Identification and Characterization of SWEET Gene Family in Peanuts and the Role of AhSWEET50 in Sugar Accumulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Methods
2.2. Identification of Members of the Peanut SWEET Gene Family
2.3. Phylogenetic Tree Construction and Prediction of Physicochemical Properties
2.4. Chromosome Localization Analysis
2.5. Conserved Motif and Gene Structure Analysis
2.6. Promoter Cis-Acting Element Analysis
2.7. GO Enrichment Analysis
2.8. Expression Pattern Analysis
2.9. RNA Extraction and cDNA Synthesis
2.10. Gene Cloning and Bioinformatic Analysis
2.11. Subcellular Localization Analysis
2.12. Determination of Soluble Sugar Content in Transgenic A. thaliana
3. Results
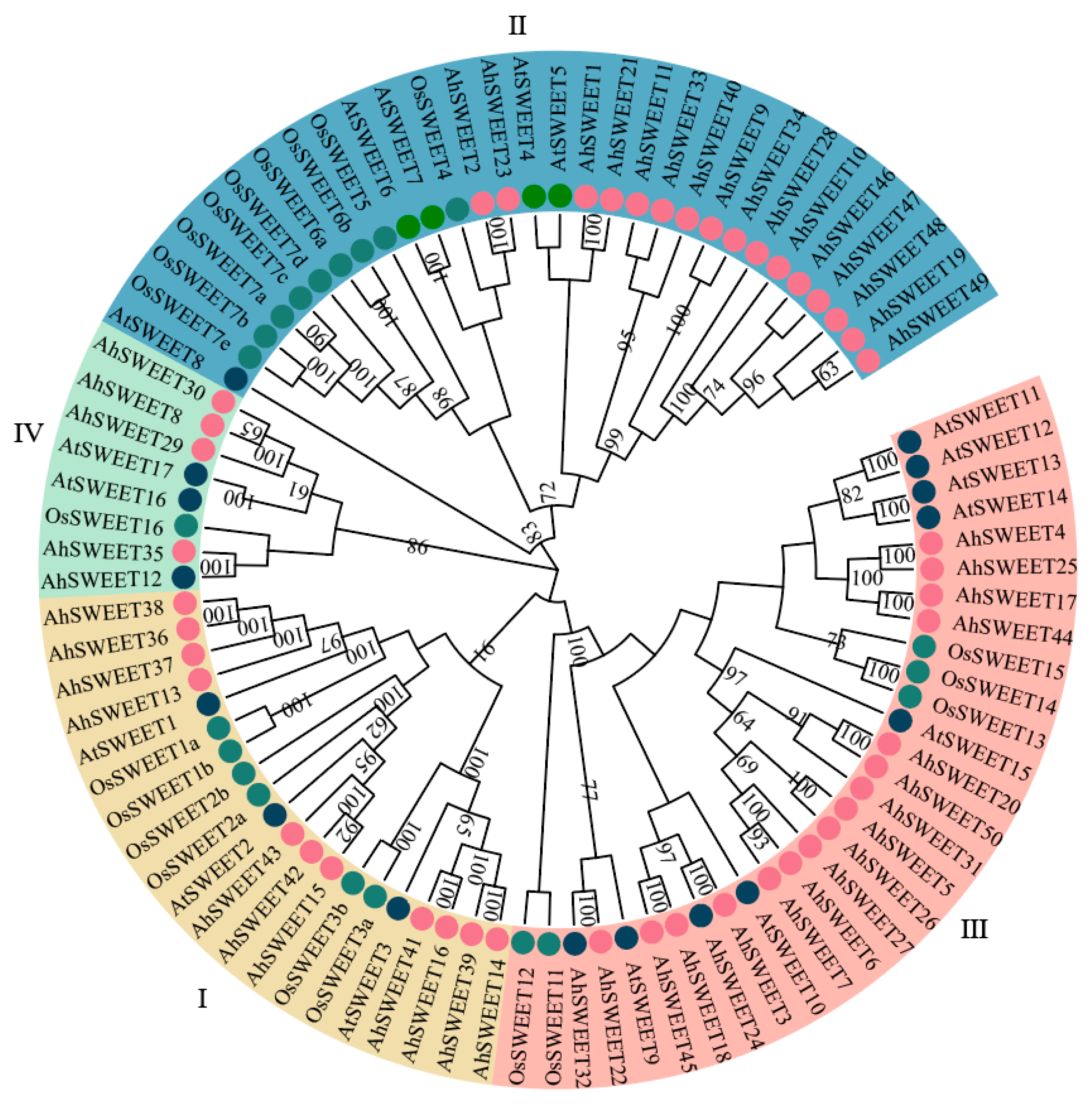
3.1. Identification, Phylogenetic Evolution, and Physicochemical Properties of AhSWEET Gene Family in Peanut
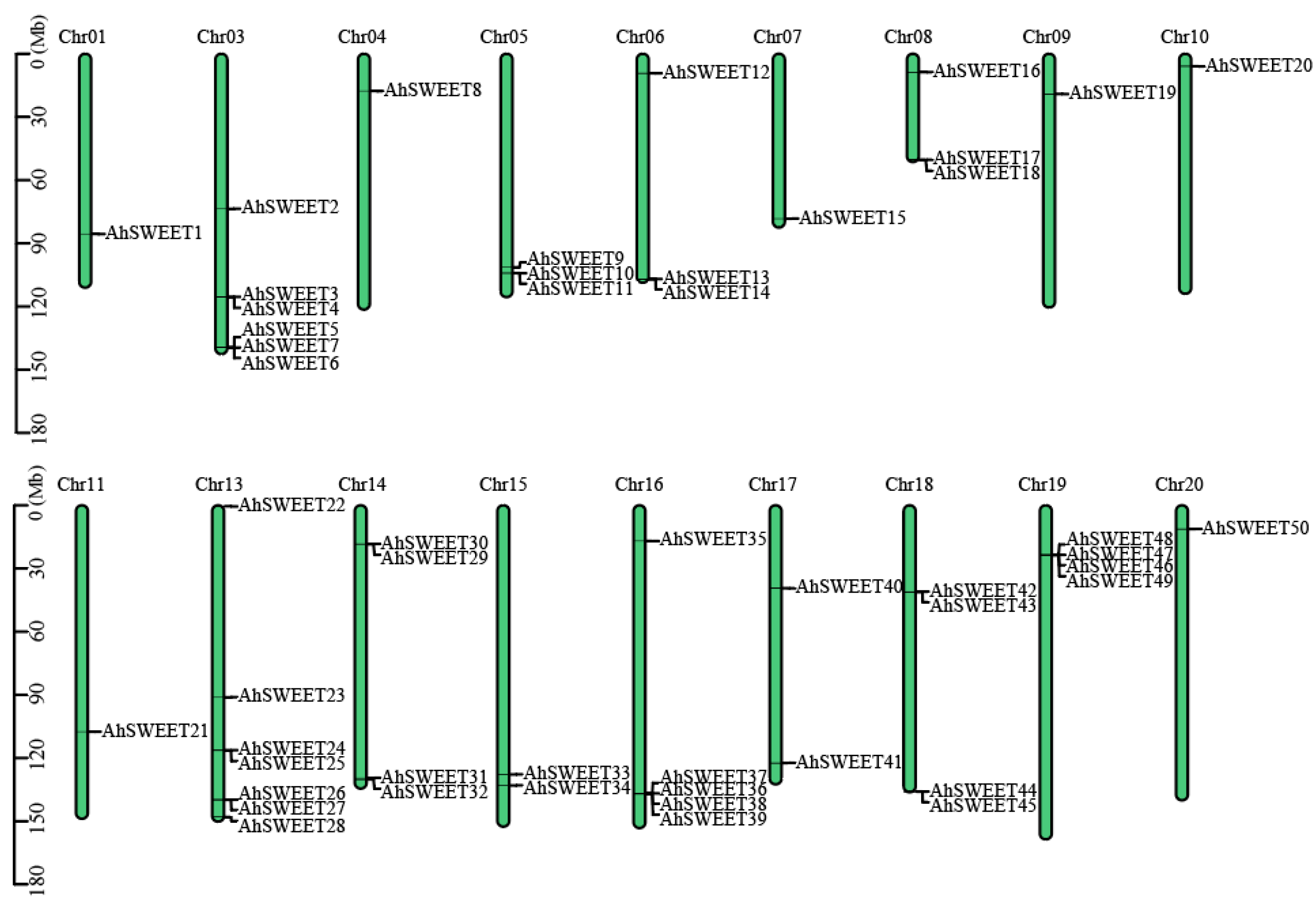
3.2. Chromosomal Localization of Peanut AhSWEET Genes
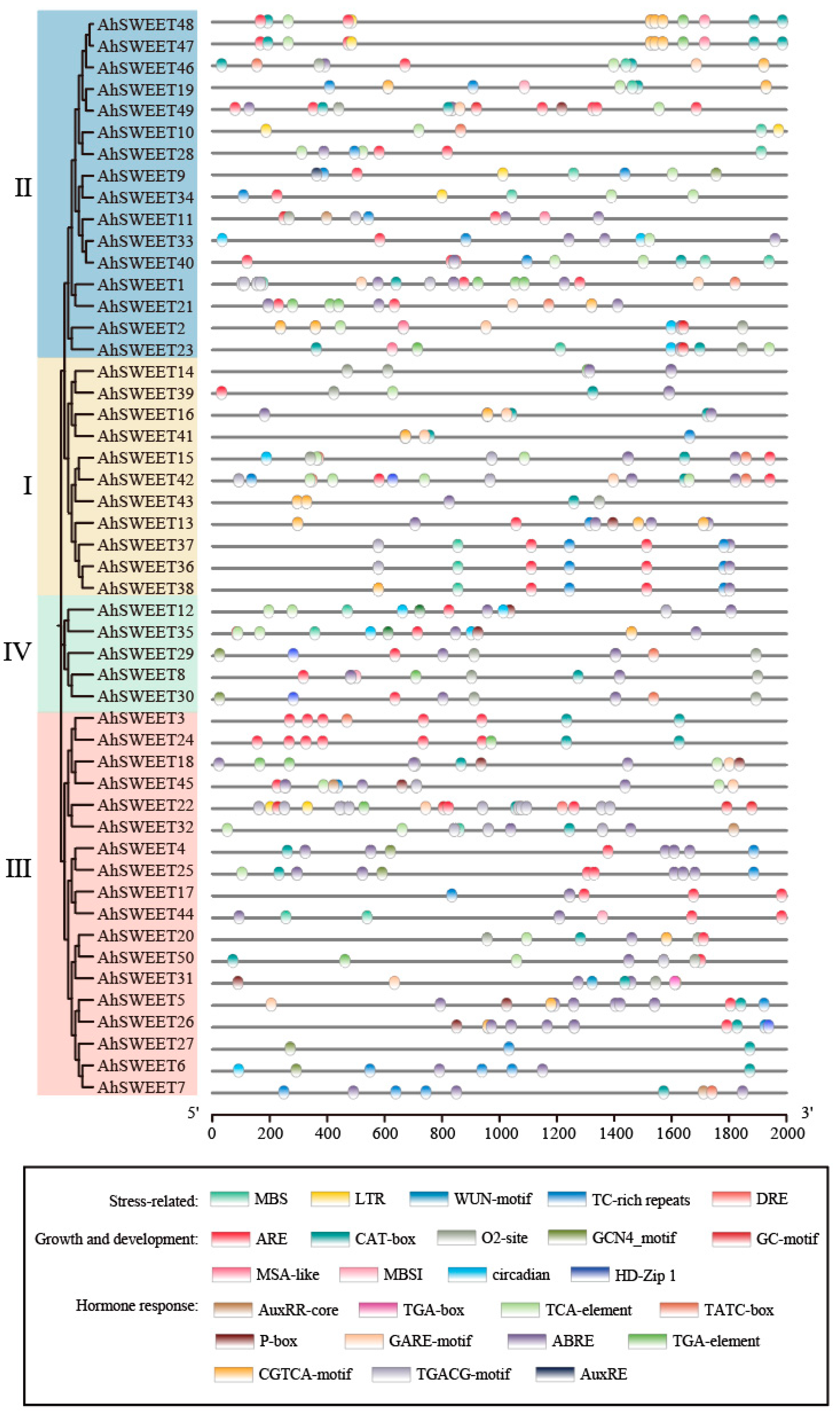
3.3. Conserved Motifs and Gene Structure of Peanut AhSWEET Genes
3.4. Promoter Cis-Acting Elements of Peanut AhSWEET Genes
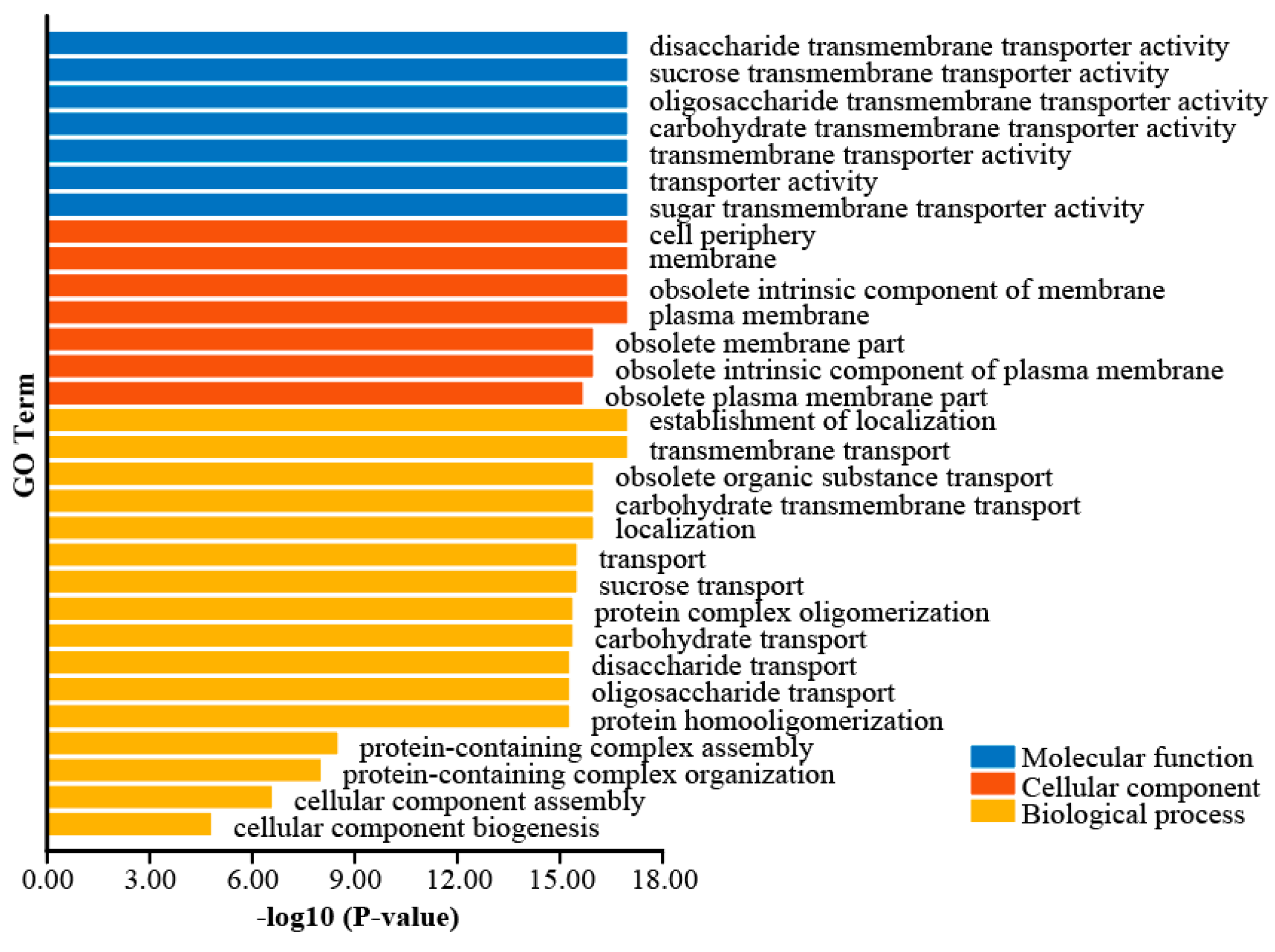
3.5. GO Enrichment Analysis of Peanut AhSWEET Gene Family
3.6. Expression Pattern of Peanut AhSWEET Genes in High- and Low-Sugar Peanut Cultivars
3.7. Characteristics of AhSWEET50 Gene in Peanut
