | Inhibit | [113] | |||||
| E9 | GmFT2a | Glyma.16G150700 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Florigen | Inhibit | [71,92,94,115] | |
| E10 | GmFT4 | Glyma.08G363100 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Inhibit | [88,89] | ||
| E11 | Unknown | Promote | [116] | |||||
| J | GmELF3 | Glyma.04G050200 | ELF3 | Circadian clock gene | Promote | [5,117] | ||
| GmFT5a | Gyma.16G044100 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Florigen | Promote | [92,93,94,95] | ||
| GmFT1a | Glyma.18G298900 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Inhibit | [90,91] | |||
| GmFT1b | Glyma.18G299000 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Inhibit | [90] | |||
| GmFT2b | Glyma.16G151000 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Promote | [93,105] | |||
| GmFT3a | Glyma.16G044200 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Promote | [93] | |||
| GmFT3b | Glyma.19G108100 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Promote | [93] | |||
| GmFT5b | Glyma.19G108200 1 | FT; TSF | At, Ho, Ag, Ph, Su, Fd, Ve | Promote | [93] | |||
| GmFT6 | Glyma.08G363200 | Inhibit | [93] | |||||
| Tof5 | GmFUL2a | Glyma.05G018800 1 | FUL, AGL8 | At, Ph, Ag, Su, Ve | Promote | [80] | ||
| Tof11 | GmPRR3a | Glyma.U034500 | PRR | Circadian clock gene | Inhibit | [8,79] | ||
| Tof12 | GmPRR3b | Glyma.12G073900 | PRR | Circadian clock gene | Inhibit | [8,78,79] | ||
| Tof16 | GmLHY1a | Glyma.16G017400 1 | CCA1 | Ph | Promote | [8,13,81] | ||
| GmLHY1b | Glyma.07G048500 | Promote | [5,81] | |||||
| GmLHY2a | Glyma.19G260900 | Promote | ||||||
| GmLHY2b | Glyma.03G261800 | Promote | ||||||
| Dt1 | GmTFL1b | Glyma.19G194300 1 | TFL1; ATC; BFT | Fd, Ph, Au | Inhibit | [97,118] | ||
| SOC1 | GmFUL | Glyma.06G205800 1 | FUL, AGL8 | At, Ph, Ag, Su, Ve | AGAMOUS-like 8 | [119] | ||
| AP1 | GmAP1a | Glyma.16G091300 1 | FUL, AGL8; AP1 | At, Ag, Ph, Su, Fd, Ve | MADS-box domain, an intervening domain, a keratin-like domain, and a C-terminal domain proteins | Transcriptional factor | Promote | [120] |
| GmAP1b | Glyma.08G269800 1 | FUL, AGL8; AP1 | At, Ag, Ph, Su, Fd, Ve | |||||
| GmAP1c | Glyma.01G064200 1 | AP1 | At, Ag, Ph, Su, Fd, Ve | |||||
| GmAP1d | Glyma.02G121600 1 | AP1 | At, Ag, Ph, Su, Fd, Ve |
1 Gene IDs highlighted represent the genes identified through genome-wide identification and analysis. Ph: photoperiod, circadian clock, and light signaling pathway. Au: autonomous pathway; Ho: hormone signaling and metabolism. Ve: vernalization. Ag: aging pathway. Su: sugar signaling. Fd: flower development/apical meristem response pathway. At: ambient temperature pathway.
Current studies primarily focus on the photoperiodic flowering pathway in soybeans. Research on other flowering regulatory pathways—including aging, hormone, autonomous, and ambient temperature pathways—remains relatively limited. To date, only a small number of genes have been associated with these pathways. Furthermore, the molecular mechanisms underlying their functions and interactions are poorly understood. For example, hormones such as GAs and auxins are known to regulate flowering in A. thaliana and many other plant species [38,42,45]. However, their specific roles and regulatory networks in soybean flowering have not been fully elucidated. Similarly, the autonomous pathway, which integrates intrinsic developmental cues, has been insufficiently studied in soybean.
Deciphering the intricate regulatory mechanisms of these understudied pathways is essential. It not only facilitates a comprehensive understanding of soybean flowering but also holds significant potential for advancing breeding strategies. By identifying and characterizing key genes within these pathways, researchers can develop innovative approaches to modulate flowering time. This, in turn, will enable the development of soybean cultivars with enhanced adaptability to fluctuating environmental conditions and diverse agricultural demands.
Against this background, the present study was designed to perform a genome-wide identification and comprehensive analysis of flowering-related genes in soybean. Three specific objectives were established. First, to identify all potential flowering-associated genes in the soybean genome through homology searches using A. thaliana flowering gene sequences as references. Second, to classify these genes based on their functional similarities to A. thaliana genes and to analyze their evolutionary relationships, gene structures, and expression profiles. Third, to elucidate the roles of these genes in the flowering regulatory pathway through functional validation experiments. Achieving these objectives will not only expand our fundamental understanding of soybean development, but also provide valuable genetic resources and theoretical support. These resources can be used to develop molecular breeding strategies aimed at optimizing flowering time and improving soybean yields. In doing so, the study will contribute to global food security and promote agricultural sustainability.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Plant materials were cultivated in a growth chamber (model AR-R1060L2F, Xunneng Instruments (Beijing) Co., Ltd., Beijing, China) under short-day conditions, following standard cultivation practices. The chamber’s light spectrum was adjusted by optimizing the proportions of red and blue light to meet the growth and developmental requirements of soybean. Two photosynthetic photon flux density (PPFD) treatments were applied: high PPFD to simulate normal light conditions and low PPFD to mimic shaded environments under dense planting. To analyze differences in hormone content and gene expression levels between varieties with distinct plant heights, Zhexian 12 (ZX12) and Zhexian 19 (ZX19) were grown under high and low PPFD, respectively. The conventional soybean cultivar Williams 82 (W82) was cultivated under high PPFD to investigate the rhythmic responses of hormones and gene expression.
2.2. Data Retrieval and Acquisition of Genomic and Expression Data
Genome annotation details for A. thaliana were obtained from the TAIR10 database (http://www.arabidopsis.org/index.jsp, accessed on 12 December 2024) [121]. For soybean, relevant datasets—including genomic sequences, coding sequences, and protein sequences—were retrieved from SoyBase (http://www.soybase.org, accessed on 2 January 2025). Soybean gene expression data were obtained from the “Expression—RNA SEQ Atlas” (RNA-seq Expression Atlas) database on the SoyBase website, with the data type specified as “Raw” (raw data). This transcriptomic dataset includes information from 14 tissues (young leaves, flowers, pod shells at 10 and 14 days after flowering (DAF), seeds at 10, 14, 21, 25, 28, 35, and 42 DAF, as well as roots and nodules), and its associated tables, figures, supplementary materials, and raw data are all available for download on the SoyBase website (http://www.soybase.org, accessed on 13 February 2025) [122].
2.3. Homology Identification
Homology identification was performed using an integrated strategy that combined synteny-dependent and similarity-dependent methods. For the similarity-dependent analysis, flowering-related genes from A. thaliana were used as query sequences, and the BLASTP method was employed to search against soybean proteins with the following parameters: e-value less than 1 × 10−20, sequence identity exceeding 60%, coverage greater than 75%, and alignment length greater than 70 amino acids [54].
2.4. Chromosomal Distribution
The distribution pattern of flowering-associated genes was determined based on their positions on chromosomes or scaffolds. Detailed chromosomal distribution data for the predicted soybean flowering-related genes were obtained from soybean genomic datasets. To map these putative flowering-associated genetic elements onto pseudomolecular chromosomes, a localization map was generated using MapChart software (MapChart 2.32; developed by Dr. ir. R.E. (Roeland) Voorrips at Wageningen University, Wageningen, The Netherlands [54].
2.5. Multiple Sequence Alignment, Phylogenetic Analysis, Gene Structure Visualization, and Conserved Motif Identification of Flowering-Associated Genes
Protein sequences of the target genes were aligned using ClustalW 1.81, a tool currently maintained by Des Higgins, Fabian Sievers, David Dineen, and Andreas Wilm at the Conway Institute, University College Dublin (Dublin, Ireland). A phylogenetic tree was constructed with MEGA 6.0 software using the maximum likelihood (ML) method with 1000 bootstrap replicates, based on the full-length protein sequences [54]. The Gene Structure Display Server 2.0 (GSDS 2.0; https://gsds.gao-lab.org/Gsds_help.php, accessed on 22 March 2025) was used to map the structure of flowering-associated genetic elements and illustrate the exon-intron organization in soybean [54]. Conserved domains within the protein sequences of key soybean flowering-associated genetic elements were identified using Multiple Em for Motif Elicitation (MEME) Version 4.11.4 (http://meme-suite.org/tools/meme, accessed on 17 February 2025). All parameters were set to default values, except for the maximum number of motifs, which was adjusted to 20 and 15.
2.6. RNA Isolation, cDNA Synthesis, and Quantitative PCR (qPCR)
Total RNA was isolated from fresh samples (≥2 g) that had been frozen in liquid nitrogen and ground into powder using the FastPure Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, China). For first-strand cDNA synthesis, 1 μg of RNA was used with the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme). Quantitative real-time PCR was performed on a QuantStudio 5 instrument (Thermo Fisher Scientific, Waltham, MA, USA) using ChamQ Universal SYBR qPCR Master Mix (Vazyme). The 10 μL reaction mixture consisted of 5 μL SYBR Mix, 0.2 μL of each primer, 1 μL cDNA, and 3.6 μL nuclease-free water. The cycling protocol was as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s, with melting curve analysis. GmTublin (Glyma.05G157300) was used as the reference gene [120], and relative expression levels were determined using the 2−ΔΔCt method, with three biological and three technical replicates. Details of the primers are provided in Supplementary Table S1.
2.7. Determination of Plant Hormones
Plant hormones were quantified using an HPLC-MS/MS system (QTRAP 5500, AB Sciex) with three biological replicates per hormone. Standards included IAA, GA1, GA4, ABA, JA, SA, and their corresponding deuterated internal standards (D-IAA, D-SA, D-ABA). Stock solutions (500 μg/mL) and mixed working solutions (5 μg/mL for standards and 0.1 μg/mL for internal standards) were prepared in methanol. Standard curves ranging from 0.1 to 200 ng/mL, each containing 20 ng/mL internal standards, were generated. For sample extraction, approximately 0.2 g of homogenized sample was mixed with 2.0 mL of acetonitrile and 30 μL of internal standard, then extracted overnight at 4 °C. The mixture was centrifuged (7000 rpm, 5 min) and re-extracted with an additional 2.0 mL of acetonitrile. The combined supernatants were purified using 200 mg of C18 sorbent, centrifuged, concentrated to dryness, re-dissolved in 150 μL of methanol, filtered through a 0.22 μm membrane, and analyzed. HPLC conditions: Waters XSelect® HSS T3 column (Waters Corporation, USA) (2.1 × 150 mm, 2.5 μm), column temperature 30 °C, injection volume 5 μL, flow rate 0.35 mL/min; mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (acetonitrile) with gradient elution. MS conditions: Multiple reaction monitoring (MRM) mode; curtain gas at 35 psi; spray voltage ± 4500 V; atomizing and auxiliary gases at 60 psi; source temperature 500 °C. MRM transitions were as follows: GA1 (negative ion mode)—347.2 → 259.2 *; GA4 (negative ion mode)—331.4 → 243.2 *; IAA (positive ion mode)—176.2 → 129.8 *; JA (negative ion mode)—209.2 → 59.1 *; SA (negative ion mode)—137 → 65 *; ABA (negative ion mode)—263.1 → 153.1 *.
2.8. Protein Structure Prediction Using ESM-Fold
To obtain the three-dimensional (3D) structures of flowering-related proteins, their amino acid sequences were submitted to the Biomedical Digital Intelligent Computing Platform WeMol (https://wemol.wecomput.com/ui/#/, accessed on 28 March 2025), utilizing the Protein Structure Prediction module (ESMFold) for structure prediction.
2.9. Calculation of Ka/Ks Ratios for Genes Associate with Flowering
For the calculation of nonsynonymous-to-synonymous substitution ratios (Ka/Ks), full-length amino acid sequences of flowering-related genes from soybean and A. thaliana were first subjected to pairwise alignment using Multiple Sequence Comparison by Log-Expectation MUSCLE 3.8.31 software [54]. Subsequently, the aligned amino acid sequences were converted into their corresponding coding nucleotide sequences using Perl scripts adapted from ParaAT_2.0 software. These coding nucleotide sequences were then used as input files to calculate Ka/Ks values with the Phylogenetic Analysis by Maximum Likelihood (PAML) program (PAML 4.9; http://www.bork.embl.de/pal2nal/index.cgi, accessed on 8 April 2025) under preset parameters [54]. All variant loci within the aligned pairs were included in the Ka/Ks computation [54].
