Transcriptome Profiling Reveals Mungbean Defense Mechanisms Against Powdery Mildew
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Methods
2.2.1. Phenotyping of SUPER5 and CN84-1 Inoculated with Sphaerotheca phaseoli
2.2.2. Sample Collection and RNA Isolation
2.2.3. RNA Library Constructing, RNA Sequencing, and Data Analysis
2.2.4. Validation of RNA Sequencing Results by Quantitative Real-Time Polymerase Chain Reaction
3. Results and Discussions
3.1. Differential Responses Between Resistant Mungbean Line SUPER5 and Susceptible Variety CN84-1 to Powdery Mildew Infection Induced by Sphaerotheca phaseoli
3.2. Transcriptome Analysis Using RNA Sequencing Technique
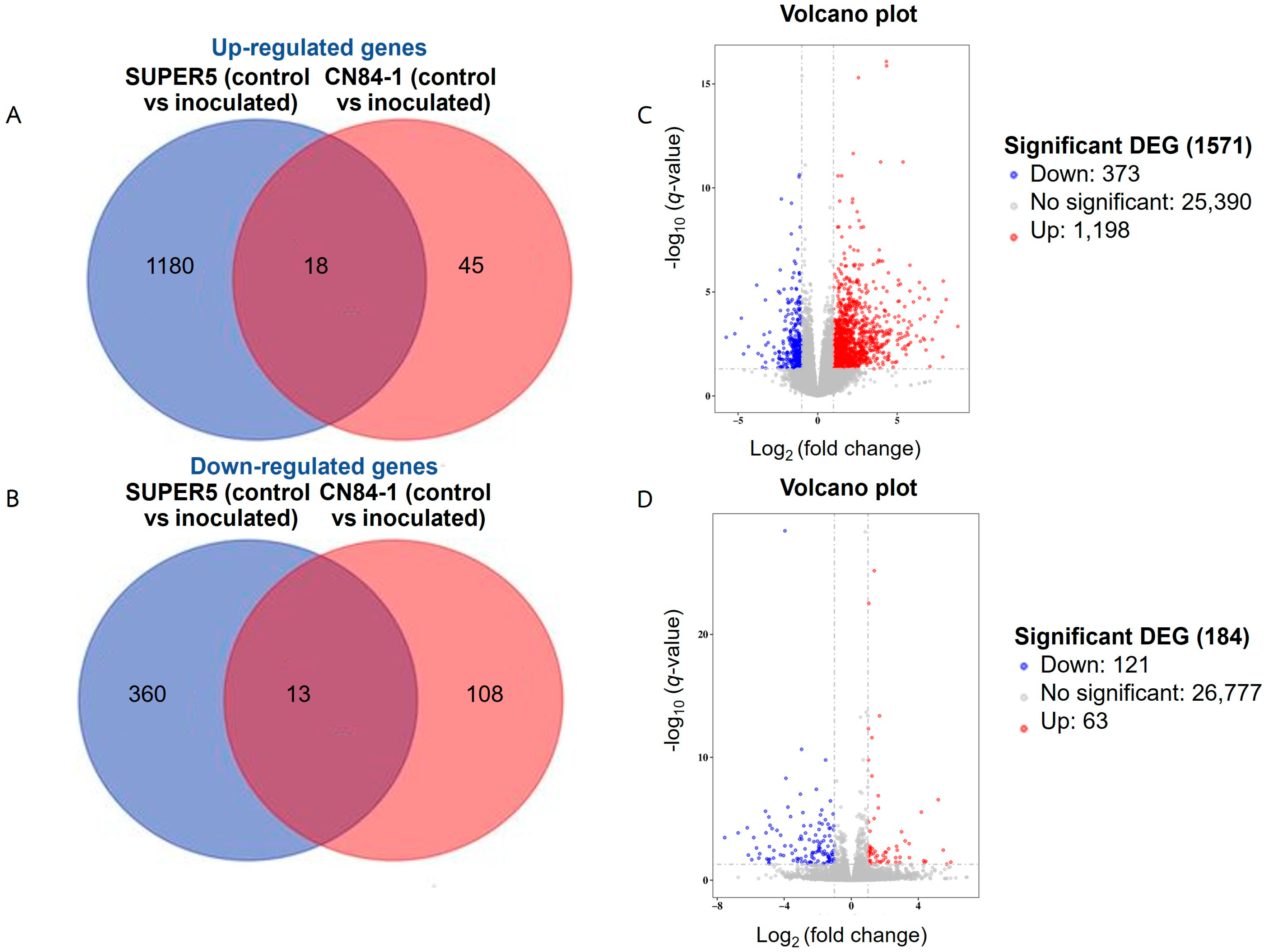
3.2.1. RNA Sequencing Analysis
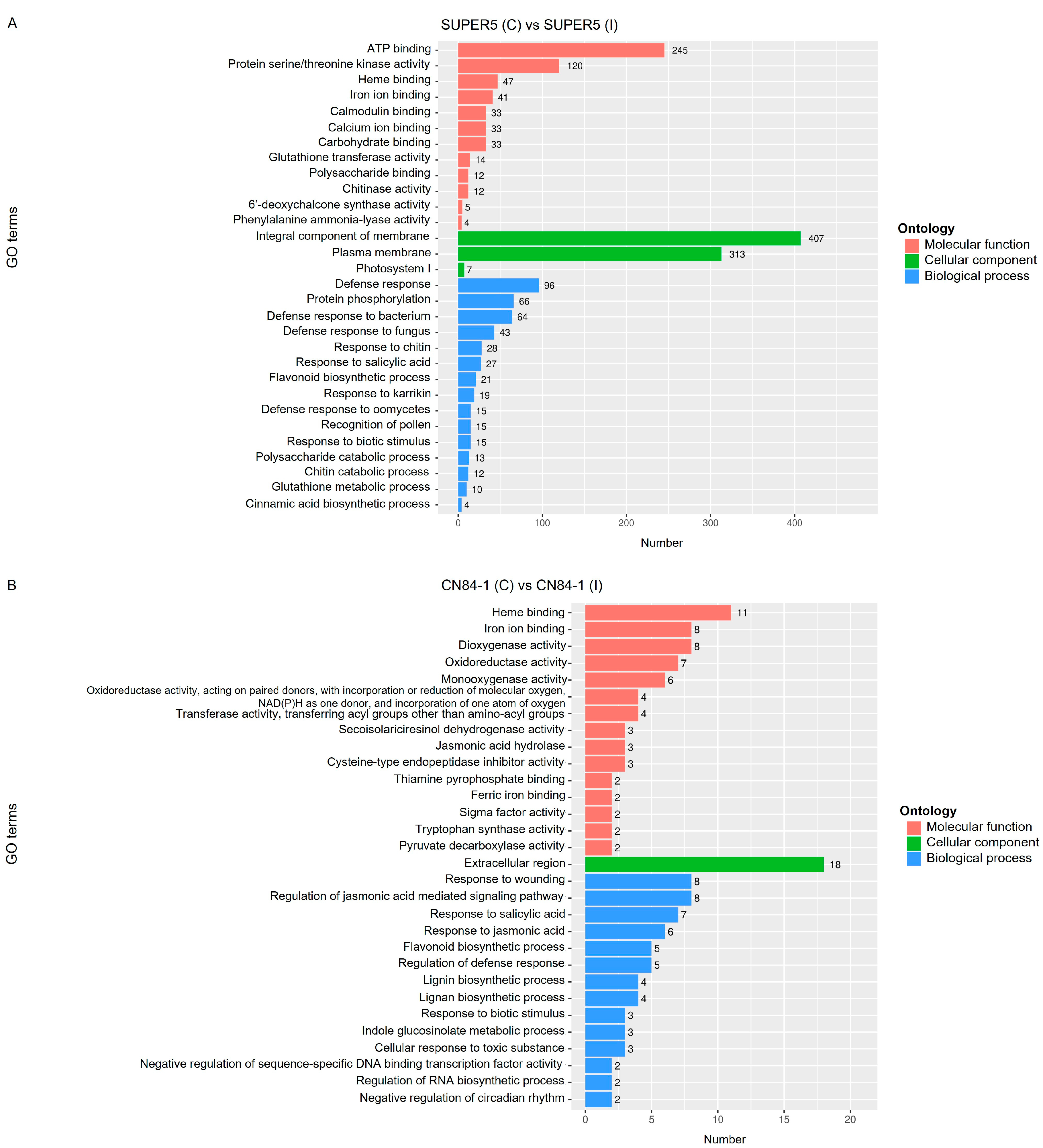
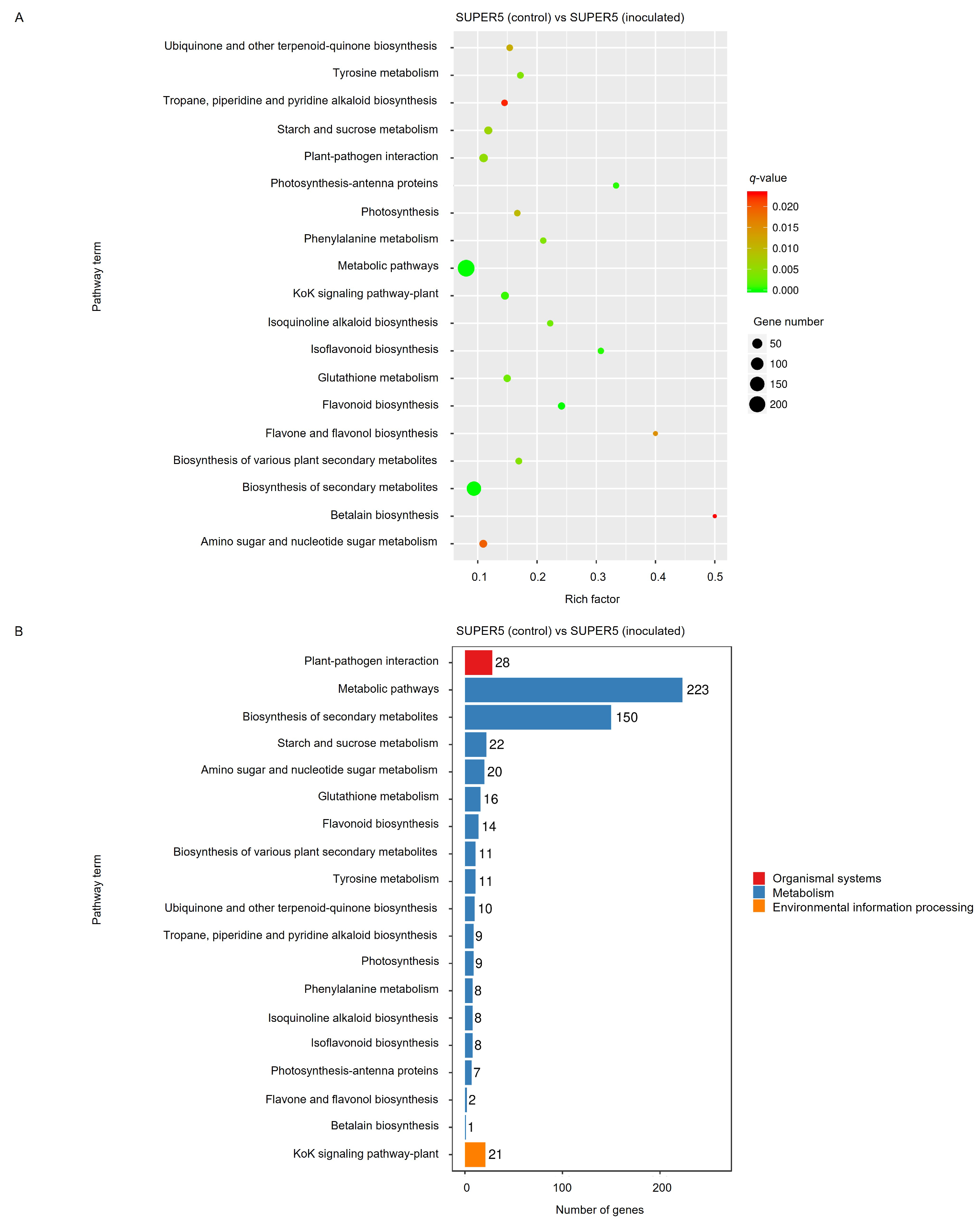
3.2.2. Analysis of Gene Ontology and Kyoto Encyclopedia of Genes and Genome Pathway Enrichment of DEGs Responding to Powdery Mildew Infection in Mungbean
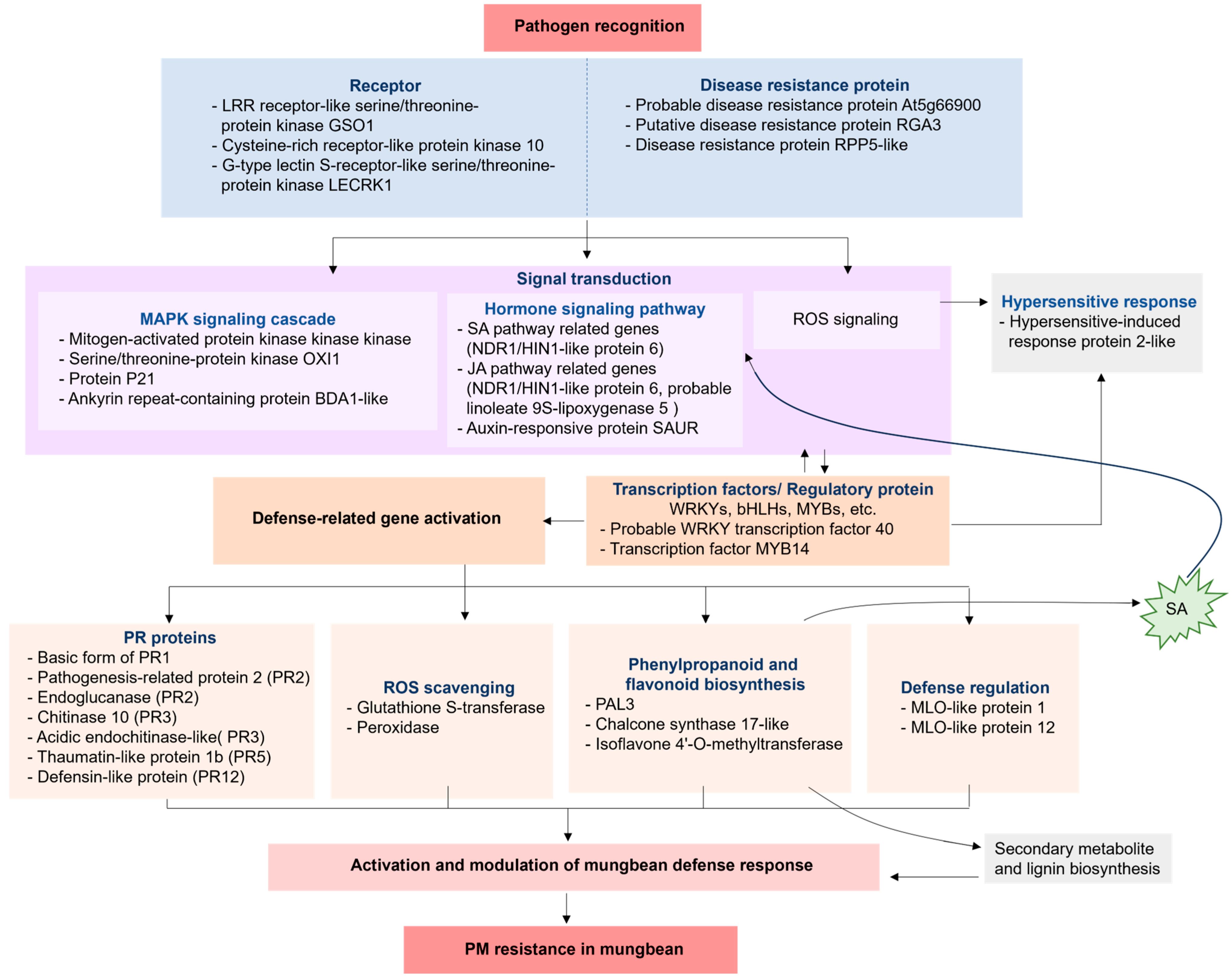
3.2.3. Co-Expression Network Analysis
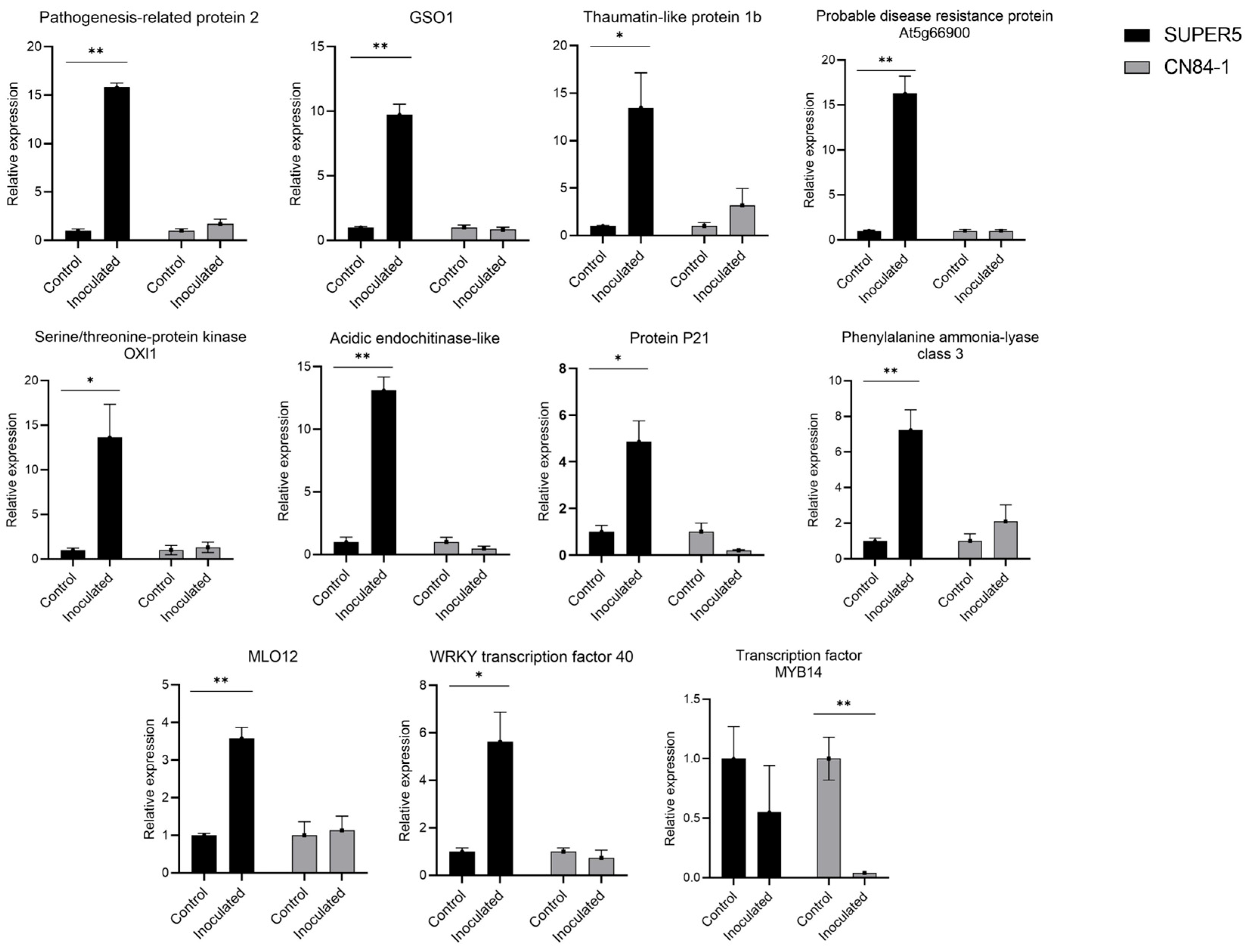
3.2.4. Gene Expression Evaluation Using Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CLS | Cercospora leaf spot |
| PM | Powdery mildew |