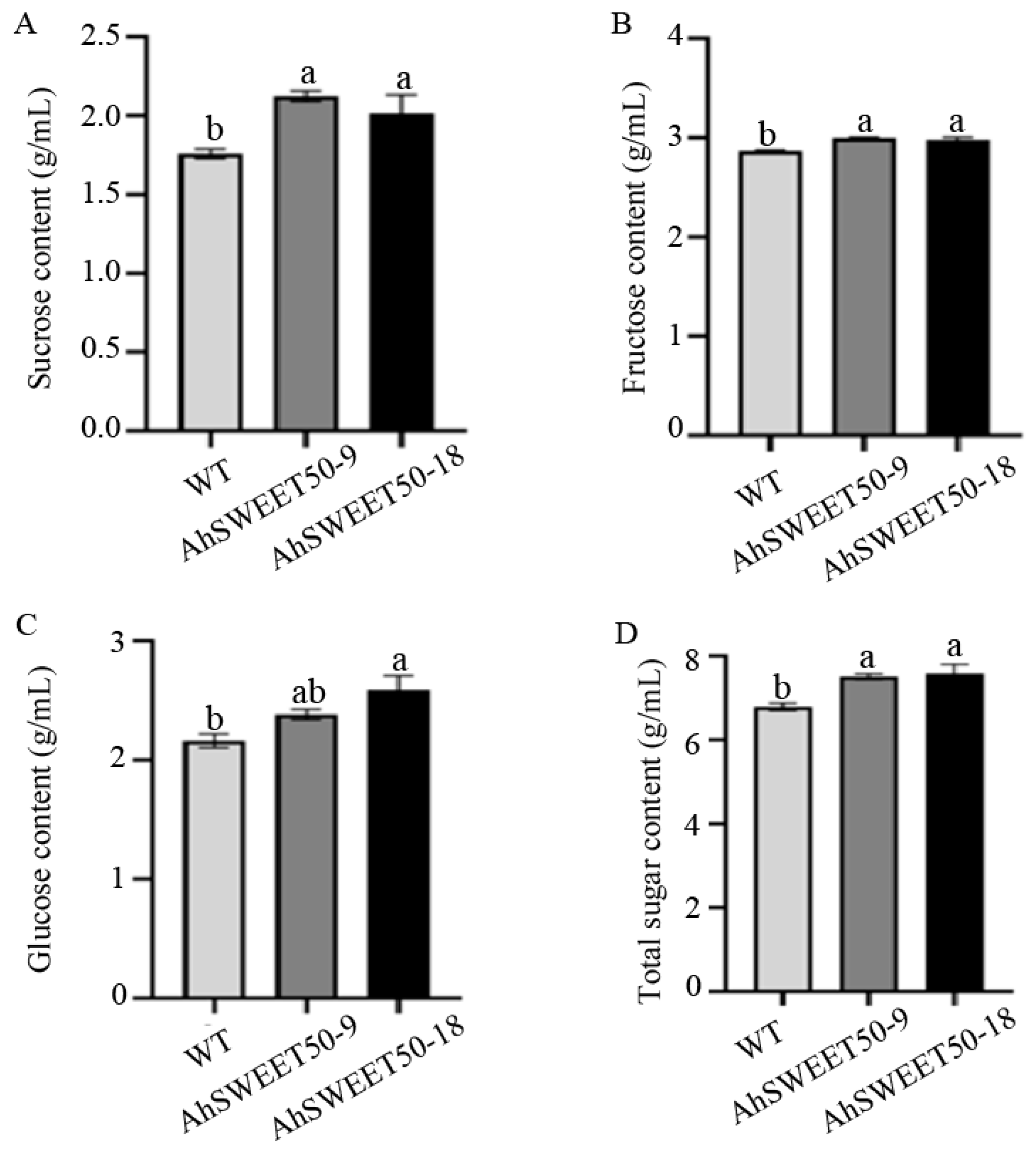
3.8. AhSWEET50 Gene Increased the Soluble Sugars
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| SWEET | sugars will eventually be exported transporter |
| MSTs | monosaccharide transporters |
| SUTs | sucrose transporters |
| FRET | fluorescence resonance energy transfer |
| TMs | transmembrane domains |
| PGR | Peanut Genome Resource |
| NCBI | National Center for Biotechnology Information |
| FPKM | fragments per kilobase of exon model per million mapped fragments |
| HPLC | high-performance liquid chromatography |
| MeJA | methyl jasmonate |
| SA | salicylic acid |
| GO | Gene Ontology |
| WT | wild-type |
References
- Çiftçi, S.; Suna, G. Functional components of peanuts (Arachis hypogaea L.) and health benefits: A review. Future Foods 2022, 5, 100140. [Google Scholar] [CrossRef]
- Pattee, H.E.; Isleib, T.G.; Giesbrecht, F.G.; McFeeters, R.F. Investigations into genotypic variations of peanut carbohydrates. J. Agric. Food Chem. 2000, 48, 750–756. [Google Scholar] [CrossRef]
- Bavnhøj, L.; Paulsen, P.A.; Flores-Canales, J.C.; Schiøtt, B.; Pedersen, B.P. Molecular mechanism of sugar transport in plants unveiled by structures of glucose/H+ symporter STP10. Nat. Plants 2021, 7, 1409–1419. [Google Scholar] [CrossRef]