2.10. Vector Construction and Genetic Transformation
The CRISPR-Cas9 system was employed for the targeted editing of gene GmSAUR46b, following the soybean transformation protocol from Boyuan Biotechnology and previous studies [123,124]. Specific sgRNAs were designed based on the GmSAUR46b sequence and cloned into the BsaI site of the PEG401 vector to generate the CRISPR-Cas9 construct (Figure S1). This construct was subsequently introduced into Agrobacterium tumefaciens strain EHA105 via electroporation.
The method for genetic transformation in soybeans is as follows: surface-sterilized seeds were aseptically germinated, and contamination-free cotyledonary node explants were obtained after 3 to 5 days. The explants were immersed in a recombinant Agrobacterium tumefaciens EHA105 inoculum (OD600 = 0.5) for 2 to 3 min, with occasional agitation. They were transferred to filter paper-lined co-cultivation plates and incubated at 25 °C in the dark for 3 to 5 days. Healthy, contamination-free explants were selected; their hypocotyl tips were excised and plated on solid callus dedifferentiation medium for 7 to 10 days under a 16 hours (h) light/8 h dark photoperiod, with subculturing on fresh medium. The formed embryoids were transferred to redifferentiation medium for 21 days of selection culture under the same photoperiod, with subculturing, then vigorous embryoids were moved to the elongation medium for another 21 days of selection under the same conditions. When shoots reached approximately 5 cm, they were transferred to the rooting medium and cultured for 21 days (16 h light/8 h dark) to promote root and shoot meristem initiation and differentiation. The transgenic plants were then analyzed using the Hi-TOM platform to assess mutation occurrence [125]. The sgRNA sequences and Hi-TOM primers used in this study are provided in Tables S2 and S3, respectively.
3. Results
3.1. Identification and Classification of Genes Related to Flowering in Soybean
A total of 1166 flowering-associated genes were identified in the soybean genome through BlastP searches, using protein sequences from 306 A. thaliana flowering-associated genes as queries (Table S4). Evolutionary analyses of these genes and their inclusion in the FLOR-ID database (http://www.flor-id.org; accessed on 3 November 2021) have been previously reported [121].
To elucidate the flowering mechanism in cultivated soybean, putative flowering-associated genes were categorized into eight gene clusters based on the A. thaliana gene classification system [54]. These clusters correspond to the following pathways: photoperiod (Ph); flower development/apical meristem response (Fd); hormone (Ho); vernalization (Ve); ambient temperature (At); aging (Ag); autonomous (Au), and sugar (Su) pathways (Figure 1). During soybean evolution, 147 flowering genes were lost (Table S5). Notably, most of these lost genes (75, accounting for 51.02%) belonged to the photoperiod pathway (Ph) (Table S6). In contrast, gene sets from three pathways were preferentially retained, as follows: flower development/apical meristem response (Fd, 11 genes lost, 7.48%), sugar (Su, 18 genes lost, 12.24%), and ambient temperature (At, 18 genes lost, 12.24%). However, over 20% of genes were lost in three other pathways, as follows: autonomous (Au, 40 genes lost, 27.21%), vernalization (Ve, 37 genes lost, 25.17%), and hormone (Ho, 30 genes lost, 20.41%) pathways (Table S6).
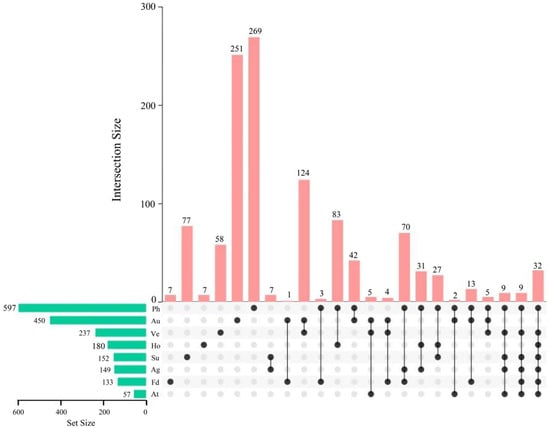
Figure 1.
Identification and classification of flowering-associated genes in Glycine max (L.) Merr. The left histogram displays the number of flowering-associated genes within each flowering regulatory pathway. In the central matrix, individual black dots represent the number of genes unique to a specific pathway, while lines connecting these dots indicate overlaps between different pathways. The vertical red histogram corresponds to the number of genes present in these intersections. Abbreviations: Ph, photoperiod, circadian clock, and light signaling pathway; Au, autonomous pathway; Ho, hormone signaling and metabolism; Ve, vernalization; Ag, aging pathway; Su, sugar signaling; Fd, flower development/apical meristem response pathway; At, ambient temperature pathway.
The photoperiod pathway (Ph) contained the largest number of genes (597), representing approximately 51.20% of the analyzed genes. It also had the highest number of unique genes (269) (Figure 1 and Figure S2).
The autonomous pathway (Au) comprises 450 genes, accounting for approximately 38.59% of the total (Figure S2). These genes regulate flowering independently of environmental cues [48,52,53]. They primarily participate in processes such as chromatin modification and RNA processing, which influence the expression of flowering-associated genes at both transcriptional and post-transcriptional levels. A total of 42 genes are uniquely shared between the photoperiod (Ph) and autonomous (Au) pathways. Additionally, the photoperiod (Ph) and flower development/apical meristem response (Fd) pathways share three common genes: GmLFY, GmFT, and GmSOC1. The Fd pathway contains seven unique genes and shares one gene with the Au pathway (Figure 1).
The hormone pathway (Ho) comprises 180 genes, representing approximately 15.44% of the total (Figure S2). It contains only 7 unique genes but shares 83 genes with the photoperiod pathway (Ph) (Figure 1). These shared genes are associated with GA biosynthesis and include those encoding enzymes involved in the GA biosynthesis pathway, such as GmGA20ox1 and GmGA3ox1, as well as genes related to signal transduction.
Moreover, this study conducted a comparative analysis between previously mapped flowering-related genes and the genome-wide identified flowering-related genes reported here (Table 1). The previously mapped genes include, for example, E1 [70,71]; E2 (GI) [73,83,84]; E3-5 (PHYA) [74,76,77,84,111]; E6 [112]; E7 [114]; E8 [113]; E9 (FT2a) [94,115]; E10 (FT4) [88,89]; E11 [116]; J (ELF3) [5,117]; FT5a [92,93,94]; FT1a/b [90,91]; FT2b [93,105]; FT3a/b, FT5b, FT6 [93]; Tof5 (FUL2a) [80]; Tof11/12 (PRR3a/b) [78,79]; Tof16 (LHY1a) [81]; GmAP1 [120]; and GmDT1 (TFL1b) [97,118].
3.2. Chromosomal Location of Flowering-Related Genes
Chromosomal location analysis revealed that flowering-related genes are distributed across all 20 soybean chromosomes (Figure 2, Table S7). However, their distribution density is uneven (Table S8).

Figure 2.
Chromosomal distribution of genes associated with flowering in soybean. The chromosomal locations of flowering-related genes were visualized, with each chromosome represented by a vertical bar labeled Gm01 to Gm20. Genes are marked on their respective chromosomes, where green indicates genes located in the upper regions and yellow indicates genes in the lower regions. Gene names are displayed alongside their positions on the chromosomes.
Chromosomes 6, 8, 11, and 13 exhibited relatively high densities of flowering-related genes (Figure 2, Table S8). For instance, chromosome 13 contained 36 flowering-related genes within a specific region spanning approximately 15 to 25 Mb. Similarly, chromosome 6 harbored 25 flowering-related genes in a region of about 5 to 50 Mb. Chromosome 8 also formed a gene-rich cluster, with multiple flowering-related genes concentrated between 3 and 40 Mb (Figure 2, Table S7). In contrast, some chromosomes exhibited relatively low densities of flowering-related genes. For instance, chromosome 14 contained only 13 flowering-related genes along its entire length (Figure 2, Table S8).
3.3. Evolutionary, Structural, and Conserved Domain Analyses of Ph Pathway Flowering-Related Genes in Soybean
To comprehensively elucidate the molecular basis of photoperiod-regulated flowering in soybean, we systematically analyzed the evolutionary relationships, expression patterns, gene structures, and conserved domains of genes associated with flowering in the photoperiod pathway.
A phylogenetic analysis was conducted to investigate the evolutionary divergence of these genes. Flowering-associated genes in the soybean photoperiod pathway were grouped into distinct clades (Figure 3A). This phylogenetic pattern suggests that functional specialization has emerged during soybean evolution. However, the core photoperiod-sensing mechanisms have been conserved.

Figure 3.
Evolutionary, structural, and conserved domain analyses of flowering-associated genes involved in the photoperiod, circadian clock, and light signaling pathways (Ph) of Glycine max (L.) Merr. (A) Phylogenetic tree of flowering-associated genes in the Ph pathway. (B) Expression level analysis of flowering-associated genes in the Ph pathway. (C) SMART domain analysis of flowering-associated genes in the Ph pathway. The legend below represents different domains. (D) Distribution of conserved motifs in flowering-associated genes of the Ph pathway.
Conserved domain analysis (Figure 3C,D and Figure S3) identified key functional motifs essential for photoperiod signaling. A high percentage of photoperiod-associated proteins contained F-box domains, which are characteristic of LOV KELCH PROTEIN2, LKP2-like proteins and critical for mediating protein–protein interactions within the circadian clock complex. The PAS domain, a hallmark of phytochrome family proteins, was detected in all homologs of GmPHYA, GmPHYB, and GmPHYE (Figure 3C). This domain is crucial for mediating light-induced transcriptional regulation, enabling these proteins to efficiently sense and transmit red/far-red light signals. In doing so, they coordinate downstream gene expression cascades that are essential for regulating photomorphogenesis and flowering in soybean. Moreover, MADS domains—characteristic of APETALA1 (AP1)-like floral integrators—were detected in downstream photoperiod effectors (Figure 3C). These domains were frequently associated with disordered regions, indicating regulatory plasticity in protein complex assembly and underscoring their role in bridging light signaling and floral meristem identity.
3.4. Expression Analysis of PH Pathway Genes in Soybean
To investigate the expression divergence of homologous genes and their functional roles in the photoperiod pathway, we systematically analyzed the expression patterns of candidate flowering-associated genes across various soybean tissues. These tissues included young leaves, flowers, pod shells at 10 and 14 days after flowering (DAF), seeds at 10, 14, 21, 25, 28, 35, and 42 DAF, as well as roots and nodules (Figure 3B, Table S9). Genes within the same phylogenetic clade often exhibited coordinated expression across tissues. A subset of genes (GmLFY, GmFKF1, GmCRY1, GmNF-YC9, GmAP1, GmSOC1, GmAHL22, GmJMJ330s, GmCPK33, GmPHYB, GmPFT1, GmMSI1, GmTPL, and GmNF-YB3) were preferentially upregulated in young leaves under inductive short-day conditions, consistent with their roles in initial photoperiod perception. In contrast, other gene clusters (GmLKP2, GmAS1, GmFKF1, GmJMJ330s, GmFTIP1, GmNF-YC9, GmSTO, GmCKB3, GmAP1, GmSVP, GmCPK33, GmPHYA, GmPHYB, GmGA2ox1, GmHDA5, GmGI, GmGA3OX1, GmMSI1, GmTPL, GmTFL1, GmLWD1, GmAFR1, and GmNF-YB3) displayed peak expression in flowers or developing pods (Figure 3B), suggesting their roles in floral transition and reproductive development. Notably, certain gene subsets were preferentially upregulated in young leaves under inductive short-day conditions and also maintained prominent expression in flowers and pods at various developmental stages (Figure 3B). For example, one gene cluster showed peak expression in young leaves during the photoperiod-sensing phase while maintaining high transcript abundance in flowers and maturing pods (Figure 3B). This dual-tissue expression profile indicates multifunctional roles, suggesting these genes may link initial photoperiod perception in leaves to subsequent floral transition and reproductive development in flowers and pods. In comparison, other gene clusters exhibited more tissue-specific peak expression; for instance, some were exclusively enriched in flowers or developing pods, implying specialized functions in floral organ formation or pod ripening.
