Bridging Genes and Sensory Characteristics in Legumes: Multi-Omics for Sensory Trait Improvement
Abstract
1. Introduction
2. Appearance
2.1. Seed-Coat Colour
2.1.1. Genetics of Legume Seed-Coat Pigmentation
2.1.2. Unveiling the Seed-Coat Colour Using a Multi-Omics Lens
2.1.3. Other Factors Affecting Seed-Coat Colour
2.2. Seed Size and Shape
2.2.1. Genes Regulating Seed Size and Shape in Legumes
2.2.2. A Multi-Omics Exploration of Legume Seed Size and Shape
3. Aroma
3.1. Genetic Regulation of Aroma in Legumes
3.2. Omics Studies to Decode Aromatic Traits in Legumes
4. Taste and Flavour
4.1. Genetic Determinants of Taste and Flavour in Legumes
4.2. Omics Studies Exploring Taste and Flavour Profiles in Legumes
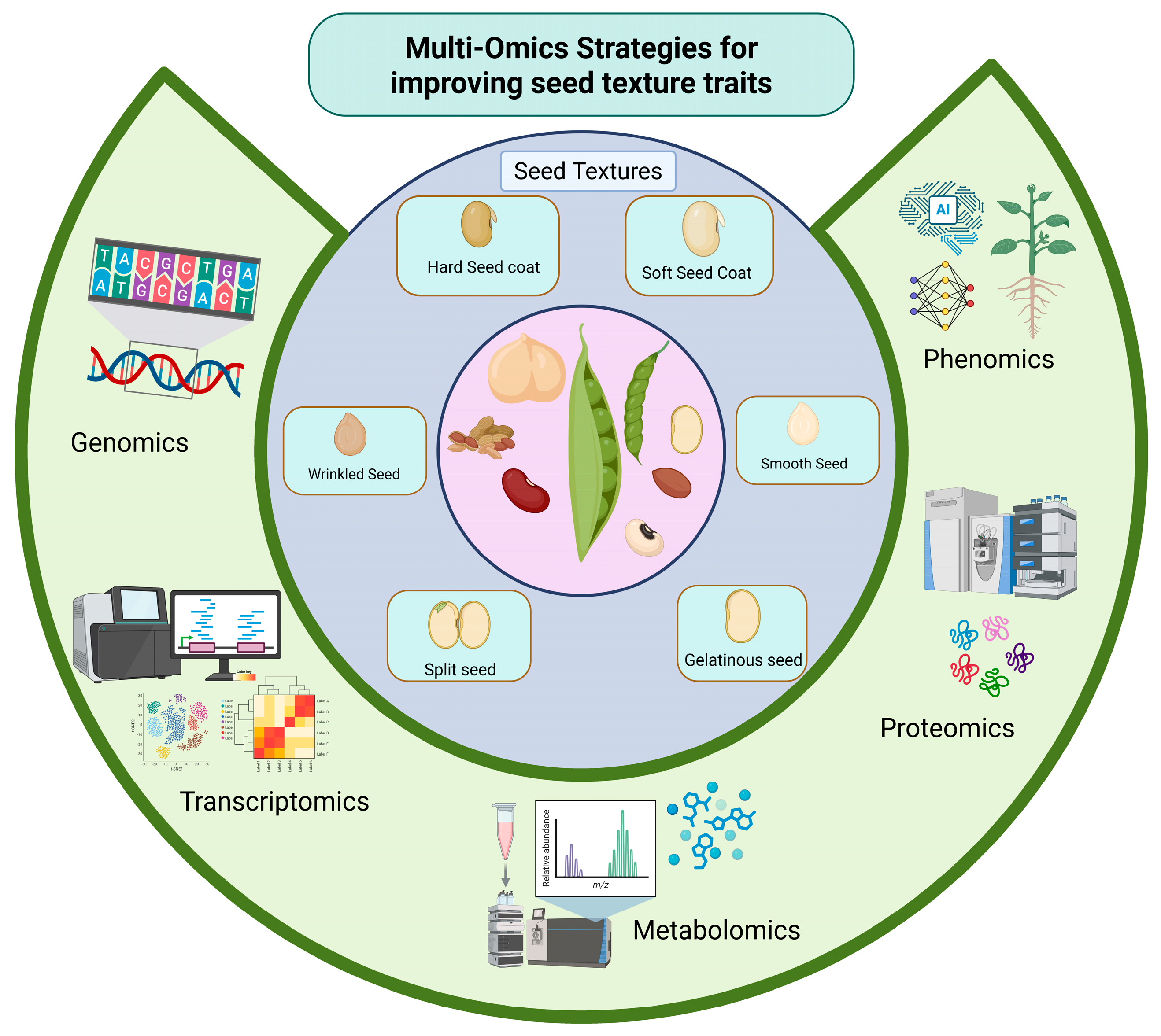
5. Texture
5.1. Genetic Basis of Seed Texture
5.2. The Multi-Omics Approaches in Legumes Reveal Seed Structure and Texture Properties
6. Palatability
Genetic Factors and Multi-Omics Approaches to Characterise Palatability
7. Current Challenges
8. Conclusions and Future Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Raina, A.; Laskar, R.A.; Khan, S.; Tomlekova, N.B.; Ravelombola, W.; Thudi, M. Editorial: Legume breeding in transition: Innovation and outlook. Front. Genet. 2023, 14, 1221551. [Google Scholar] [CrossRef]
- Geraldo, R.; Santos, C.S.; Pinto, E.; Vasconcelos, M.W. Widening the Perspectives for Legume Consumption: The Case of Bioactive Non-nutrients. Front. Plant Sci. 2022, 13, 772054. [Google Scholar] [CrossRef]
- Polak, R.; Phillips, E.M.; Campbell, A. Legumes: Health Benefits and Culinary Approaches to Increase Intake. Clin. Diabetes 2015, 33, 198–205. [Google Scholar] [CrossRef]
- Rizkalla, S.W.; Bellisle, F.; Slama, G. Health benefits of low glycaemic index foods, such as pulses, in diabetic patients and healthy individuals. Br. J. Nutr. 2002, 88, S255–S262. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.K.; Dakora, F.D. Seed-Coat Pigmentation Plays a Crucial Role in Partner Selection and N(2) Fixation in Legume-Root-Microbe Associations in African Soils. Plants 2024, 13, 1464. [Google Scholar] [CrossRef]
- Mathesius, U. Are legumes different? Origins and consequences of evolving nitrogen fixing symbioses. J. Plant Physiol. 2022, 276, 153765. [Google Scholar] [CrossRef]
- Majidian, P. Legume breeding: From conventional method to modern technique. In Legumes Research-Volume 1; IntechOpen: London, UK, 2022. [Google Scholar]
- Biswas, M.K.; Patil, A.; Sunkad, G. Enhancing Legume Cultivars through Agronomy, Breeding, and Genetics. Agronomy 2023, 13, 1035. [Google Scholar] [CrossRef]
- Ahn, E.; Botkin, J.; Curtin, S.J.; Zsogon, A. Ideotype breeding and genome engineering for legume crop improvement. Curr. Opin. Biotechnol. 2023, 82, 102961. [Google Scholar] [CrossRef]