- Slewinski, T.L. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: A physiological perspective. Mol. Plant 2011, 4, 641–662. [Google Scholar] [CrossRef] [PubMed]
- Kühn, C.; Grof, C.P. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014, 201, 1150–1155. [Google Scholar] [CrossRef]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Zhu, Y.; Tian, Y.; Han, S.; Wang, J.; Liu, Y.; Yin, J. Structure, evolution, and roles of SWEET proteins in growth and stress responses in plants. Int. J. Biol. Macromol. 2024, 263, 130441. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Z.Y.; Kumar, V.; Xu, X.F.; Yuan, P.; Zhu, X.F.; Li, T.Y.; Jia, B.; Xuan, Y.H. Genome-wide identification of the SWEET gene family in wheat. Gene 2018, 5, 284–292. [Google Scholar] [CrossRef]
- Han, X.; Han, S.; Zhu, Y.; Liu, Y.; Gao, S.; Yin, J.; Wang, F.; Yao, M. Genome-wide identification and expression analysis of the SWEET gene family in Capsicum annuum L. Int. J. Mol. Sci. 2023, 24, 17508. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Yan, P.; He, S.; Jia, L.; Wang, Y.; Liu, Q.; Zhai, H.; Zhao, N.; Gao, S.; Zhang, H. Genome-wide identification and expression analysis of SWEET family genes in sweet potato and its two diploid relatives. Int. J. Mol. Sci. 2022, 23, 15848. [Google Scholar] [CrossRef]