3.5. Evolutionary, Expression, Structural, and Conserved Domain Analyses of HO, AT, and FD Pathways in Flowering-Related Genes of Soybean
A considerable number of flowering-associated genes are shared among the hormone, ambient temperature, flower development/apical meristem response, and photoperiod pathways. Some genes participate in multiple flowering pathways simultaneously. Notably, the hormone and photoperiod pathways share the greatest number of flowering-related genes (Figure 1 and Figure S2). We analyzed genes in three pathways separately: hormone (Figures S4 and S5), flower development/apical meristem response (Figures S6 and S7), and ambient temperature (Figures S8 and S9). For each pathway, we examined gene evolution, expression patterns (Tables S10–S12), gene structures, and conserved functional domains. Phylogenetic analysis grouped soybean flowering-related genes into distinct clades (Figures S4A, S6A and S8A). Core regulators, including GmFT, GmLFY, GmSVP, and GmSOC1, were conserved across the photoperiod, ambient temperature, and flower development/apical meristem response pathways (Figure 3, Figures S6 and S8). This conservation reflects the retention of core flowering regulatory mechanisms that underpin functional crosstalk between pathways. Protein GmSOC1 integrates signals from four pathways—photoperiod, ambient temperature, hormone, and flower development/apical meristem response—and thereby regulates flowering. Additionally, genes GmGA20ox1, GmGA3OX1, GmGA2ox1, GmGA2ox2, and GmGI are specifically shared between the photoperiod and hormone pathways; they were not detected in the flower development/apical meristem response or ambient temperature pathways (Figure 3, Figures S4, S6 and S8).
Expression heatmaps revealed tissue- and stage-specific expression dynamics. Shared genes exhibited coordinated expression patterns. For example, GmSOC1 showed peak expression in two key sites: young leaves (where photoperiod and ambient temperature are sensed) and floral meristems (where flower development occurs). GmLFY was enriched in young leaves (Figure 3B, Figures S4B, S6B and S8B). GmFT was enriched in floral buds and developing pods (Figure 3, Figures S6 and S8). GmGI was preferentially expressed in young leaves and developing pods (Figure 3B and Figure S4B). Genes specific to individual pathways exhibited tissue-restricted expression. In contrast, shared genes functioned as “hubs” for signal integration [17,91,107,126,127]. Protein structure predictions and motif analyses (Figures S4C,D, S6C,D and S8C,D) revealed functional domains shared across pathways. For instance, MADS-box domains in GmAP1 and GmSOC1 mediated DNA binding and transcriptional regulation. These processes are involved in photoperiodic signal transduction, floral meristem identity specification, and temperature-responsive flowering.
3.6. Protein Structure Prediction of Flowering-Associated Genes in the Ph Pathway
To investigate the functional mechanisms of photoperiod pathway-related flowering genes in soybean, we analyzed the protein structures of key genes involved in this pathway. As shown in Figure 4, these photoperiod pathway genes display a diverse array of three-dimensional protein structures.
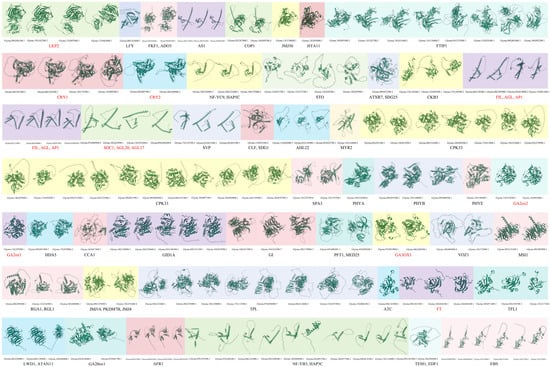
Figure 4.
Protein structure prediction of flowering-related genes involved in the photoperiod, circadian clock, and light signaling (Ph) pathways. Different shaded colors represent different genes.
For instance, proteins involved in light perception—such as LKP2 and CRY1/2—exhibit unique structural folds. The LKP2 protein features a compact, multi-domain architecture, which likely supports its role in mediating protein–protein interactions within the circadian clock machinery. CRY1 and CRY2 proteins possess characteristic β-barrel structures in their photolyase-homology regions (Figure 4). These structures are essential for blue light absorption and play a key role in initiating light signaling.
Transcription factors in the photoperiod pathway include CO family proteins and AP1-like proteins containing MADS domains. These proteins exhibit structural features characteristic of their functional classes. AP1-like proteins possess a MADS-box domain, which forms a conserved DNA-binding motif. This motif consists of a helix–loop–helix structure (Figure 4) that facilitates specific binding to target gene promoters. In this way, it regulates floral meristem identity.
Enzymes involved in hormone biosynthesis and signaling within the photoperiod pathway include GA20ox and GA3OX, which are GA-related enzymes (Figure 4). These enzymes feature catalytic domains with active-site architectures consistent with their roles in GA metabolism. The catalytic domains contain conserved amino acid residues that are likely involved in substrate binding and enzymatic reactions. These functions are essential for promoting flowering under appropriate photoperiodic conditions.
3.7. Selection Pressure on Flowering Pathway Gene Sets
As a major legume crop, soybean exhibits well-documented variations in flowering time among domesticated germplasm. This trait is essential for modern agricultural production; however, its significance during domestication remains unclear [8].
Selection pressure was analyzed using the Ka/Ks ratio, a metric that reflects evolutionary selection patterns in genomes [128]. Calculations of the Ka/Ks ratio for orthologous pairs of flowering-related genes between the G. max and A. thaliana genomes revealed that all flowering-associated genes had a Ka/Ks ratio below 1 (Figure 5, Table S13). This finding indicates that these genes are under purifying selection.
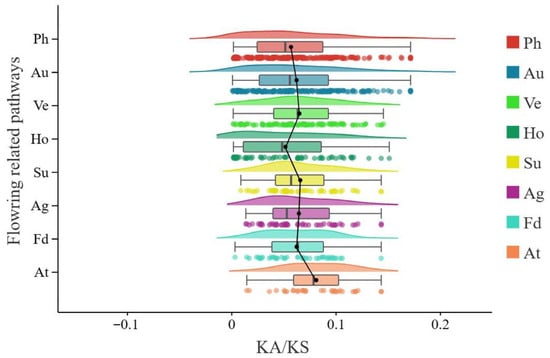
Figure 5.
Direction and magnitude of natural selection acting on distinct sets of flowering-associated genes. Ka/Ks ratios of flowering-associated genes in Glycine max (L.) Merr. Abbreviations: Ph, photoperiod, circadian clock, and light signaling pathway; Au, autonomous pathway; Ho, hormone signaling and metabolism; Ve, vernalization; Ag, aging pathway; Su, sugar signaling; Fd, flower development/apical meristem response pathway; At, ambient temperature pathway.
Selection pressure analysis revealed that the average Ka/Ks ratios of flowering-associated gene sets between A. thaliana and soybean ranged from 0.001 to 0.200 (Figure 5, Table S13). Notably, genes involved in the photoperiod and hormone pathways experienced weaker negative selection pressure compared to those in other pathways (Figure 5). Additionally, the selection pressure on genes in the photoperiod and autonomous pathways exhibited a broader distribution range than that observed in other flowering pathways (Figure 5). This suggests that a substantial number of genes in these two pathways must maintain functional conservation to ensure the proper execution of soybean’s fundamental physiological processes. In contrast, genes in the ambient temperature pathway experienced the strongest selection pressure, indicating that flowering-related genes in this pathway are subject to relatively intense selective constraints (Figure 5). These genes have undergone strong selective pressure during evolution, with many experiencing selection followed by evolutionary divergence. This implies that genes in these pathways retain conserved functions throughout evolution. Furthermore, among all flowering-related pathway genes, those in the photoperiod pathway exhibited the greatest variation in selection pressure during soybean evolution (Figure 5, Table S13).
3.8. Expression Patterns of Flowering Pathway Genes in Various Tissues and Their Relationships with Plant Height and Hormone Levels
ZX12 (dwarf, early-maturing) and ZX19 (tall, late-maturing) were used to investigate gene responses in different flowering pathways related to plant height. Under low-PPFD conditions (Figure 6A), ZX19 exhibited significantly greater plant height and internode length than ZX12 (Figure S10A,B). In contrast, ZX12 had a thicker stem diameter compared to ZX19 (Figure S10C). These morphological differences suggest that internal genetic and hormonal factors may play regulatory roles in determining plant height.
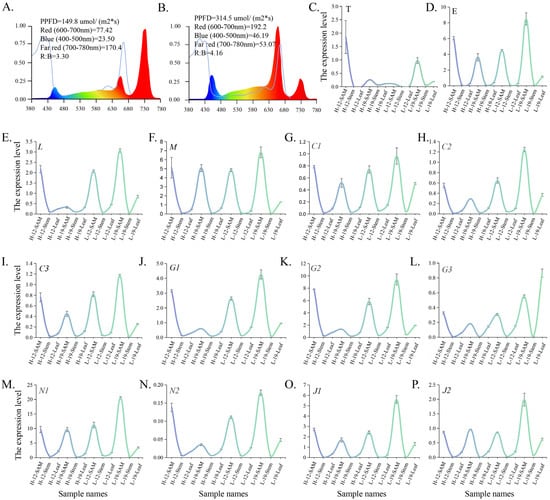
Figure 6.
Analysis of the expression levels of flowering-related genes involved in different flowering pathways in young soybean leaves. (A,B) Spectra of low PPFD (A) and high PPFD (B) under short-day (SD; 8 h light/16 h dark) conditions. (C–P) Young leaves were harvested for qRT-PCR analysis of the expression levels of T (C), E (D), L (E), M (F), C1 (G), C2 (H), C3 (I), G1 (J), G2 (K), G3 (L), N1 (M), N2 (N), J1 (O), and J2 (P) in soybean cultivars ZX12 and ZX19 grown under SD conditions with high and low PPFD. Relative expression levels were normalized to GmTublin (Glyma.05G157300). Data are presented as means ± SD of three replicates. Gene abbreviations: T, GmTPL; E, GmEBS; L, GmLWD1; M, GmMSI1; C1, GmCPK33a; C2, GmCPK33b; C3, GmCPK33c; G1, GmGA20ox1; G2, GA3OX1; G3, GA2ox1; N1, GmNF-YC9a; N2, GmNF-YC9b; J1, GmJMJ330a; J2, GmJMJ330b. Abbreviations: PPFD, photosynthetic photon flux density; H, high PPFD; L, low PPFD; SD, short-day; 12, Zhexian 12; 19, Zhexian 19; SAM, shoot apical meristem.
The expression patterns of genes in ZX12 and ZX19 were analyzed in stems, leaves, and shoot apical meristems (SAMs) under both high- (Figure 6B) and low-PPFD conditions (Figure 6A). The results show that genes associated with various flowering pathways were preferentially expressed in SAMs, with the lowest expression levels observed in stems. GmTPL, which participates in the Ag, Ph, and Fd flowering pathways, reached peak expression in ZX12’s SAMs under high PPFD and in ZX19’s SAMs under low PPFD (Figure 6C). GmEBS, GmLWD1, and GmCPK33, all specific to the Ph pathway, exhibited peak expression in the SAMs of both ZX12 and ZX19 under both low- and high-PPFD conditions. A similar pattern was observed for GmMSI1, which functions in the photoperiod, flower development/apical meristem response, and autonomous pathways (Figure 6D–I). Genes GmGA20ox1 and GmGA3OX1, involved in both photoperiod and hormone pathways, showed the highest expression in ZX19’s SAMs under low PPFD. In contrast, GmGA2ox1 was most highly expressed in ZX19’s leaves under low PPFD (Figure 6J–L). Additionally, GmNF-YC9, involved in both photoperiod and hormone pathways, and GmJMJ330, present in the photoperiod, autonomous, and ambient temperature pathways, reached their highest expression levels in ZX19’s SAMs under low PPFD (Figure 6M–P). The SAM is a critical site for plant growth and development, involved in processes such as flowering initiation and the regulation of plant architecture. This preferential expression in SAMs suggests that the functional activity of these flowering-related genes is concentrated in this tissue.
The determination of hormone content in the leaves of ZX12 and ZX19 revealed that ZX19 had significantly higher levels of IAA, JA, and ABA than ZX12. In contrast, ZX12 contained significantly greater amounts of GA-1 and SA than ZX19. Additionally, ZX12 exhibited significantly higher GA-4 levels compared to ZX19 (Figure 7). These hormonal differences strongly suggest that hormones play a crucial role in regulating soybean plant height. ZX19, which is tall-stemmed, and has elevated levels of IAA, JA, and ABA, whereas ZX12, which is short-stemmed, has higher concentrations of GA-1, GA-4, and SA (Figure 7).