- Appleton, K.M. The importance of enjoyment, sensory properties and perceived cooking abilities in legume and pulse consumption: A questionnaire study. Public Health Nutr. 2024, 27, e138. [Google Scholar] [CrossRef] [PubMed]
- Lawless, H.T.; Heymann, H. Sensory Evaluation of Food: Principles and Practices; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Sadohara, R.; Long, Y.F.; Izquierdo, P.; Urrea, C.A.; Morris, D.; Cichy, K. Seed coat color genetics and genotype x environment effects in yellow beans via machine-learning and genome-wide association. Plant Genome 2022, 15, e20173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jia, R.; Liu, L.; Shen, W.; Fang, Z.; Zhou, B.; Liu, B. Seed coat colour and structure are related to the seed dormancy and overwintering ability of crop-to-wild hybrid soybean. AoB Plants 2023, 15, plad081. [Google Scholar] [CrossRef]
- Saffarionpour, S. Off-flavors in pulses and grain legumes and processing approaches for controlling flavor-plant protein interaction: Application prospects in plant-based alternative foods. Food Bioprocess. Technol. 2024, 17, 1141–1182. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.P.; Saliba, A.J.; Carr, B.T.; Blanchard, C.L.; Wood, J.A.; Prenzler, P.D. Sensory profiling and preference mapping of Australian puffed desi chickpeas. LWT-Food Sci. Technol. 2018, 89, 229–236. [Google Scholar] [CrossRef]
- Kou, X.H.; Gao, J.; Xue, Z.H.; Zhang, Z.J.; Wang, H.; Wang, X. Purification and identification of antioxidant peptides from chickpea (Cicer arietinum L.) albumin hydrolysates. LWT-Food Sci. Technol. 2013, 50, 591–598. [Google Scholar] [CrossRef]
- Huamaní-Perales, C.; Vidaurre-Ruiz, J.; Salas-Valerio, W.; Cabezas, D.M.; Repo-Carrasco-Valencia, R. A review of techno-functional properties of legume proteins and their potential for development of new products. Eur. Food Res. Technol. 2024, 250, 2069–2092. [Google Scholar] [CrossRef]
- Cichonska, P.; Kostyra, E.; Piotrowska, A.; Scibisz, I.; Roszko, M.; Ziarno, M. Enhancing the sensory and nutritional properties of bean-based and lentil-based beverages through fermentation and germination. LWT-Food Sci. Technol. 2024, 199, 116140. [Google Scholar] [CrossRef]
- Duxbury, D. Laboratory-Sensory Evaluation Provides Value. Food Technol. 2005, 59, 68–73. [Google Scholar]
- Schmidt, H.D.; de Oliveira, V.R. Overview of the Incorporation of Legumes into New Food Options: An Approach on Versatility, Nutritional, Technological, and Sensory Quality. Foods 2023, 12, 2586. [Google Scholar] [CrossRef] [PubMed]
- Montejano-Ramírez, V.; Valencia-Cantero, E. The Importance of Lentils: An Overview. Agriculture 2024, 14, 103. [Google Scholar] [CrossRef]
- Roorkiwal, M.; Bharadwaj, C.; Barmukh, R.; Dixit, G.P.; Thudi, M.; Gaur, P.M.; Chaturvedi, S.K.; Fikre, A.; Hamwieh, A.; Kumar, S.; et al. Integrating genomics for chickpea improvement: Achievements and opportunities. Theor. Appl. Genet. 2020, 133, 1703–1720. [Google Scholar] [CrossRef]