| PTI | Pattern-triggered immunity |
| ETI | Effector-triggered immunity |
| RNA-seq | RNA sequencing |
| PR protein | Pathogenesis-related protein |
| SA | Salicylic acid |
| JA | Jasmonic acid |
| ET | Ethylene |
| ROS | Reactive oxygen species |
| DEGs | Differentially expressed genes |
| MYMV | Mungbean Yellow Mosaic Virus |
| DAI | Days after inoculation |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| cDNA | Complementary DNA |
| Ct | Threshold cycle |
| PAL | Phenylalanine ammonia lyase |
| CHS | Chalcone synthase |
| MAPKKK | Mitogen-activated protein kinase kinase kinase |
| GST | Glutathione S-transferases |
| RLKs | Receptor-like kinases |
| PR2 | Pathogenesis-related protein 2 |
| PAMPs | Pathogen-associated molecular patterns |
| TLPs | Thaumatin-like proteins |
| MLO12 | MLO-like protein 12 |
References
- Singh, B.; Singh, J.P.; Shevkani, K.; Singh, N.; Kaur, A. Bioactive constituents in pulses and their health benefits. J. Food Sci. Technol. 2017, 54, 858–870. [Google Scholar] [CrossRef]
- Office of Agricultural Economics. Information of Agricultural Economics 2024 Database. Available online: https://oae.go.th/uploads/files/2025/05/06/13e8089a3f69ea96.pdf. (accessed on 21 June 2025).