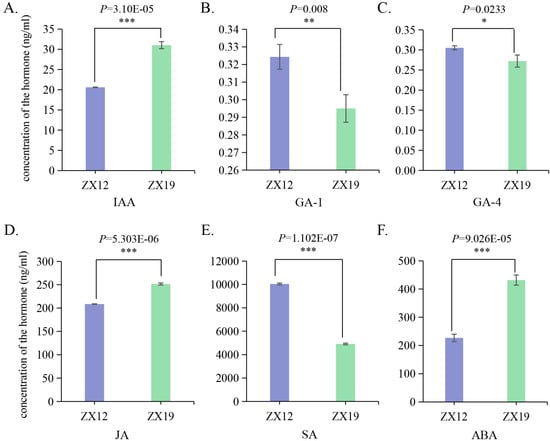
Figure 7.
The levels of hormones IAA (A), GA-1 (B), GA-4 (C), JA (D), SA (E), and ABA (F) were measured in young leaves of soybean cultivars ZX12 and ZX19 under short day conditions with high photosynthetic photon flux density (PPFD). Data are presented as means ± standard deviations of three replicates (*** p ≤ 0.001, ** 0.001 < p ≤ 0.01; * 0.01 < p ≤ 0.05). ZX12, Zhexian 12; ZX19, Zhexian 19.
3.9. Analysis of Expression Patterns and Hormonal Content Responses to Rhythms and Light Signals
To investigate whether genes involved in different flowering pathways and leaf hormones respond to light signals, we analyzed two aspects. First, we examined the diurnal rhythmic expression patterns of flowering pathway genes under dark treatment. Second, we monitored dynamic changes in leaf hormone content in response to photoperiod and circadian rhythms.
As shown in Figure 8A, genes from various flowering pathways (GmTPL, GmMSI1, GmEBS, GmLWD1, GmCPK33a/b/c, GmGA20ox1, GA3OX1, GA2ox1, GmNF-YC9a/b) exhibited coordinated responses to dark-induced rhythmicity. Their expression slightly increased after 4 h of dark treatment, peaked at 28 h, declined to the lowest level at 36 h, and then showed a minor upward trend. In contrast, GmJMJ330a/b peaked at 32 and 36 h, respectively, with elevated expression maintained for 4 to 6 h. This rhythmic expression pattern indicates that genes across different flowering pathways are sensitive to light-related rhythmic signals and suggests their involvement in integrating photoperiodic information.
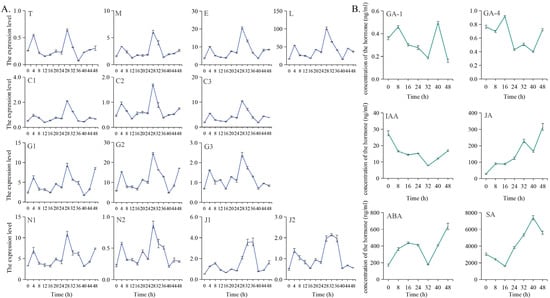
Figure 8.
Analysis of the expression levels of flowering-related genes in different flowering pathways and the contents of various hormones in young soybean leaves. (A) Young leaves were harvested for qRT-PCR analysis to measure the expression levels of T, E, L, M, C1, C2, C3, G1, G2, G3, N1, N2, J1, and J2 at multiple time points (0 h, 4 h, 8 h, 12 h, 16 h, 20 h, 24 h, 28 h, 32 h, 36 h, 40 h, 44 h, and 48 h) following the dark treatment of soybean cultivar W82 under short-day (SD) conditions with high photosynthetic photon flux density (PPFD). (B) Hormone contents of IAA, GA-1, GA-4, JA, SA, and ABA were measured in young leaves at various time points (0 h, 8 h, 16 h, 24 h, 32 h, 40 h, and 48 h) after the dark treatment of soybean cultivar W82 under SD conditions with high PPFD. Relative gene expression levels were normalized to GmTublin. Data are presented as means ± SD of three replicates. Abbreviations: T, GmTPL; E, GmEBS; L, GmLWD1; M, GmMSI1; C1, GmCPK33a; C2, GmCPK33b; C3, GmCPK33c; G1, GmGA20ox1; G2, GA3OX1; G3, GA2ox1; N1, GmNF-YC9a; N2, GmNF-YC9b; J1, GmJMJ330a; J2, GmJMJ330b; W82, Williams 82; h, hour.
Figure 8B illustrates that leaf hormone levels also responded to photoperiod and rhythmic changes. GA-1 increased at 8 h, then decreased, followed by a substantial rise to a maximum at 40 h—exhibiting a 4 h delay compared to the peak of gene expression. GA-4 peaked at 16 h, then declined with fluctuations, reflecting its response to rhythmic variations. IAA showed a downward trend after dark treatment but gradually increased at 32 h, with its peak also delayed by 4 h relative to the gene expression peak. JA content rose rapidly following dark treatment. ABA exhibited a parabolic pattern: it increased to a peak at 16 h, declined to its lowest point at 32 h, then rose rapidly again. SA responded to rhythmic signals by gradually decreasing during the first 16 h, reaching its lowest level at 16 h—as opposed to ABA, which peaked at that time. After 16 h, SA content increased rapidly, reaching a maximum at 40 h before declining.
3.10. GmSAUR46b Not Only Regulates the Plant Height of Soybeans but Also Controls Flowering
To further validate the correlation between plant height and flowering time, we generated transgenic soybean lines carrying targeted mutations in GmSAUR46b using the CRISPR-Cas9 system (Figure 9A,B). Phenotypic analysis of homozygous mutant lines revealed pleiotropic effects (Figure 9C,D).
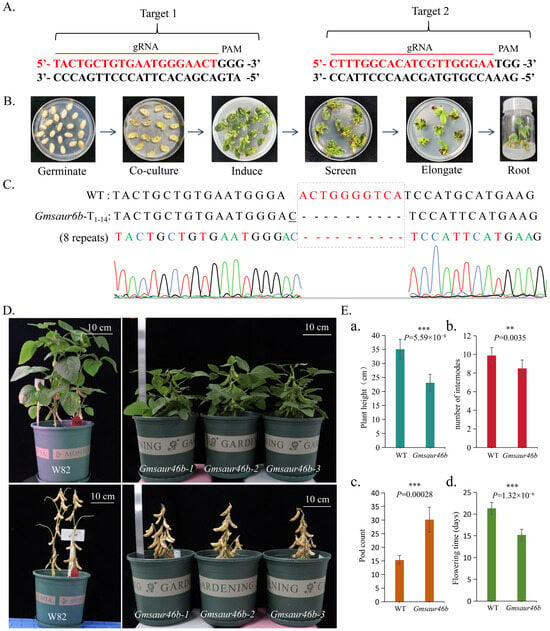
Figure 9.
GmSAUR46b not only regulates soybean plant height but also controls flowering. (A) The red bar indicates the sequence of the single guide RNA (sgRNA). (B) The process of CRISPR/Cas9 genetic transformation in soybeans. (C) Detection of CRISPR/Cas9 editing status in soybean gene-edited materials using Sanger sequencing. Curves of different colors represent different bases. (D,E) Phenotypic analysis (D) and plant height (a), number of internodes (b), pod count (c), and flowering time (d) statistical evaluation (E) of plant height, number of internodes, pod count, and flowering time in the edited Gmsaur46b lines compared to wild-type plants (WT), specifically Williams 82 (W82). The plants were grown under short-day (SD) conditions (8 h light/16 h dark). Scale bar, 10 cm. *** p ≤ 0.001, ** 0.001 < p ≤ 0.01.
Compared to wild-type plants, Gmsaur46b mutant lines exhibited significant reductions in several traits, including reduced plant height, number of internodes, pod count, and flowering time (Figure 9D,E). These findings indicate that GmSAUR46b is a key pleiotropic regulator controlling both soybean flowering and plant height development. Statistical analysis revealed that the average height of mutant plants was 23.10 cm, approximately 34.11% shorter than that of wild-type plants (p < 0.001) (Figure 9E), suggesting that GmSAUR46b plays a critical role in regulating stem elongation. Additionally, mutant plants had significantly fewer internodes, averaging 8.50 per plant compared to wild-type plants (p < 0.001) (Figure 9E). This further confirms its role in internode development. Furthermore, flowering time was significantly advanced in Gmsaur46b mutants (Figure 9E). Mutant plants initiated flowering at 15.20 days after sowing, whereas control plants flowered at 21.30 days. This indicates that flowering time in mutants was shortened by 28.64% compared with controls (p < 0.001) (Figure 9E).
4. Discussion
4.1. Evolutionary Conservation and Functional Divergence of Flowering Pathways
Our research results show that a substantial proportion of the candidate genes associated with flowering correspond to known mapped genes. Of all the flowering-related genes, the photoperiod pathway exhibited the highest gene retention, consistent with current findings; most mapping and functional analyses of soybean flowering genes focus on this pathway [2,3,5,6,8,68,80,89,91,97,98,127,129]. This pathway is crucial for soybean flowering as soybeans are short-day plants [107], and the photoperiod-related genes play a key role in sensing and responding to changes in day-length. For example, genes encoding photoreceptors such as phytochromes (GmPHYA, GmPHYB) and cryptochromes (GmCRY1, GmCRY2) were identified (Table S4). These photoreceptors can sense the light signals and transmit them to downstream regulatory factors. Notably, some photoperiod pathway genes were lost during soybean evolution, while new flowering-related genes have emerged. Examples include E1, ELF3, FT6, PRR3a/b, LHY1b, and LHY2a/b (Table 1 and Table S4). This diversification is likely due to whole-genome duplication (WGD) events and domestication in soybean [5,13,78]. Additionally, novel mutations have played a role, facilitating adaptation to environmental changes throughout evolution [5,6,12,13,78,80,96,107,130,131,132,133,134]. These processes increased the number of flowering-related homologous genes and generated new functions and regulatory mechanisms. In contrast, the flower development/apical meristem response and ambient temperature pathways showed higher gene retention, suggesting conserved roles in meristem identity and temperature sensing [19,20,21,22,23,97].
Key integrators—such as GmFT, GmLFY, and GmSOC1—are co-retained across the photoperiod, ambient temperature, and flower development/apical meristem response pathways, indicating the preservation of core regulatory modules. For example, GmSOC1 integrates signals from photoperiod, temperature, hormonal cues, and meristem activity, demonstrating how conserved genes function as “hubs” for cross-pathway crosstalk [126]. Protein structure predictions further support this, showing that the MADS-box domains in GmAP1 and GmSOC1 facilitate DNA binding across multiple pathways [135]. These structural features are tightly associated with their respective functions, including light perception, signal transduction, transcriptional regulation, and hormone-related processes. This provides a structural basis for enhancing our understanding of how these genes coordinate to regulate soybean flowering in response to photoperiodic cues.
Chromosomal location analysis revealed that flowering-related genes exhibit relatively high density in certain chromosomes, forming gene-rich clusters. Owing to their close physical proximity, these clustered genes may be co-regulated, potentially sharing common cis-regulatory elements or being influenced by the same trans-acting factors. Such coordinated regulation is likely to play a critical role in the synchronized control of soybean flowering. Furthermore, the uneven chromosomal distribution of these genes may be linked to the evolutionary history of soybeans [4,5]. Gene duplication and translocation events during evolution presumably facilitated the accumulation of flowering-related genes in specific chromosomal regions, whereas other regions remained relatively gene-poor.
Conserved domain analysis identified functional motifs critical for pathway crosstalk. Selection pressure analysis revealed that all pathways are under purifying selection (Ka/Ks < 1). However, the photoperiod and hormone pathways experienced relatively relaxed constraints, suggesting that while core functions are conserved, adaptive evolution in these pathways has enabled soybean to fine-tune flowering in response to diverse environmental conditions [15,68,95]. The ambient temperature pathway exhibited the strongest selection pressure, indicating rapid evolution to adapt to temperature fluctuations [29,95].