- Bohra, A.; Tiwari, A.; Kaur, P.; Ganie, S.A.; Raza, A.; Roorkiwal, M.; Mir, R.R.; Fernie, A.R.; Smykal, P.; Varshney, R.K. The Key to the Future Lies in the Past: Insights from Grain Legume Domestication and Improvement Should Inform Future Breeding Strategies. Plant Cell Physiol. 2022, 63, 1554–1572. [Google Scholar] [CrossRef]
- Zhao, P.; Chu, L.; Wang, K.; Zhao, B.; Li, Y.; Yang, K.; Wan, P. Analyses on the pigment composition of different seed coat colors in adzuki bean. Food Sci. Nutr. 2022, 10, 2611–2619. [Google Scholar] [CrossRef]
- Sivasakthi, K.; Marques, E.; Kalungwana, N.; Carrasquilla-Garcia, N.; Chang, P.L.; Bergmann, E.M.; Bueno, E.; Cordeiro, M.; Sani, S.G.A.S.; Udupa, S.M.; et al. Functional Dissection of the Chickpea Stay-Green Phenotype Associated with Molecular Variation at an Ortholog of Mendel’s I Gene for Cotyledon Color: Implications for Crop Production and Carotenoid Biofortification. Int. J. Mol. Sci. 2019, 20, 5562. [Google Scholar] [CrossRef] [PubMed]
- Pal, L.; Dwivedi, V.; Gupta, S.K.; Saxena, S.; Pandey, A.; Chattopadhyay, D. Biochemical analysis of anthocyanin and proanthocyanidin and their regulation in determining chickpea flower and seed coat colour. J. Exp. Bot. 2023, 74, 130–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, W.; Huang, Y.; Zhao, P.; Yang, K.; Wan, P.; Chu, L. Progress in Adzuki Bean Seed Coat Colour Studies. Plants 2023, 12, 3242. [Google Scholar] [CrossRef]
- McClean, P.E.; Roy, J.; Colbert, C.L.; Osborne, C.; Lee, R.; Miklas, P.N.; Osorno, J.M. T and Z, partial seed coat patterning genes in common bean, provide insight into the structure and protein interactions of a plant MBW complex. G3 (Bethesda) 2024, 14, jkae184. [Google Scholar] [CrossRef]
- McClean, P.E.; Lee, R.K.; Otto, C.; Gepts, P.; Bassett, M.J. Molecular and phenotypic mapping of genes controlling seed coat pattern and color in common bean (Phaseolus vulgaris L.). J. Hered. 2002, 93, 148–152. [Google Scholar] [CrossRef]
- Parker, T.; Bolt, T.; Williams, T.; Penmetsa, R.V.; Mulube, M.; Celebioglu, B.; Palkovic, A.; Jochua, C.N.; Del Mar Rubio Wilhelmi, M.; Lo, S.; et al. Seed color patterns in domesticated common bean are regulated by MYB-bHLH-WD40 transcription factors and temperature. Plant J. 2024, 119, 2765–2781. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, J.H.; Zabala, G.; Varala, K.; Hudson, M.; Vodkin, L.O. Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in Glycine max seed coats. Plant Cell 2009, 21, 3063–3077. [Google Scholar] [CrossRef]
- Tuteja, J.H.; Clough, S.J.; Chan, W.C.; Vodkin, L.O. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 2004, 16, 819–835. [Google Scholar] [CrossRef]