- Nair, R.M.; Pandey, A.K.; War, A.R.; Hanumantharao, B.; Shwe, T.; Alam, A.; Pratap, A.; Malik, S.R.; Karimi, R.; Mbeyagala, E.K.; et al. Biotic and abiotic constraints in mungbean production-progress in genetic improvement. Front. Plant Sci. 2019, 10, 1340. [Google Scholar] [CrossRef]
- Sharma, A.; Mehandi, S.; Paudel, P.; Subedi, M. Abiotic and biotic stresses in green gram (Vigna radiata L. Wilczek): A comprehensive review. J. Adv. Biol. Biotechnol. 2024, 27, 940–953. [Google Scholar] [CrossRef]
- Somta, P.; Laosatit, K.; Yuan, X.; Chen, X. Thirty years of mungbean genome research: Where do we stand and what have we learned? Front. Plant Sci. 2022, 13, 944721. [Google Scholar] [CrossRef]
- Pandey, A.K.; Burlakoti, R.R.; Kenyon, L.; Nair, R.M. Perspectives and challenges for sustainable management of fungal diseases of mungbean [Vigna radiata (L.) R. Wilczek var. radiata]: A review. Front. Environ. Sci. 2018, 6, 53. [Google Scholar] [CrossRef]
- Kasettranan, W.; Somta, P.; Srinives, P. Genetics of the resistance to powdery mildew disease in mungbean (Vigna radiata L. Wilczek). J. Crop Sci. Biotechnol. 2009, 12, 37–42. [Google Scholar] [CrossRef]
- Khajudparn, P.; Wongkaew, S.; Thipyapong, P. Mungbean powdery mildew resistance identification of genes for resistance to powdery mildew in mungbean. Afr. Crop Sci. Conf. Proc. 2007, 8, 743–745. [Google Scholar]
- Sulima, A.S.; Zhukov, V.A. War and peas: Molecular bases of resistance to powdery mildew in pea (Pisum sativum L.) and other legumes. Plants 2022, 11, 339. [Google Scholar] [CrossRef]
- Yundaeng, C.; Somta, P.; Chen, J.; Yuan, X.; Chankaew, S.; Srinives, P.; Chen, X. Candidate gene mapping reveals VrMLO12 (MLO Clade II) is associated with powdery mildew resistance in mungbean (Vigna radiata [L]. Wilczek). Plant Sci. 2020, 298, 110594. [Google Scholar] [CrossRef] [PubMed]
- Waengwan, P.; Laosatit, K.; Lin, Y.; Yimram, T.; Yuan, X.; Chen, X.; Somta, P. A cluster of Peronospora parasitica 13-like (NBS-LRR) genes is associated with powdery mildew (Erysiphe polygoni) resistance in mungbean (Vigna radiata). Plants 2024, 13, 1230. [Google Scholar] [CrossRef] [PubMed]
- Papan, P.; Chueakhunthod, W.; Khairum, A.; Siwapitakpong, K.; Chaiyapan, C.; Inthaisong, S.; Jinagool, W.; Tharapreuksapong, A.; Masari, A.; Kaewkasi, C.; et al. Marker-assisted gene pyramiding for powdery mildew resistance in Thai mungbean variety SUT1 by backcross breeding. Plant Mol. Biol. Rep. 2025, 43, 90–102. [Google Scholar] [CrossRef]
- Gan, C.-M.; Tang, T.; Zhang, Z.-Y.; Li, M.; Zhao, X.-Q.; Li, S.-Y.; Yan, Y.-W.; Chen, M.-X.; Zhou, X. Unraveling the intricacies of powdery mildew: Insights into colonization, plant defense mechanisms, and future strategies. Int. J. Mol. Sci. 2025, 26, 3513. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Garg, M. Chapter 5—RNA sequencing: A revolutionary tool for transcriptomics. In Advances in Animal Genomics; Academic Press: Cambridge, MA, USA, 2021; pp. 61–73. [Google Scholar] [CrossRef]
- Cao, Y.; Diao, Q.; Lu, S.; Zhang, Y.; Yao, D. Comparative transcriptomic analysis of powdery mildew resistant and susceptible melon inbred lines to identify the genes involved in the response to Podosphaera xanthii infection. Sci. Hortic. 2022, 304, 111305. [Google Scholar] [CrossRef]
- Zhao, Z.; Dong, Y.; Wang, J.; Zhang, G.; Zhang, Z.; Zhang, A.; Wang, Z.; Ma, P.; Li, Y.; Zhang, X.; et al. Comparative transcriptome analysis of melon (Cucumis melo L.) reveals candidate genes and pathways involved in powdery mildew resistance. Sci. Rep. 2022, 12, 4936. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J.; et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Basak, P.; Gurjar, M.S.; Kumar, T.P.J.; Kashyap, N.; Singh, D.; Jha, S.K.; Saharan, M.S. Transcriptome analysis of Bipolaris sorokiniana–Hordeum vulgare provides insights into mechanisms of host-pathogen interaction. Front. Microbiol. 2024, 15, 1360571. [Google Scholar] [CrossRef]
- Poretti, M.; Sotiropoulos, A.G.; Graf, J.; Jung, E.; Bourras, S.; Krattinger, S.G.; Wicker, T. Comparative transcriptome analysis of wheat lines in the field reveals multiple essential biochemical pathways suppressed by obligate pathogens. Front. Plant Sci. 2021, 12, 720462. [Google Scholar] [CrossRef]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef]