4.2. Cross-Pathway Interactions and Molecular Signal Integration in the Regulation of Soybean Flowering
The genetic networks regulating soybean flowering demonstrate significant crosstalk between the photoperiod and hormone regulatory pathways. These two pathways share 83 genes, the highest number among all pathway pairs, underscoring their close interaction. Genes such as GmGA20ox1, GmGA3OX1, and GmGI are present in both pathways (Figure 3 and Figure S4), coordinating GA metabolism with photoperiod signaling. For example, key photoperiodic regulators like E1, GmFT2a, and GmGI intersect with GA metabolism and circadian clock pathways, forming integrated regulatory hubs [6,92,95]. E1 acts as a major repressor in the photoperiod pathway by directly inhibiting the expression of GA biosynthesis genes GmGA20ox1 and GmGA3ox1, thereby linking day-length sensing to GA-driven stem elongation and floral transition [136].
Such interconnections form a complex regulatory network in which environmental cues (e.g., photoperiod, temperature) are integrated to precisely regulate flowering time [13,80,95] (Figure 10). Under short-day conditions, the photoperiodic induction of GmFT2a triggers flowering [98,104]. Elevated temperatures can accelerate soybean growth and development, sometimes leading to earlier flowering by transcriptionally reprogramming flowering-associated genes within the photoperiod pathway [20]. Specifically, high temperatures induce the expression of soybean flowering activator genes, including GmFT2a, GmFT5a, and several GmCOL, while simultaneously suppressing the expression of homologs of the flowering repressor genes E1 and E2. This regulatory pattern ultimately promotes flowering in soybean [20] (Figure 10). Furthermore, under short-day conditions, soybean plants exhibit accelerated flowering at 30 °C but delayed flowering at 35 °C. The upregulation of FT2a and FT5a induced by a condition of 30 °C results in early flowering, indicating that high temperatures influence soybean flowering time, with effects varying across different high-temperature ranges [21]. This finding confirms that high temperatures can influence soybean flowering time and promote early flowering. The complexity of these interactions further underscores the adaptive advantage of integrated regulatory mechanisms. In various eco-geographic regions, genetic variations in cross-pathway genes (e.g., GmFT, GmAP1, and J) have undergone positive selection. This enables soybean to balance photoperiod sensitivity with other adaptive traits [5,80,97,120] (Figure 10). Such functional overlap illustrates how pleiotropic genes serve as nodal points for cross-pathway communication, allowing soybean to synchronize flowering with developmental and environmental cues.
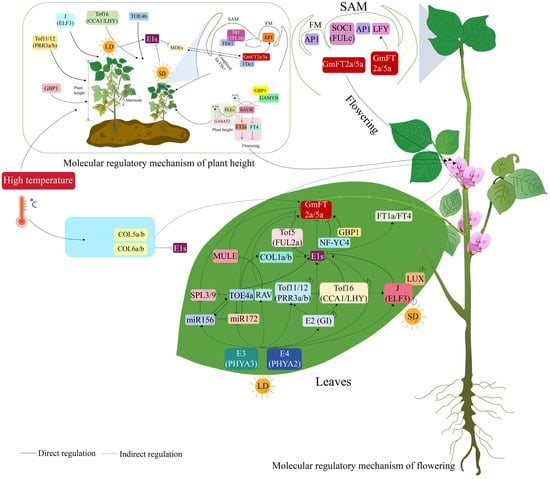
Figure 10.
Molecular mechanisms regulating soybean flowering and plant height, with a molecular model diagram illustrating their connection.
4.3. Breeding Implications of Genome-Wide Identification and Characterization of Flowering-Related Genes for Regulating Flowering Time and Plant Height in Soybean
Our findings collectively reveal a close connection between the flowering regulatory pathways and plant height regulatory network in soybean. This conclusion is supported by three key observations: genes involved in various flowering pathways are influenced by rhythmic and light signals (Figure 8A); leaf hormone levels respond to rhythmic signals (Figure 8B); and GmSAUR46b regulates both plant height and flowering (Figure 9).
Prior studies have highlighted the intricate interactions between soybean flowering-regulatory genes and those controlling plant height, underscoring a key aspect of soybean developmental plasticity. For example, E1 is a core regulatory factor influencing both plant height and flowering [17,137]. Under short-day conditions, the suppression of E1 expression is a pivotal regulatory event that initiates a cascade of molecular interactions governing soybean development. Reduced inhibition by E1 releases GmMDEs (MADS-box transcription factor), which then promote the expression of GmFT2a and GmFT5a in SAM [17]. The upregulation of GmFT2a and GmFT5a in the SAM triggers two interconnected pathways that together modulate both flowering time and plant height. This highlights the integration of these developmental processes [17]. On one hand, GmFT2a and GmFT5a act as key signals to induce GmAP1 expression [17,97,118]. Increased GmAP1 then inhibits GmDt1, suppressing soybean stem elongation [96,97] (Figure 10). On the other hand, GmFT2a and GmFT5a promote the expression of GmAP1, GmSOC1, and GmLFY in the SAM. This promotes soybean flowering [2,3,6,8,12,91,94,97] (Figure 10). Notably, recent studies show that while both GmFT2a and GmFT5a induce flowering, they differ in their control of post-flowering stem growth. GmFT5a is markedly more effective than GmFT2a in terminating post-flowering stem growth by downregulating GmTFL1b and upregulating GmAP1 [97,120,138] (Figure 10). This dual role of GmFT2a and GmFT5a—coordinating floral initiation and height suppression—underscores their central role in mediating photoperiodic responses. This ensures soybean plants align reproductive timing and morphological development adaptively with environmental cues. Moreover, GmTFL1b (the candidate gene for the GmDt1 locus) interacts with GmAP1 in the SAM, regulating soybean stem growth habit [118,120,139] (Figure 10). Based on the observation that genes involved in different flowering pathways are predominantly expressed in the SAM, we hypothesize that the SAM acts as a key tissue for integrating regulatory signals governing plant height and flowering. Moreover, the coordinated interaction between these genes and hormones determines the ultimate plant height and flowering traits of soybean cultivars.
Several other genes regulate both flowering and plant height, thereby influencing yield and environmental adaptability. These genes include J (GmELF3) [5], GmTof11/12 (GmPRR3a/b) [8], GmTof16 (CCA1/LHY) [81,120], and GmTOE4b [2,11]. The MADS-box transcription factor GmFULc participates in regulating flowering time, maturity [119,140,141], and plant height [142]. GmFULc modulates plant height by binding to the promoter of the GA-responsive gene GmGASA. This enhances the expression of GmGASA14 and GmGASA32 [142]. The R2R3-MYB transcription factor GmGAMYB positively regulates both flowering and plant height. It is induced by long-day conditions and GAs [143]. GmGBP1, a soybean transcriptional co-regulator, interacts with GmGAMYB. This interaction enhances the transcription of GmSAUR genes. These GmSAUR genes subsequently upregulate flowering-promoting factors such as GmFT2a and GmFDL19, resulting in early flowering [144]. Additionally, the GmGBP1/GmGAMYB complex contributes to the GA-mediated promotion of soybean plant height and hypocotyl elongation by facilitating GmSAUR expression [145]. Furthermore, GmGAMYB interacts with GmGBP1 and upregulates GmFULc to promote flowering; it also enhances GA sensitivity and upregulates GmGA20ox to increase plant height [143]. This gene may function by integrating environmental signals, such as photoperiod, and endogenous hormonal cues. In doing so, it coordinates the transition from vegetative to reproductive growth while influencing plant height.
The temporal asynchrony between gene expression and hormone dynamics suggests complex crosstalk between genetic regulatory networks and hormonal signaling pathways in response to photoperiodic cues (Figure 8). Such interactions may link flowering pathway genes to the regulation of plant height. The integration of photoperiodic and hormonal signals is crucial for plant growth and development, including flowering and the determination of plant height. These findings provide a foundation for further exploration of the molecular mechanisms by which flowering pathway genes regulate plant height though the integration of light and hormonal signals.
Current research on soybean flowering-related genes has predominantly focused on those involved in the Ph and Fd pathways. Examples include genes J, GmFTs, GmPRR3s, GmLHYs, and GmAP1s [5,92,104,107,120]. However, there is a scarcity of studies investigating the molecular mechanisms regulating flowering through other pathways, such as the hormone, autonomous, sugar, ambient temperature, and vernalization pathways. This limits our comprehensive understanding of the complex regulatory networks governing soybean flowering and underscores the need for further investigation into these understudied pathways.
In addition to flowering, soybean plant height is a critical agronomic trait that significantly influences planting density, lodging resistance, and ultimately yield [120,145,146]. However, the molecular mechanisms regulating soybean plant height remain relatively poorly understood. This study provides a comprehensive review of key genes involved in soybean plant height regulation in recent years, and summarizes their roles in soybean flowering. These traits are influenced by both genetic factors and external environmental cues, such as light signals and temperature, which together form a complex regulatory network. This complexity underscores the need for integrated research approaches to unravel the interplay between genetic and environmental factors shaping soybean development. By comparing the flowering-related genes identified in this analysis with those previously characterized, we confirmed the reliability of our findings. These results thus serve as a valuable reference for research on the molecular mechanisms underlying soybean flowering and plant height regulation.
Overall, this study provides valuable insights into the regulation of soybean flowering and plant height. The comprehensive analysis of flowering-related candidate genes, combined with the review of genes associated with plant height, establishes a solid foundation for future research in these fields.
5. Conclusions
This study systematically identified 1166 soybean flowering-related genes. These genes are distributed across eight pathways, with the photoperiod pathway being the most prominent (51.20%). Core regulators such as GmFTs and GmSOC1s are conserved across pathways. They are preferentially expressed in SAMs, where they integrate developmental signals. Flowering genes and hormones exhibit rhythmic responses to light. Additionally, GmSAUR46b was validated as a pleiotropic regulator. It influences both flowering time and plant height. The findings show a 67.74% overlap with previously mapped genes. This confirms the reliability of the results. The remaining 32.26% of undetected genes may represent novel networks. These networks likely arose from genome duplication and adaptive evolution. Notably, only 10.12% of the identified genes have been mapped to date. These results reveal thus reveal interconnected regulatory networks governing flowering and plant height. They provide a genetic framework for molecular breeding. This framework can be used to optimize key agronomic traits in soybean.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15092204/s1, Figure S1: Schematic diagram of the CRISPR/Cas9 gene editing vector for GmSAUR46b; Figure S2: Statistics of flowering-related genes in soybeans across different flowering pathways. (A) Distribution of soybean flowering-associated genes across various flowering pathways. (B) The number of flowering genes with different flowering pathways. Ph: photoperiodism pathway. Au: autonomous pathway. Ho: hormone pathway. Ve: vernalization. Ag: ageing pathway. Su: sugar signaling. Fd: flower development/pical meristem response pathway. At: ambient temperature pathway; Figure S3: The identified motifs (20) of photoperiod, circadian clock, and light signaling pathway (Ph) flowering-related genes in Glycine max (L.) Merr; Figure S4: Distribution of conserved motifs and exon–intron structures of hormone signaling and metabolism (Ho) pathway flowering-related genes. (A) Phylogenetic tree of Ho pathway flowering-related genes in Glycine max (L.) Merr. (B) Expression level analysis of Ho pathway flowering-related genes. (C) SMART domain analysis of flowering-related genes in the Ho pathway. (D) Distribution of conserved motifs in Ho pathway flowering-related genes; Figure S5: The identified motifs (15) of hormone signaling and metabolism (Ho) pathway flowering-related genes in Glycine max (L.) Merr; Figure S6: Evolutionary, structural, and conserved domain analyses of flower development/apical meristem response pathway (Fd) flowering-related genes in Glycine max (L.) Merr. (A) Phylogenetic tree of Fd pathway flowering-related genes. (B) Expression level analysis of Fd pathway flowering-related genes. (C) SMART domain analysis of flowering-related genes in the Fd pathway. (D) Distribution of conserved motifs in Fd pathway flowering-related genes; Figure S7: The identified motifs (15) of flower development/apical meristem response pathway (Fd) flowering-related genes in Glycine max (L.) Merr; Figure S8: Evolutionary, structural, and conserved domain analyses of ambient temperature (At) pathway flowering-related genes in Glycine max (L.) Merr. (A) Phylogenetic tree of At pathway flowering-related genes in Glycine max (L.) Merr. (B) Expression level analysis of At pathway flowering-related genes. (C) SMART domain analysis of flowering-related genes in the At pathway. (D) Distribution of conserved motifs in flowering-related genes of the At pathway; Figure S9: The 15 identified motifs of ambient temperature (At) pathway flowering-related genes in Glycine max (L.) Merr; Figure S10: The statistics and analysis of the internode length and plant height (A,B), and the stem diameter (C) of soybean cultivars ZX12 and ZX19 grown under SD conditions with low PPFD. ZX12, Zhexian 12; ZX19, Zhexian 19. Table S1: Flowering-related genes in Glycine max L. identified using A. thaliana flowering genes as queries via BLASTP; Table S2: Presence or absence of homologous flowering genes in Glycine max L. compared to A. thaliana; Table S3: Statistics on flowering-related genes lost in soybean; Table S4: Distribution of flowering-related genes on Glycine max L. chromosomes; Table S5: Count of flowering-related genes on each chromosome; Table S6: FPKM of the Ph flowering pathway genes; Table S7: FPKM values of the Ho flowering pathway genes; Table S8: FPKM values of the Fd flowering pathway genes; Table S9: FPKM values of the At flowering pathway genes; Table S10: Direction and magnitude of natural selection acting on different flowering-related genes for Glycine max L.; Table S11: Primers used in qRT-PCR; Table S12: The sgRNA sequences of GmSAUR46b; Table S13: Primers for detection of gene editing events in mutant lines.