- McClean, P.E.; Bett, K.E.; Stonehouse, R.; Lee, R.; Pflieger, S.; Moghaddam, S.M.; Geffroy, V.; Miklas, P.; Mamidi, S. White seed color in common bean (Phaseolus vulgaris) results from convergent evolution in the P (pigment) gene. New Phytol. 2018, 219, 1112–1123. [Google Scholar] [CrossRef]
- Penmetsa, R.V.; Carrasquilla-Garcia, N.; Bergmann, E.M.; Vance, L.; Castro, B.; Kassa, M.T.; Sarma, B.K.; Datta, S.; Farmer, A.D.; Baek, J.M.; et al. Multiple post-domestication origins of chickpea through allelic variation in a diversification-associated transcription factor. New Phytol. 2016, 211, 1440–1451. [Google Scholar] [CrossRef]
- Mirali, M.; Purves, R.W.; Stonehouse, R.; Song, R.; Bett, K.; Vandenberg, A. Genetics and Biochemistry of Zero-Tannin Lentils. PLoS ONE 2016, 11, e0164624. [Google Scholar] [CrossRef]
- Gutierrez, N.; Torres, A.M. Characterization and diagnostic marker for TTG1 regulating tannin and anthocyanin biosynthesis in faba bean. Sci. Rep. 2019, 9, 16174. [Google Scholar] [CrossRef]
- Gutierrez, N.; Avila, C.M.; Torres, A.M. The bHLH transcription factor VfTT8 underlies zt2, the locus determining zero tannin content in faba bean (Vicia faba L.). Sci. Rep. 2020, 10, 14299. [Google Scholar] [CrossRef]
- Chu, L.W.; Zhao, P.; Huang, X.Q.; Zhao, B.; Li, Y.S.; Yang, K.; Wan, P. Genetic analysis of seed coat colour in adzuki bean (Vigna angularis L.). Plant Genet Resour.-C 2021, 19, 67–73. [Google Scholar] [CrossRef]
- Garcia-Fernandez, C.; Campa, A.; Ferreira, J.J. Dissecting the genetic control of seed coat color in a RIL population of common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2021, 134, 3687–3698. [Google Scholar] [CrossRef]
- Herniter, I.A.; Muñoz-Amatriaín, M.; Lo, S.S.; Guo, Y.N.; Lonardi, S.; Close, T.J. Identification of Candidate Genes Controlling Red Seed Coat Color in Cowpea (Vigna unguiculata [L.] Walp). Horticulturae 2024, 10, 161. [Google Scholar] [CrossRef]
- Lay, L.; Khan, W.; Jo, H.; Kim, S.H.; Kim, Y. Genome-Wide Association Study on Cowpea seed coat color using RGB images. Mol. Breed. 2024, 44, 80. [Google Scholar] [CrossRef]
- Zhang, K.; Yuan, M.; Xia, H.; He, L.; Ma, J.; Wang, M.; Zhao, H.; Hou, L.; Zhao, S.; Li, P. BSA-seq and genetic mapping reveals AhRt2 as a candidate gene responsible for red testa of peanut. Theor. Appl. Genet. 2022, 135, 1529–1540. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Zhang, K.; Ma, J.; Yuan, M.; Zhao, S.; Wang, M.; Deng, L.; Ren, L.; Gangurde, S.S.; Pan, J.; et al. Transcriptional networks orchestrating red and pink testa color in peanut. BMC Plant Biol. 2023, 23, 44. [Google Scholar] [CrossRef]
- Campa, A.; Rodriguez Madrera, R.; Jurado, M.; Garcia-Fernandez, C.; Suarez Valles, B.; Ferreira, J.J. Genome-wide association study for the extractable phenolic profile and coat color of common bean seeds (Phaseolus vulgaris L.). BMC Plant Biol. 2023, 23, 158. [Google Scholar] [CrossRef]