- Bi, Q.; Lu, F.; Wu, J.; Liu, X.; Han, X.; Zhao, J. The control effect and induced disease resistance mechanism of Bacillus tequilensis on wheat powdery mildew. Biol. Control 2025, 201, 105698. [Google Scholar] [CrossRef]
- Sabet, H.; Asadi-Gharneh, H.A.; Nasr-Esfahani, M. Transcriptome-oxidoreductases analysis associated with resistance to pumkin powdery mildew. Sci. Hortic. 2025, 345, 114122. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef] [PubMed]
- Siwapithakpong, K.; Chueakhunthod, W.; Khairum, A.; Tharapreuksapong, A.; Chantakot, T.; Woramit, T.; Tantasawat, P.A. Evaluation of powdery mildew resistance and agronomic performance of pyramided mungbean lines from marker assisted selection. Chiang Mai J. Sci. 2023, 50, e2023037. [Google Scholar] [CrossRef]
- Reyad, N.E.-H.A.; Azoz, S.N.; Ali, A.M.; Sayed, E.G. Mitigation of powdery mildew disease by integrating biocontrol agents and shikimic acid with modulation of antioxidant defense system, anatomical characterization, and improvement of squash plant productivity. Horticulturae 2022, 8, 1145. [Google Scholar] [CrossRef]
- Levesque, R.; SPSS Inc. Programming and Data Management, 3rd ed.; SPSS Institute: Somers, NY, USA, 2006. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Piromyou, P.; Nguyen, H.P.; Songwattana, P.; Boonchuen, P.; Teamtisong, K.; Tittabutr, P.; Boonkerd, N.; Tantasawat, P.A.; Göttfert, M.; Okazaki, S.; et al. The Bradyrhizobium diazoefficiens type III effector NopE modulates the regulation of plant hormones towards nodulation in Vigna radiata. Sci. Rep. 2021, 11, 16604. [Google Scholar] [CrossRef] [PubMed]
- Jaree, P.; Boonchuen, P.; Thawonsuwan, J.; Kondo, H.; Hirono, I.; Somboonwiwat, K. Transcriptome profiling reveals the novel immunometabolism-related genes against WSSV infection from Fenneropenaeus merguiensis. Fish Shellfish Immunol. 2022, 120, 31–44. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.L.; Yang, X.M.; Cao, X.L.; Wang, Y.G.; Ma, X.F.; Chandran, V.; Fan, J.; Yang, H.; Shang, J.; et al. Transcriptome analysis reveals pathways facilitating the growth of tobacco powdery mildew in Arabidopsis. Phytopathol. Res. 2019, 1, 7. [Google Scholar] [CrossRef]
- Huang, X.; Li, Y.; Du, J.; Liang, Y.; Han, H.; Gao, C.; Zhao, Y. Transcriptomic analysis and identification of candidate genes involved in rhizome developmentin Agropyron michnoi. Agronomy 2025, 15, 674. [Google Scholar] [CrossRef]
- Inthaisong, S.; Boonchuen, P.; Jaichopsanthia, T.; Songwattana, P.; Khairum, A.; Chueakhunthod, W.; Tharapreuksapong, A.; Tittabutr, P.; Teaumroong, N.; Tantasawat, P.A. Insights into mungbean defense response to Cercospora leaf spot based on transcriptome analysis. Sci. Rep. 2025, 15, 1334. [Google Scholar] [CrossRef]
- Mackey, D.; Holt, B.F.; Wiig, A.; Dangl, J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhu, Y.; Leng, Y.; Yang, L.; Zhang, B.; Yang, J.; Zhang, X.; Lan, H.; Tang, H.; Chen, J.; et al. Transcriptomic analysis reveals candidate genes responding maize gray leaf spot caused by Cercospora zeina. Plants 2021, 10, 2257. [Google Scholar] [CrossRef] [PubMed]
- Binagwa, P.H.; Traore, S.M.; Egnin, M.; Bernard, G.C.; Ritte, I.; Mortley, D.; Kamfwa, K.; He, G.; Bonsi, C. Genome-wide identification of powdery mildew resistance in common bean (Phaseolus vulgaris L.). Front. Genet. 2021, 12, 673069. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ma, X.; Pan, Z.; Kale, S.D.; Song, Y.; King, H.; Zhang, Q.; Presley, C.; Deng, S.; Wei, C.-I.; et al. Comparative genome analyses reveal sequence features reflecting distinct modes of host adaptation between dicot and monocot powdery mildew. BMC Genom. 2018, 19, 705. [Google Scholar] [CrossRef]
- Wu, P.; Hu, J.; Zou, J.; Qiu, D.; Qu, Y.; Li, Y.; Li, T.; Zhang, H.; Yang, L.; Liu, H.; et al. Fine mapping of the wheat powdery mildew resistance gene Pm52 using comparative genomics analysis and the Chinese Spring reference genomic sequence. Theor. Appl. Genet. 2019, 132, 1451–1461. [Google Scholar] [CrossRef]
- Li, H.; Dong, Z.; Ma, C.; Tian, X.; Xiang, Z.; Xia, Q.; Ma, P.; Liu, W. Discovery of powdery mildew resistance gene candidates from Aegilops biuncialis chromosome 2Mb based on transcriptome sequencing. PLoS ONE 2019, 14, e0220089. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, C.; Zhang, L.; Huang, J.; Dang, C.; Xie, C.; Wang, Z. TaRPP13-3, a CC-NBS-LRR-like gene located on chr 7D, promotes disease resistance to wheat powdery mildew in Brock. J. Phytopathol. 2020, 168, 688–699. [Google Scholar] [CrossRef]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar] [CrossRef]