Author Contributions
Conceptualization, F.Y., J.Z. and M.Z.; methodology, X.L., H.D. and Y.C.; software, X.L. and H.Y.; validation, X.L., H.W. and B.L.; formal analysis, X.L., H.W. and H.Y.; investigation, X.L., Y.Y. and H.G.; resources, F.Y.; data curation, X.L., H.D. and Y.C.; writing—original draft preparation, X.L. and B.L.; writing—review and editing, F.Y., J.Z. and M.Z.; visualization, X.L., H.G. and Y.Y.; supervision, X.L. and F.Y.; project administration, F.Y.; funding acquisition, F.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2024YFD1201400).
Data Availability Statement
The datasets used and/or analyzed in the current study are available from the corresponding author (Professor Fengjie Yuan) on reasonable request.
Acknowledgments
We would like to express our sincere gratitude to Xiaochao Chen from Xianghu Laboratory (Agricultural Laboratory of Zhejiang Province) for offering valuable insights into the overall structure and logical flow of this manuscript. We also wish to acknowledge Tianwang Wen from Jiangxi Agricultural University for his substantial contributions to the revision of this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fang, C.; Kong, F. Soybean. Curr. Biol. 2022, 32, R902–R904. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Du, H.; Wang, L.; Liu, B.; Kong, F. Mechanisms underlying key agronomic traits and implications for molecular breeding in soybean. J. Genet. Genom. 2024, 51, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, S.; Wang, Z.; Yuan, Y.; Zhang, Z.; Liang, Q.; Yang, X.; Duan, Z.; Liu, Y.; Kong, F.; et al. Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. 2021, 20, 256–282. [Google Scholar] [CrossRef] [PubMed]
- Sedivy, E.J.; Wu, F.; Hanzawa, Y. Soybean domestication: The origin, genetic architecture and molecular bases. New Phytol. 2017, 214, 539–553. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar] [CrossRef]
- Lin, X.; Liu, B.; Weller, J.L.; Abe, J.; Kong, F. Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 2021, 63, 981–994. [Google Scholar] [CrossRef]
- Lai, B.; Chen, L.; Lu, S. The current status of photoperiod adaptability in soybean. Yi Chuan 2023, 45, 793–800. [Google Scholar] [CrossRef]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef]
- Liu, S.; He, M.; Lin, X.; Kong, F. Epigenetic regulation of photoperiodic flowering in plants. Plant Genome 2023, 16, e20320. [Google Scholar] [CrossRef]
- Lin, X.; Dong, L.; Tang, Y.; Li, H.; Cheng, Q.; Li, H.; Zhang, T.; Ma, L.; Xiang, H.; Chen, L.; et al. Novel and multifaceted regulations of photoperiodic flowering by phytochrome A in soybean. Proc. Natl. Acad. Sci. USA 2022, 119, e2208708119. [Google Scholar] [CrossRef]
- Li, H.; Du, H.; Huang, Z.; He, M.; Kong, L.; Fang, C.; Chen, L.; Yang, H.; Zhang, Y.; Liu, B.; et al. The AP2/ERF transcription factor TOE4b regulates photoperiodic flowering and grain yield per plant in soybean. Plant Biotechnol. J. 2023, 21, 1682–1694. [Google Scholar] [CrossRef]
- Kou, K.; Yang, H.; Li, H.; Fang, C.; Chen, L.; Yue, L.; Nan, H.; Kong, L.; Li, X.; Wang, F.; et al. A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 2022, 32, 1728–1742.e6. [Google Scholar] [CrossRef]
- Dong, L.; Fang, C.; Cheng, Q.; Su, T.; Kou, K.; Kong, L.; Zhang, C.; Li, H.; Hou, Z.; Zhang, Y.; et al. Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 2021, 12, 5445. [Google Scholar] [CrossRef]
- Cao, D.; Takeshima, R.; Zhao, C.; Liu, B.; Jun, A.; Kong, F. Molecular mechanisms of flowering under long days and stem growth habit in soybean. J. Exp. Bot. 2017, 68, 1873–1884. [Google Scholar] [CrossRef]
- Bu, T.; Lu, S.; Wang, K.; Dong, L.; Li, S.; Xie, Q.; Xu, X.; Cheng, Q.; Chen, L.; Fang, C.; et al. A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. USA 2021, 118, e2010241118. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, C.; Sun, J.; Dong, L.; Li, M.; Liu, Y.; Wang, J.; Zhang, X.; Li, D.; Sun, J.; et al. GmRAV confers ecological adaptation through photoperiod control of flowering time and maturity in soybean. Plant Physiol. 2021, 187, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Wan, Z.; Jiao, S.; Zhou, J.; Xu, K.; Nan, H.; Liu, Y.; Xiong, S.; Fan, R.; Zhu, J.; et al. GmMDE genes bridge the maturity gene E1 and florigens in photoperiodic regulation of flowering in soybean. Plant Physiol. 2022, 189, 1021–1036. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.R.; Wang, H.; Wang, Z.; Ren, Y.; Niu, L.; Liu, J.; Liu, B. Photoperiodism dynamics during the domestication and improvement of soybean. Sci. China Life Sci. 2017, 60, 1416–1427. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, L.; Su, T.; Wang, Q.; Gao, Y.; Zhang, S.; Jia, Q.; Yu, G.; Fu, Y.; Cheng, Q.; et al. Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 2020, 43, 637–648. [Google Scholar] [CrossRef]
- No, D.H.; Baek, D.; Lee, S.H.; Cheong, M.S.; Chun, H.J.; Park, M.S.; Cho, H.M.; Jin, B.J.; Lim, L.H.; Lee, Y.B.; et al. High-Temperature Conditions Promote Soybean Flowering through the Transcriptional Reprograming of Flowering Genes in the Photoperiod Pathway. Int. J. Mol. Sci. 2021, 22, 1314. [Google Scholar] [CrossRef]
- Tang, Y.; Lu, S.; Fang, C.; Liu, H.; Dong, L.; Li, H.; Su, T.; Li, S.; Wang, L.; Cheng, Q.; et al. Diverse flowering responses subjecting to ambient high temperature in soybean under short-day conditions. Plant Biotechnol. J. 2023, 21, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Cai, Z.; Li, Y.; Suo, H.; Yi, R.; Zhang, S.; Nian, H. The Floral Repressor GmFLC-like Is Involved in Regulating Flowering Time Mediated by Low Temperature in Soybean. Int. J. Mol. Sci. 2020, 21, 1322. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Li, W.; Fan, C.; Liu, M.; Liu, J.; Liang, W.; Wang, L.; Di, S.; Fang, C.; Li, H.; et al. Genome-wide association study uncovers major genetic loci associated with flowering time in response to active accumulated temperature in wild soybean population. BMC Genom. 2022, 23, 749. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, X.; Zheng, N.; Song, C.; He, Y. Molecular Mechanisms of Temperature-Mediated Flowering Regulation: From Arabidopsis to Short-Day Crops. Plant Cell Environ. 2025, 48, 7020–7037. [Google Scholar] [CrossRef]
- Fisher, J.E.; Loomis, W.E. Auxin-florigen Balance in Flowering of Soybean. Science 1954, 119, 71–73. [Google Scholar] [CrossRef]
- Wong, C.E.; Singh, M.B.; Bhalla, P.L. Floral initiation process at the soybean shoot apical meristem may involve multiple hormonal pathways. Plant Signal. Behav. 2009, 4, 648–651. [Google Scholar] [CrossRef]
- Qin, F.; Shen, Y.; Li, Z.; Qu, H.; Feng, J.; Kong, L.; Teri, G.; Luan, H.; Cao, Z. Shade Delayed Flowering Phenology and Decreased Reproductive Growth of Medicago sativa L. Front. Plant Sci. 2022, 13, 835380. [Google Scholar] [CrossRef]
- Sreeharsha, R.V.; Sekhar, K.M.; Reddy, A.R. Delayed flowering is associated with lack of photosynthetic acclimation in Pigeon pea (Cajanus cajan L.) grown under elevated CO2. Plant Sci. 2015, 231, 82–93. [Google Scholar] [CrossRef]
- Weller, J.L.; Ortega, R.L. Genetic control of flowering time in legumes. Front. Plant Sci. 2015, 6, 207. [Google Scholar] [CrossRef]
- Ren, C.; He, L.; Rosa, L. Integrated irrigation and nitrogen optimization is a resource-efficient adaptation strategy for US maize and soybean production. Nat. Food 2025, 6, 389–400. [Google Scholar] [CrossRef]
- Siamabele, B.; Moral, M.T. The significance of soybean production in the face of changing climates in Africa. Cogent Food Agric. 2021, 7, 1933745. [Google Scholar] [CrossRef]
- Feng, L.; Wang, H.; Ma, X.; Peng, H.; Shan, J. Modeling the current land suitability and future dynamics of global soybean cultivation under climate change scenarios. Field Crops Res. 2021, 263, 108069. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Giakountis, A.; Coupland, G. Phloem transport of flowering signals. Curr. Opin. Plant Biol. 2008, 11, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Ito, S.; Imaizumi, T. Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013, 18, 575–583. [Google Scholar] [CrossRef]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef]
- Sawa, M.; Kay, S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef]
- Shrestha, R.; Go’mez-Ariza, J.; Brambilla, V.; Fornara, F. Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 2014, 114, 1445–1458. [Google Scholar] [CrossRef]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef]
- Lee, B.D.; Cha, J.Y.; Kim, M.R.; Paek, N.C.; Kim, W.Y. Photoperiod sensing system for timing of flowering in plants. BMB Rep. 2018, 51, 163–164. [Google Scholar] [CrossRef]
- Christie, J.M.; Salomon, M.; Nozue, K.; Wada, M.; Briggs, W.R. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 1999, 96, 8779–8783. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Wu, M.F.; Winter, C.M.; Berns, M.C.; Nole-Wilson, S.; Yamaguchi, A.; Coupland, G.; Krizek, B.A.; Wagner, D. A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 2013, 24, 271–282. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Winter, C.M.; Wu, M.-F.; Kanno, Y.; Yamaguchi, A.; Seo, M.; Wagner, D. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 2014, 344, 638–641. [Google Scholar] [CrossRef]
- Jokela, V.; Virkajarvi, P.; Tanskanen, J.; Seppanen, M.M. Vernalization, gibberellic acid and photo period are important signals of yield formation in timothy (Phleum pratense). Physiol. Plant 2014, 152, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Campos-Rivero, G.; Osorio-Montalvo, P.; Sánchez-Borges, R.; Us-Camas, R.; Duarte-Aké, F.; De-la-Peña, C. Plant hormone signaling in flowering: An epigenetic point of view. J. Plant Physiol. 2017, 214, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Galvao, V.C.; Zhang, Y.C.; Horrer, D.; Zhang, T.Q.; Hao, Y.H.; Feng, Y.Q.; Wang, S.; Schmid, M.; Wang, J.W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef]
- Marquardt, S.; Boss, P.K.; Hadfield, J.; Dean, C. Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. J. Exp. Bot. 2006, 57, 3379–3386. [Google Scholar] [CrossRef]