- Li, W.; Huang, L.; Liu, N.; Pandey, M.K.; Chen, Y.; Cheng, L.; Guo, J.; Yu, B.; Luo, H.; Zhou, X.; et al. Key regulators of sucrose metabolism identified through comprehensive comparative transcriptome analysis in peanuts. Int. J. Mol. Sci. 2021, 22, 7266. [Google Scholar] [CrossRef] [PubMed]
- Ninan, A.S.; Grant, J.; Song, J.; Jameson, P.E. Expression of genes related to sugar and amino acid transport and cytokinin metabolism during leaf development and senescence in Pisum sativum L. Plants 2019, 8, 76. [Google Scholar] [CrossRef]
- Klemens, P.A.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef]
- Li, P.; Wang, L.; Liu, H.; Yuan, M. Impaired SWEET-mediated sugar transportation impacts starch metabolism in developing rice seeds. Crop J. 2022, 10, 98–108. [Google Scholar] [CrossRef]
- Radchuk, V.; Belew, Z.M.; Gündel, A.; Mayer, S.; Hilo, A.; Hensel, G.; Sharma, R.; Neumann, K.; Ortleb, S.; Wagner, S.; et al. SWEET11b transports both sugar and cytokinin in developing barley grains. Plant Cell 2023, 35, 2186–2207. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, C.; Wang, M.; Li, T.; Liu, X.; Jiang, J. Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits. Hortic Res. 2021, 8, 186. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, H.; Wang, T.; Zhang, J.; Liu, W.; Fang, H.; Zhang, Z.; Peng, F.; Chen, X.; Wang, N. Abscisic acid and regulation of the sugar transporter gene MdSWEET9b promote apple sugar accumulation. Plant Physiol. 