- Liu, C.; Peng, H.; Li, X.; Liu, C.; Lv, X.; Wei, X.; Zou, A.; Zhang, J.; Fan, G.; Ma, G.; et al. Genome-wide analysis of NDR1/HIN1-like genes in pepper (Capsicum annuum L.) and functional characterization of CaNHL4 under biotic and abiotic stresses. Hortic. Res. 2020, 7, 93. [Google Scholar] [CrossRef]
- Vellosillo, T.; Aguilera, V.; Marcos, R.; Bartsch, M.; Vicente, J.; Cascón, T.; Hamberg, M.; Castresana, C. Defense activated by 9-lipoxygenase-derived oxylipins requires specific mitochondrial proteins. Plant Physiol. 2013, 161, 617–627. [Google Scholar] [CrossRef]
- Mo, F.; Cui, J.; Li, C.; Zhang, Y.; Xue, X.; Cheng, M.; Lv, R.; Meng, F.; He, X.; Chen, X.; et al. Identification of the small auxin up-regulated RNA in tomato and investigation of SlSAUR50 as a positive regulator of tomato resistance to Botrytis cinerea. Int. J. Biol. Macromol. 2025, 306, 141738. [Google Scholar] [CrossRef]
- Rentel, M.C.; Lecourieux, D.; Ouaked, F.; Usher, S.L.; Lindsay, P.; Okamoto, H.; Knight, H.; Peck, S.C.; Grierson, C.S.; Hirt, H.; et al. OXI1 kinase is necessary for oxidative burst-mediated signaling in Arabidopsis. Nature 2004, 427, 858–861. [Google Scholar] [CrossRef]
- Adhikari, B.; Pradhan, B.; Parajuli, S.; Nepal, M.P. Genome-wide identification of WRKY transcription factors in Amborella trichopoda, the basal flowering plant species. Monocytomics 2024, 1, 2890. Available online: https://www.researchgate.net/publication/381769204_Genome-wide_identification_of_WRKY_transcription_factors_in_Amborella_trichopoda_the_basal_flowering_plant_species (accessed on 1 December 2023).
- Zeng, Y.; Song, H.; Xia, L.; Yang, L.; Zhang, S. The responses of poplars to fungal pathogens: A review of the defensive pathway. Front. Plant Sci. 2023, 14, 1107583. [Google Scholar] [CrossRef]
- Mapuranga, J.; Chang, J.; Yang, W. Combating powdery mildew: Advances in molecular interactions between Blumeria graminis f. sp. tritici and wheat. Front. Plant Sci. 2022, 13, 1102908. [Google Scholar] [CrossRef]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef]
- Ferreira, R.B.; Monteiro, S.; Freitas, R.; Santos, C.N.; Chen, Z.; Batista, L.M.; Duarte, J.; Borges, A.; Teixeira, A.R. The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 2007, 8, 677–700. [Google Scholar] [CrossRef]
- Meyer, J.; Berger, D.K.; Christensen, S.A.; Murray, S.L. RNA-seq analysis of resistant and susceptible sub-tropical maize lines reveals a role for kauralexins in resistance to grey leaf spot disease, caused by Cercospora zeina. BMC Plant Biol. 2017, 17, 197. [Google Scholar] [CrossRef]
- Feng, L.; Wei, S.; Li, Y. Thaumatin-like proteins in legumes: Functions and potential applications-A Review. Plants 2024, 13, 1124. [Google Scholar] [CrossRef]
- Chakraborty, J.; Ghosh, P.; Sen, S.; Das, S. Epigenetic and transcriptional control of chickpea WRKY40 promoter activity under Fusarium stress and its heterologous expression in Arabidopsis leads to enhanced resistance against bacterial pathogen. Plant Sci. 2018, 276, 250–267. [Google Scholar] [CrossRef]
- Yang, J.; Wang, Q.; Luo, H.; He, C.; An, B. HbWRKY40 plays an important role in the regulation of pathogen resistance in Hevea brasiliensis. Plant Cell Rep. 2020, 39, 1095–1107. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef]
- Devoto, A.; Hartmann, H.A.; Piffanelli, P.; Elliott, C.; Simmons, C.; Taramino, G.; Goh, C.S.; Cohen, F.E.; Emerson, B.C.; Schulze- Lefert, P.; et al. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 2003, 56, 77–88. [Google Scholar] [CrossRef]
- Höll, J.; Vannozzi, A.; Czemmel, S.; D’Onofrio, C.; Walker, A.R.; Rausch, T.; Lucchin, M.; Boss, P.K.; Dry, I.B.; Bogs, J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 2013, 25, 4135–4149. [Google Scholar] [CrossRef]
- Saha, B.; Nayak, J.; Srivastava, R.; Samal, S.; Kumar, D.; Chanwala, J.; Dey, N.; Giri, M.K. Unraveling the involvement of WRKY TFs in regulating plant disease defense signaling. Planta 2024, 259, 7. [Google Scholar] [CrossRef]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic acid biosynthesis in plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef]