- Kim, D.H.; Doyle, M.R.; Sung, S.; Amasino, R.M. Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 2009, 25, 277–299. [Google Scholar] [CrossRef]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Krober, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes. Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Finnegan, E.J.; Peacock, W.J.; Dennis, E.S. Mechanisms of gene repression by vernalization in Arabidopsis. Plant J. 2009, 59, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G. The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 2004, 7, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-Z.; Zhou, Y.-P.; Lv, T.-X.; Xie, C.-P.; Tian, C.-E. Research progress on the autonomous flowering time pathway in Arabidopsis. Physiol. Mol. Biol. Plants 2017, 23, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, Y.; Chi, H.; Wei, H.; Wang, H.; Yu, S. Genomewide Identification and Characterization of the Genes Involved in the Flowering of Cotton. Int. J. Mol. Sci. 2022, 23, 7940. [Google Scholar] [CrossRef]
- Cho, L.H.; Yoon, J.; An, G. The control of flowering time by environmental factors. Plant J. 2017, 90, 708–719. [Google Scholar] [CrossRef]
- Castillejo, C.; Pelaz, S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 2008, 18, 1338–1343. [Google Scholar] [CrossRef]
- Galvão, V.C.; Horrer, D.; Küttner, F.; Schmid, M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 2012, 139, 4072–4082. [Google Scholar] [CrossRef]
- Osnato, M.; Castillejo, C.; Matias-Hernandez, L.; Pelaz, S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 2012, 3, 808. [Google Scholar] [CrossRef]
- Andres, F.; Porri, A.; Torti, S.; Mateos, J.; Romera-Branchat, M.; Garcia-Martinez, J.L.; Fornara, F.; Gregis, V.; Kater, M.M.; Coupland, G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 2014, 111, E2760–E2769. [Google Scholar] [CrossRef]
- Notaguchi, M.; Abe, M.; Kimura, T.; Daimon, Y.; Kobayashi, T.; Yamaguchi, A.; Tomita, Y.; Dohi, K.; Mori, M.; Araki, T. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 2008, 49, 1645–1658. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Wood, C.C.; Robertson, M.; James Peacock, W.; Dennis, E.S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006, 46, 183–192. [Google Scholar] [CrossRef]
- Jung, J.-H.; Ju, Y.; Seo, P.J.; Lee, J.-H.; Park, C.-M. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012, 69, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Ying, H.; Helliwell, C.A.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 6680–6685. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, C.; Dean, C. The FLC Locus: A Platform for Discoveries in Epigenetics and Adaptation. Annu. Rev. Cell Dev. Biol. 2017, 33, 555–575. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Dent, C.; Liang, H.; Lv, J.; Shang, G.; Liu, Y.; Feng, F.; Wang, F.; Pang, J.; Li, X.; et al. CRY2 interacts with CIS1 to regulate thermosensory flowering via FLM alternative splicing. Nat. Commun. 2022, 13, 7045. [Google Scholar] [CrossRef]
- Kong, X.; Luo, L.; Zhao, J.; Chen, Q.; Chang, G.; Huang, J.; Yang, Y.; Hu, X.; Zhang, D. Expression of FRIGIDA in root inhibits flowering in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 5101–5114. [Google Scholar] [CrossRef]
- Jiang, D.; Gu, X.; He, Y. Establishment of the Winter-Annual Growth Habit via FRIGIDA-Mediated Histone Methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 2009, 21, 1733–1746. [Google Scholar] [CrossRef]
- Hou, Z.; Liu, B.; Kong, F. Regulation of flowering and maturation in soybean. Adv. Bot. Res. 2022, 102, 43–75. [Google Scholar] [CrossRef]
- Virág, E.; Hegedűs, G.; Nagy, Á.; Pallos, J.P.; Kutasy, B. Temporal Shifts in Hormone Signaling Networks Orchestrate Soybean Floral Development Under Field Conditions: An RNA-Seq Study. Int. J. Mol. Sci. 2025, 26, 6455. [Google Scholar] [CrossRef]
- Bernard, R. Two major genes for time of flowering and maturity in soybeans. Crop Sci. 1971, 11, 242–244. [Google Scholar] [CrossRef]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lü, S.; et al. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yamagishi, N.; Zhao, C.; Takeshima, R.; Kasai, M.; Watanabe, S.; Kanazawa, A.; Yoshikawa, N.; Liu, B.; Yamada, T.; et al. The Soybean-Specific Maturity Gene E1 Family of Floral Repressors Controls Night-Break Responses through Down-Regulation of FLOWERING LOCUS T Orthologs. Plant Physiol. 2015, 168, 1735–1746. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Saindon, G.; Voldeng, H.D.; Beversdorf, W.D.; Buzzell, R.I. Genetic Control of Long Daylength Response in Soybean. Crop Sci. 1989, 29, 1436–1439. [Google Scholar] [CrossRef]
- Watanabe, S.; Hideshima, R.; Nakamoto, Y.; Yamanaka, N.; Takahashi, R.; Tabata, S.; Xia, Z.; Tsubokura, Y.; Sato, S.; Ishimoto, M.; et al. Map-Based Cloning of the Gene Associated with the Soybean Maturity Locus E3. Genetics 2009, 182, 1251–1262. [Google Scholar] [CrossRef]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic Redundancy in Soybean Photoresponses Associated with Duplication of the Phytochrome A Gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef]
- Buzzell, R.; Voldeng, H. Inheritance of insensitivity to long daylength. Soybean Genet. Newsl 1980, 7, 13. [Google Scholar]
- Li, C.; Li, Y.-H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A Domestication-Associated Gene GmPRR3b Regulates the Circadian Clock and Flowering Time in Soybean. Mol. Plant 2020, 13, 745–759. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Y.; Wu, H.; Hu, B.; Zhai, H.; Yang, J.; Xia, Z. Positional Cloning of the Flowering Time QTL qFT12-1 Reveals the Link Between the Clock Related PRR Homolog with Photoperiodic Response in Soybeans. Front. Plant Sci. 2019, 10, 1303. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, Q.; Fang, C.; Kong, L.; Yang, H.; Hou, Z.; Li, Y.; Nan, H.; Zhang, Y.; Chen, Q.; et al. Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol. Plant 2022, 15, 308–321. [Google Scholar] [CrossRef]
- Cheng, Q.; Dong, L.; Su, T.; Li, T.; Gan, Z.; Nan, H.; Lu, S.; Fang, C.; Kong, L.; Li, H.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 2019, 19, 562. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, S.; Wu, T.; Liu, L.; Sun, X.; Cai, Y.; Li, J.; Jia, H.; Yuan, S.; Chen, L.; et al. Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 2020, 18, 1869–1881. [Google Scholar] [CrossRef] [PubMed]
- Buzzell, R. Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can. J. Genet. Cytol. 1971, 13, 703–707. [Google Scholar] [CrossRef]
- McBlain, B.A.; Bernard, R.L. A new gene affecting the time of flowering and maturity in soybeans. J. Hered. 1987, 78, 160–162. [Google Scholar] [CrossRef]
- Li, H.; Du, H.; He, M.; Wang, J.; Wang, F.; Yuan, W.; Huang, Z.; Cheng, Q.; Gou, C.; Chen, Z.; et al. Natural variation of FKF1 controls flowering and adaptation during soybean domestication and improvement. New Phytol. 2023, 238, 1671–1684. [Google Scholar] [CrossRef]
- Somers, D.E.; Wu, F.; Price, B.W.; Haider, W.; Seufferheld, G.; Nelson, R.; Hanzawa, Y. Functional and Evolutionary Characterization of the CONSTANS Gene Family in Short-Day Photoperiodic Flowering in Soybean. PLoS ONE 2014, 9, e85754. [Google Scholar] [CrossRef]
- González, A.M.; Vander Schoor, J.K.; Fang, C.; Kong, F.; Wu, J.; Weller, J.L.; Santalla, M. Ancient relaxation of an obligate short-day requirement in common bean through loss of CONSTANS-like gene function. Curr. Biol. 2021, 31, 1643–1652.e1642. [Google Scholar] [CrossRef]
- Samanfar, B.; Molnar, S.J.; Charette, M.; Schoenrock, A.; Dehne, F.; Golshani, A.; Belzile, F.; Cober, E.R. Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 2016, 130, 377–390. [Google Scholar] [CrossRef]
- Zhai, H.; Lu, S.; Liang, S.; Wu, H.; Zhang, X.; Liu, B.; Kong, F.; Yuan, X.; Li, J.; Xia, Z. GmFT4, a Homolog of FLOWERING LOCUS T, Is Positively Regulated by E1 and Functions as a Flowering Repressor in Soybean. PLoS ONE 2014, 9, e89030. [Google Scholar] [CrossRef]
- Guo, G.; Xu, K.; Zhang, X.; Zhu, J.; Lu, M.; Chen, F.; Liu, L.; Xi, Z.-Y.; Bachmair, A.; Chen, Q.; et al. Extensive Analysis of GmFTL and GmCOL Expression in Northern Soybean Cultivars in Field Conditions. PLoS ONE 2015, 10, e0136601. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, B.; Ma, L.; Zhang, S.; Zhai, H.; Xu, X.; Hou, W.; Xia, Z.; Wu, C.; Sun, S.; et al. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 2017, 217, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Liu, B.; Xia, Z.; Sato, S.; Kim, B.M.; Watanabe, S.; Yamada, T.; Tabata, S.; Kanazawa, A.; Harada, K.; et al. Two Coordinately Regulated Homologs of FLOWERING LOCUS T Are Involved in the Control of Photoperiodic Flowering in Soybean. Plant Physiol. 2010, 154, 1220–1231. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Hu, R.; Zhang, X.; Wang, X.; Zhang, W.; Zhang, Q.; Ma, J.; Fu, Y.-F. Conserved CO-FT regulons contribute to the photoperiod flowering control in soybean. BMC Plant Biol. 2014, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Wang, L.; Chen, L.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; Yao, W.; Jiang, B.; Yuan, S.; et al. Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 2020, 18, 298–309. [Google Scholar] [CrossRef]
- Li, X.; Fang, C.; Yang, Y.; Lv, T.; Su, T.; Chen, L.; Nan, H.; Li, S.; Zhao, X.; Lu, S.; et al. Overcoming the genetic compensation response of soybean florigens to improve adaptation and yield at low latitudes. Curr. Biol. 2021, 31, 3755–3767.e3754. [Google Scholar] [CrossRef]
- Takeshima, R.; Hayashi, T.; Zhu, J.; Zhao, C.; Xu, M.; Yamaguchi, N.; Sayama, T.; Ishimoto, M.; Kong, L.; Shi, X.; et al. A soybean quantitative trait locus that promotes flowering under long days is identified as FT5a, a FLOWERING LOCUS T ortholog. J. Exp. Bot. 2016, 67, 5247–5258. [Google Scholar] [CrossRef]
- Yue, L.; Li, X.; Fang, C.; Chen, L.; Yang, H.; Yang, J.; Chen, Z.; Nan, H.; Chen, L.; Zhang, Y.; et al. FT5a interferes with the Dt1-AP1 feedback loop to control flowering time and shoot determinacy in soybean. J. Integr. Plant Biol. 2021, 63, 1004–1020. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Liu, X.; Yao, W.; Hou, W. GmNF-YC4 delays soybean flowering and maturation by directly repressing GmFT2a and GmFT5a expression. J. Integr. Plant Biol. 2024, 66, 1370–1384. [Google Scholar] [CrossRef]
- Cao, D.; Li, Y.; Wang, J.; Nan, H.; Wang, Y.; Lu, S.; Jiang, Q.; Li, X.; Shi, D.; Fang, C.; et al. GmmiR156b overexpression delays flowering time in soybean. Plant Mol. Biol. 2015, 89, 353–363. [Google Scholar] [CrossRef]
- Ding, X.; Guo, J.; Lv, M.; Wang, H.; Sheng, Y.; Liu, Y.; Gai, J.; Yang, S. The miR156b–GmSPL2b module mediates male fertility regulation of cytoplasmic male sterility-based restorer line under high-temperature stress in soybean. Plant Biotechnol. J. 2023, 21, 1542–1559. [Google Scholar] [CrossRef]
- Sun, Z.; Su, C.; Yun, J.; Jiang, Q.; Wang, L.; Wang, Y.; Cao, D.; Zhao, F.; Zhao, Q.; Zhang, M.; et al. Genetic improvement of the shoot architecture and yield in soya bean plants via the manipulation of GmmiR156b. Plant Biotechnol. J. 2019, 17, 50–62. [Google Scholar] [CrossRef]