2023, 192, 2081–2101. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, W.; Zang, S.; Qin, L.; Liang, Y.; Lin, P.; Su, Y.; Que, Y. Sugarcane transcription factor ScWRKY4 negatively regulates resistance to pathogen infection through the JA signaling pathway. Crop J. 2024, 12, 164–176. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, Y.; Wang, L.; Liu, L.; Zhong, X.; Chu, P.; Gao, M.; Chen, H.; Cai, T.; Xiong, F.; et al. Genome-wide identification of papain-like cysteine protease family genes in cultivated peanut (Arachis hypogaea L.) and functional characterization of AhRD21B in response to chilling stress. Environ. Exp. Bot. 2023, 209, 105272. [Google Scholar] [CrossRef]
- Anjali, A.; Fatima, U.; Manu, M.S.; Ramasamy, S.; Senthil-Kumar, M. Structure and regulation of SWEET transporters in plants: An update. Plant Physiol. Biochem. 2020, 156, 1–6. [Google Scholar] [CrossRef]
- Jeena, G.S.; Kumar, S.; Shukla, R.K. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol. Biol. 2019, 100, 351–365. [Google Scholar] [CrossRef]
- Gautam, T.; Saripalli, G.; Gahlaut, V.; Kumar, A.; Sharma, P.K.; Balyan, H.S.; Gupta, P.K. Further studies on sugar transporter (SWEET) genes in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2019, 46, 2327–2353. [Google Scholar] [CrossRef]