- Xu, Z.; Guo, Z.; Wu, F.; Zhang, Y.; Zhao, Y.; Han, L.; Gao, P.; Zhu, H.; Xu, Q.; Zhao, X.; et al. Transcriptomic analysis reveals the molecular defense mechanisms of Poa pratensis against powdery mildew fungus Blumeria graminis f. sp. Poae. Agron 2024, 14, 2543. [Google Scholar] [CrossRef]
- Ticli, G.; Cazzalini, O.; Stivala, L.A.; Prosperi, E. Revisiting the function of p21CDKN1A in DNA repair: The influence of protein interactions and stability. Int. J. Mol. Sci. 2022, 23, 7058. [Google Scholar] [CrossRef]



| Days Post Inoculation | SUPER5 | CN84-1 | ||
|---|---|---|---|---|
| Severity Score | Response 3 | Severity Score | Response | |
| 0 | 1.00 ± 0.00 2 | R | 1.00 ± 0.00 c 2 | R |
| 3 | 1.00 ± 0.00 | R | 1.00 ± 0.00 c | R |
| 6 | 1.00 ± 0.00 | R | 1.00 ± 0.00 c | R |
| 11 | 1.00 ± 0.00 | R | 2.50 ± 0.29 b | MR |
| 16 | 1.33 ± 0.33 | R | 5.00 ± 0.00 a | S |
| F-test 1 | ns | ** | ||
| Samples | Total Sequences | Sequence Length | GC Content (%) | |||
|---|---|---|---|---|---|---|
| Raw Read | Clean Read | Raw Read | Clean Read | Raw Read | Clean Read | |
| CN84-1 (control)-1 | 51,054,240 | 50,532,360 | 150 | 144.12 | 44.18 | 43.86 |
| CN84-1 (control)-2 | 43,313,750 | 42,895,648 | 150 | 144.83 | 44.00 | 43.75 |
| CN84-1 (control)-3 | 43,260,030 | 42,713,682 | 150 | 144.38 | 44.22 | 43.92 |
| CN84-1 (inoculated)-1 | 42,844,270 | 42,383,432 | 150 | 145.22 | 44.14 | 43.87 |
| CN84-1 (inoculated)-2 | 43,655,424 | 43,103,460 | 150 | 144.26 | 44.29 | 43.94 |
| CN84-1 (inoculated)-3 | 45,585,534 | 45,125,966 | 150 | 144.20 | 44.13 | 43.79 |
| SUPER5 (control)-1 | 43,307,702 | 42,866,758 | 150 | 144.70 | 43.62 | 43.31 |
| SUPER5 (control)-2 | 40,001,488 | 39,660,530 | 150 | 145.72 | 43.65 | 43.43 |
| SUPER5 (control)-3 | 47,896,438 | 47,366,050 | 150 | 145.23 | 43.93 | 43.65 |
| SUPER5 (inoculated)-1 | 43,388,166 | 42,902,528 | 150 | 143.91 | 44.21 | 43.87 |
| SUPER5 (inoculated)-2 | 41,040,500 | 40,562,014 | 150 | 143.13 | 43.75 | 43.33 |
| SUPER5 (inoculated)-3 | 50,155,254 | 49,657,154 | 150 | 144.32 | 44.19 | 43.91 |
| Total | 535,502,796 | 529,769,582 | - | - | - | - |
| Gene ID | Description | Gene Expression (Log2 Fold Change) | |
|---|---|---|---|
| SUPER5 (C) vs. SUPER5 (I) | CN84-1 (C) vs. CN84-1 (I) | ||
| LOC106773636 | Pathogenesis-related protein 2 | 7.8936 | ns |
| LOC106761498 | LRR receptor-like serine/threonine-protein kinase GSO1 | 6.4630 | ns |
| LOC106771405 | Thaumatin-like protein 1b | 5.7996 | ns |
| LOC106766245 | Probable disease resistance protein At5g66900 | 5.7650 | ns |
| LOC106758248 | Serine/threonine-protein kinase OXI1 | 5.5646 | ns |
| LOC106755727 | Acidic endochitinase-like | 5.4741 | ns |
| LOC106776559 | Cysteine-rich receptor-like protein kinase 10 | 5.0755 | ns |
| LOC106777376 | Protein P21 | 5.0029 | ns |
| LOC106777264 | G-type lectin S-receptor-like serine/threonine-protein kinase LECRK1 | 4.4129 | ns |
| LOC106770081 | Putative disease resistance protein RGA3 | 4.1250 | ns |
| LOC106764951 | Phenylalanine ammonia lyase class 3 | 3.6704 | ns |
| LOC106761315 | Ankyrin repeat-containing protein BDA1-like | 3.4511 | ns |
| LOC106780426 | MLO-like protein 12 | 3.0627 | ns |
| LOC106764381 | Probable WRKY transcription factor 40 | 3.8125 | ns |
| LOC106755803 | Transcription factor MYB14 | ns | −4.7250 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inthaisong, S.; Boonchuen, P.; Tharapreuksapong, A.; Tittabutr, P.; Teaumroong, N.; Tantasawat, P.A. Transcriptome Profiling Reveals Mungbean Defense Mechanisms Against Powdery Mildew. Agronomy 2025, 15, 1871. https://doi.org/10.3390/agronomy15081871
Inthaisong S, Boonchuen P, Tharapreuksapong A, Tittabutr P, Teaumroong N, Tantasawat PA. Transcriptome Profiling Reveals Mungbean Defense Mechanisms Against Powdery Mildew. Agronomy. 2025; 15(8):1871. https://doi.org/10.3390/agronomy15081871
Chicago/Turabian StyleInthaisong, Sukanya, Pakpoom Boonchuen, Akkawat Tharapreuksapong, Panlada Tittabutr, Neung Teaumroong, and Piyada Alisha Tantasawat. 2025. "Transcriptome Profiling Reveals Mungbean Defense Mechanisms Against Powdery Mildew" Agronomy 15, no. 8: 1871. https://doi.org/10.3390/agronomy15081871
APA StyleInthaisong, S., Boonchuen, P., Tharapreuksapong, A., Tittabutr, P., Teaumroong, N., & Tantasawat, P. A. (2025). Transcriptome Profiling Reveals Mungbean Defense Mechanisms Against Powdery Mildew. Agronomy, 15(8), 1871. https://doi.org/10.3390/agronomy15081871