- Lee, S.; Singh, M.B.; Bhalla, P.L. Functional analysis of soybean miR156 and miR172 in tobacco highlights their role in plant morphology and floral transition. Plant Physiol. Biochem. 2023, 196, 393–401. [Google Scholar] [CrossRef]
- Zhao, X.; Cao, D.; Huang, Z.; Wang, J.; Lu, S.; Xu, Y.; Liu, B.; Kong, F.; Yuan, X. Dual functions of GmTOE4a in the regulation of photoperiod-mediated flowering and plant morphology in soybean. Plant Mol. Biol. 2015, 88, 343–355. [Google Scholar] [CrossRef]
- Nan, H.; Cao, D.; Zhang, D.; Li, Y.; Lu, S.; Tang, L.; Yuan, X.; Liu, B.; Kong, F. GmFT2a and GmFT5a Redundantly and Differentially Regulate Flowering through Interaction with and Upregulation of the bZIP Transcription Factor GmFDL19 in Soybean. PLoS ONE 2014, 9, e97669. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cai, Y.; Qu, M.; Wang, L.; Sun, H.; Jiang, B.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; et al. Soybean adaption to high-latitude regions is associated with natural variations of GmFT2b, an ortholog of FLOWERING LOCUS T. Plant Cell Environ. 2020, 43, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Qin, C.; Zhao, T.; Liu, B.; Li, H.-Y.; Liu, J. Function analysis of GmELF3s in regulating soybean flowering time and circadian rhythm. Acta Agron. Sin. 2022, 48, 812–824. [Google Scholar] [CrossRef]
- Fang, C.; Liu, J.; Zhang, T.; Su, T.; Li, S.; Cheng, Q.; Kong, L.; Li, X.; Bu, T.; Li, H.; et al. A recent retrotransposon insertion of J caused E6 locus facilitating soybean adaptation into low latitude. J. Integr. Plant Biol. 2021, 63, 995–1003. [Google Scholar] [CrossRef]
- Qin, C.; Li, H.; Zhang, S.; Lin, X.; Jia, Z.; Zhao, F.; Wei, X.; Jiao, Y.; Li, Z.; Niu, Z.; et al. GmEID1 modulates light signaling through the Evening Complex to control flowering time and yield in soybean. Proc. Natl. Acad. Sci. USA 2023, 120, e2212468120. [Google Scholar] [CrossRef]
- Zhao, X.; Li, H.; Wang, L.; Wang, J.; Huang, Z.; Du, H.; Li, Y.; Yang, J.; He, M.; Cheng, Q.; et al. A critical suppression feedback loop determines soybean photoperiod sensitivity. Dev. Cell 2024, 59, 1750–1763.e1754. [Google Scholar] [CrossRef]
- Bao, A.; Chen, H.; Chen, L.; Chen, S.; Hao, Q.; Guo, W.; Qiu, D.; Shan, Z.; Yang, Z.; Yuan, S.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 2019, 19, 131. [Google Scholar] [CrossRef]
- Dissanayaka, A.; Rodriguez, T.O.; Di, S.; Yan, F.; Githiri, S.M.; Rodas, F.R.; Abe, J.; Takahashi, R. Quantitative trait locus mapping of soybean maturity gene E5. Breed. Sci. 2016, 66, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Bonato, E.R.; Vello, N.A. E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 1999, 22, 2229–2232. [Google Scholar] [CrossRef]
- Cober, E.R.; Molnar, S.J.; Charette, M.; Voldeng, H.D. A new locus for early maturity in soybean. Crop Sci. 2010, 50, 524–527. [Google Scholar] [CrossRef]
- Cober, E.R.; Voldeng, H.D. Low R:FR Light Quality Delays Flowering of E7E7 Soybean Lines. Crop Sci. 2001, 41, 1823–1826. [Google Scholar] [CrossRef]
- Zhao, C.; Takeshima, R.; Zhu, J.; Xu, M.; Sato, M.; Watanabe, S.; Kanazawa, A.; Liu, B.; Kong, F.; Yamada, T.; et al. A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 2016, 16, 20. [Google Scholar] [CrossRef]
- Wang, F.; Nan, H.; Chen, L.; Fang, C.; Zhang, H.; Su, T.; Li, S.; Cheng, Q.; Dong, L.; Liu, B.; et al. A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol. Breed. 2019, 39, 70. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, N.; Jiang, B.; Li, M.; Wang, H.; Jiang, Z.; Pan, H.; Xia, Q.; Ma, Q.; Han, T.; et al. A Single Nucleotide Deletion in J Encoding GmELF3 Confers Long Juvenility and Is Associated with Adaption of Tropic Soybean. Mol. Plant 2017, 10, 656–658. [Google Scholar] [CrossRef]
- Liu, B.; Watanabe, S.; Uchiyama, T.; Kong, F.; Kanazawa, A.; Xia, Z.; Nagamatsu, A.; Arai, M.; Yamada, T.; Kitamura, K.; et al. The Soybean Stem Growth Habit Gene Dt1 Is an Ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 2010, 153, 198–210. [Google Scholar] [CrossRef]
- Sun, J.; Liu, Y.; Zheng, Y.; Xue, Y.; Fan, Y.; Ma, X.; Ji, Y.; Liu, G.; Zhang, X.; Li, Y.; et al. The MADS-box transcription factor GmFULc promotes GmZTL4 gene transcription to modulate maturity in soybean. J. Integr. Plant Biol. 2024, 66, 1603–1619. [Google Scholar] [CrossRef]
- Chen, L.; Nan, H.; Kong, L.; Yue, L.; Yang, H.; Zhao, Q.; Fang, C.; Li, H.; Cheng, Q.; Lu, S.; et al. Soybean AP1 homologs control flowering time and plant height. J. Integr. Plant Biol. 2020, 62, 1868–1879. [Google Scholar] [CrossRef]
- Bouche, F.; Lobet, G.; Tocquin, P.; Perilleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016, 44, D1167–D1171. [Google Scholar] [CrossRef]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qiu, Z.; Wang, X.; Gong, P.; Xu, Q.; Yu, Q.B.; Guan, Y. Pooled CRISPR/Cas9 reveals redundant roles of plastidial phosphoglycerate kinases in carbon fixation and metabolism. Plant J. 2019, 98, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, H.; Chen, J.; Yan, L.; Xia, L. Toward Precision Genome Editing in Crop Plants. Mol. Plant 2020, 13, 811–813. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef]
- Liang, Q.; Chen, L.; Yang, X.; Yang, H.; Liu, S.; Kou, K.; Fan, L.; Zhang, Z.; Duan, Z.; Yuan, Y.; et al. Natural variation of Dt2 determines branching in soybean. Nat. Commun. 2022, 13, 6429. [Google Scholar] [CrossRef]
- Fang, C.; Sun, Z.; Li, S.; Su, T.; Wang, L.; Dong, L.; Li, H.; Li, L.; Kong, L.; Yang, Z.; et al. Subfunctionalisation and self-repression of duplicated E1 homologues finetunes soybean flowering and adaptation. Nat. Commun. 2024, 15, 6184. [Google Scholar] [CrossRef]
- Wang, J.; Qiu, Y.; Cheng, F.; Chen, X.; Zhang, X.; Wang, H.; Song, J.; Duan, M.; Yang, H.; Li, X. Genome-wide identification, characterization, and evolutionary analysis of flowering genes in radish (Raphanus sativus L.). BMC Genom. 2017, 18, 981. [Google Scholar] [CrossRef]
- Zhang, J.; Song, Q.; Cregan, P.B.; Nelson, R.L.; Wang, X.; Wu, J.; Jiang, G.-L. Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genom. 2015, 16, 217. [Google Scholar] [CrossRef]
- Liu, W.; Kim, M.Y.; Kang, Y.J.; Van, K.; Lee, Y.-H.; Srinives, P.; Yuan, D.L.; Lee, S.-H. QTL identification of flowering time at three different latitudes reveals homeologous genomic regions that control flowering in soybean. Theor. Appl. Genet. 2011, 123, 545–553. [Google Scholar] [CrossRef]
- Zhu, J.; Takeshima, R.; Harigai, K.; Xu, M.; Kong, F.; Liu, B.; Kanazawa, A.; Yamada, T.; Abe, J. Loss of Function of the E1-Like-b Gene Associates with Early Flowering Under Long-Day Conditions in Soybean. Front. Plant Sci. 2018, 9, 1867. [Google Scholar] [CrossRef]
- Dong, L.; Li, S.; Wang, L.; Su, T.; Zhang, C.; Bi, Y.; Lai, Y.; Kong, L.; Wang, F.; Pei, X.; et al. The genetic basis of high-latitude adaptation in wild soybean. Curr. Biol. 2023, 33, 252–262.e4. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Zhang, L.; Wang, J.; Wang, X.; Guo, S.; Xu, Z.J.; Li, D.; Liu, Z.; Li, Y.H.; Liu, B.; et al. Flowering time regulator qFT13-3 involved in soybean adaptation to high latitudes. Plant Biotechnol. J. 2023, 22, 1164–1176. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, H.; He, M.; Dong, L.; Huang, Z.; Chen, L.; Nan, H.; Kong, F.; Liu, B.; Zhao, X. GIGANTEA orthologs, E2 members, redundantly determine photoperiodic flowering and yield in soybean. J. Integr. Plant Biol. 2023, 65, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Vega-Léon, R.; Hugouvieux, V.; Blanc-Mathieu, R.; van der Wal, F.; Lucas, J.; Silva, C.S.; Jourdain, A.; Muino, J.M.; Nanao, M.H.; et al. The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 2021, 12, 4760. [Google Scholar] [CrossRef]
- Zhang, C.; Ogilvie, H.A.; Drummond, A.J.; Stadler, T. Bayesian Inference of Species Networks from Multilocus Sequence Data. Mol. Biol. Evol. 2018, 35, 504–517. [Google Scholar] [CrossRef]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farré, E.M.; Kay, S.A. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef]
- Takeshima, R.; Nan, H.; Harigai, K.; Dong, L.; Zhu, J.; Lu, S.; Xu, M.; Yamagishi, N.; Yoshikawa, N.; Liu, B.; et al. Functional divergence between soybean FLOWERING LOCUS T orthologues FT2a and FT5a in post-flowering stem growth. J. Exp. Bot. 2019, 70, 3941–3953. [Google Scholar] [CrossRef]
- Ping, J.; Liu, Y.; Sun, L.; Zhao, M.; Li, Y.; She, M.; Sui, Y.; Lin, F.; Liu, X.; Tang, Z.; et al. Dt2Is a Gain-of-Function MADS-Domain Factor Gene That Specifies Semideterminacy in Soybean. Plant Cell 2014, 26, 2831–2842. [Google Scholar] [CrossRef]
- Sun, J.; Wang, M.; Zhao, C.; Liu, T.; Liu, Z.; Fan, Y.; Xue, Y.; Li, W.; Zhang, X.; Zhao, L. GmFULc Is Induced by Short Days in Soybean and May Accelerate Flowering in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 333. [Google Scholar] [CrossRef]
- Jia, Z.; Jiang, B.; Gao, X.; Yue, Y.; Fei, Z.; Sun, H.; Wu, C.; Sun, S.; Hou, W.; Han, T. GmFULa, a FRUITFULL homolog, functions in the flowering and maturation of soybean. Plant Cell Rep. 2014, 34, 121–132. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, X.; Feng, J.; Ma, X.; Ji, Y.; Chen, S.; Li, J.; Li, D.; Wang, X.; Zhao, L. The transcription factor GmFULc regulates soybean plant height by binding the promoter of a gibberellin-responsive gene. Plant Physiol. 2025, 197, kiaf021. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, X.; Shan, J.; Li, Y.; Zhang, Y.; Wang, Y.; Li, W.; Zhao, L. Overexpression of GmGAMYB Accelerates the Transition to Flowering and Increases Plant Height in Soybean. Front. Plant Sci. 2021, 12, 667242. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, M.; Xu, C.; Yang, X.; Li, D.; Zhao, X.; Wang, K.; Li, Y.; Zhang, X.; Liu, L.; et al. Natural variation in GmGBP1 promoter affects photoperiod control of flowering time and maturity in soybean. Plant J. 2018, 96, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zheng, Y.; Guo, J.; Zhang, Y.; Liu, Y.; Tao, Y.; Wang, M.; Liu, T.; Liu, Y.; Li, X.; et al. GmGAMYB-BINDING PROTEIN 1 promotes small auxin-up RNA gene transcription to modulate soybean maturity and height. Plant Physiol. 2023, 193, 775–791. [Google Scholar] [CrossRef]
- Lyu, X.; Cheng, Q.; Qin, C.; Li, Y.; Xu, X.; Ji, R.; Mu, R.; Li, H.; Zhao, T.; Liu, J.; et al. GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 2021, 14, 298–314. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).