- Feng, C.Y.; Han, J.X.; Han, X.X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Valifard, M.; Khan, A.; Berg, J.; Le Hir, R.; Pommerrenig, B.; Neuhaus, H.E.; Keller, I. Carbohydrate distribution via SWEET17 is critical for Arabidopsis inflorescence branching under drought. J. Exp. Bot. 2024, 75, 3903–3919. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Li, H.; Xia, X.; Liu, X.; Yang, L. Functional and evolution characterization of SWEET sugar transporters in Ananas comosus. Biochem. Biophys. Res. Commun. 2018, 496, 407–414. [Google Scholar] [CrossRef]
- Ji, J.; Yang, L.; Fang, Z.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y. Plant SWEET family of sugar transporters: Structure, evolution and biological functions. Biomolecules 2022, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, R.; Jiang, Y.; Dong, X.; Xu, J.; Xu, Q.; Kan, Q.; Luo, Z.; Springer, N.M.; Li, Q. A novel active transposon creates allelic variation through altered translation rate to influence protein abundance. Nucleic Acids Res. 2023, 51, 595–609. [Google Scholar] [CrossRef]
- Fatima, U.; Balasubramaniam, D.; Khan, W.A.; Kandpal, M.; Vadassery, J.; Arockiasamy, A.; Senthil-Kumar, M. AtSWEET11 and AtSWEET12 transporters function in tandem to modulate sugar flux in plants. Plant Direct. 2023, 7, e481. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, D.; Miao, Q.; Yang, J.; Xuan, Y.; Hu, Y. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.L.; Ma, J.F.; Shou, H. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, T.; Pan, Y.; Zhang, C.; Chen, L.; Ji, B.; Yang, Q.; Xiong, F.; Zhuang, W. Identification and Characterization of SWEET Gene Family in Peanuts and the Role of AhSWEET50 in Sugar Accumulation. Agronomy 2025, 15, 1149. https://doi.org/10.3390/agronomy15051149
Cai T, Pan Y, Zhang C, Chen L, Ji B, Yang Q, Xiong F, Zhuang W. Identification and Characterization of SWEET Gene Family in Peanuts and the Role of AhSWEET50 in Sugar Accumulation. Agronomy. 2025; 15(5):1149. https://doi.org/10.3390/agronomy15051149
Chicago/Turabian StyleCai, Tiecheng, Yijing Pan, Chong Zhang, Lang Chen, Biaojun Ji, Qiang Yang, Faqian Xiong, and Weijian Zhuang. 2025. "Identification and Characterization of SWEET Gene Family in Peanuts and the Role of AhSWEET50 in Sugar Accumulation" Agronomy 15, no. 5: 1149. https://doi.org/10.3390/agronomy15051149
APA StyleCai, T., Pan, Y., Zhang, C., Chen, L., Ji, B., Yang, Q., Xiong, F., & Zhuang, W. (2025). Identification and Characterization of SWEET Gene Family in Peanuts and the Role of AhSWEET50 in Sugar Accumulation. Agronomy, 15(5), 1149. https://doi.org/10.3390/agronomy15051149