- Plestenjak, E.; Neji, M.; Sinkovic, L.; Meglic, V.; Pipan, B. Genomic insights into genetic diversity and seed coat color change in common bean composite populations. Front. Plant Sci. 2025, 15, 1523745. [Google Scholar] [CrossRef]
- Elessawy, F.M.; Vandenberg, A.; El-Aneed, A.; Purves, R.W. An Untargeted Metabolomics Approach for Correlating Pulse Crop Seed Coat Polyphenol Profiles with Antioxidant Capacity and Iron Chelation Ability. Molecules 2021, 26, 3833. [Google Scholar] [CrossRef]
- Jha, A.B.; Purves, R.W.; Elessawy, F.M.; Zhang, H.X.; Vandenberg, A.; Warkentin, T.D. Polyphenolic Profile of Seed Components of White and Purple Flower Pea Lines. Crop Sci. 2019, 59, 2711–2719. [Google Scholar] [CrossRef]
- Elessawy, F.M.; Wright, D.; Vandenberg, A.; El-Aneed, A.; Purves, R.W. Mass Spectrometry-Based Untargeted Metabolomics Reveals the Importance of Glycosylated Flavones in Patterned Lentil Seed Coats. J. Agric. Food Chem. 2023, 71, 3541–3549. [Google Scholar] [CrossRef]
- Jeon, S.; Kim, B.C.; Ha, J. Tissue-Specific Metabolic Profiling of Mungbean (Vigna radiata L.) Genotypes with Different Seed Coat Colors. J. Food Qual. 2023, 2023, 7555915. [Google Scholar] [CrossRef]
- Desta, K.T.; Hur, O.S.; Lee, S.; Yoon, H.; Shin, M.J.; Yi, J.; Lee, Y.; Ro, N.Y.; Wang, X.; Choi, Y.M. Origin and seed coat color differently affect the concentrations of metabolites and antioxidant activities in soybean (Glycine max (L.) Merrill) seeds. Food Chem. 2022, 381, 132249. [Google Scholar] [CrossRef]
- Zhang, K.; Ma, J.; Gangurde, S.S.; Hou, L.; Xia, H.; Li, N.; Pan, J.; Tian, R.; Huang, H.; Wang, X.; et al. Targeted metabolome analysis reveals accumulation of metabolites in testa of four peanut germplasms. Front. Plant Sci. 2022, 13, 992124. [Google Scholar] [CrossRef] [PubMed]
- Kafer, J.M.; Molinari, M.D.C.; Henning, F.A.; Koltun, A.; Marques, V.V.; Marin, S.R.R.; Nepomuceno, A.L.; Mertz-Henning, L.M. Transcriptional Profile of Soybean Seeds with Contrasting Seed Coat Color. Plants 2023, 12, 1555. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, X.; Wang, W.; Hu, X.; Han, W.; He, Q.; Yang, H.; Xiang, S.; Gai, J. Identifying Wild Versus Cultivated Gene-Alleles Conferring Seed Coat Color and Days to Flowering in Soybean. Int. J. Mol. Sci. 2021, 22, 1559. [Google Scholar] [CrossRef]
- Li, C.; Gao, Z.; Hu, W.; Zhu, X.; Li, Y.; Li, N.; Ma, C. Integration of comparative transcriptomics and WGCNA characterizes the regulation of anthocyanin biosynthesis in mung bean (Vigna radiata L.). Front. Plant Sci. 2023, 14, 1251464. [Google Scholar] [CrossRef]
- Ma, C.; Feng, Y.L.; Zhou, S.; Zhang, J.; Guo, B.B.; Xiong, Y.; Wu, S.W.; Li, Y.; Li, Y.J.; Li, C.X. Metabolomics and transcriptomics provide insights into the molecular mechanisms of anthocyanin accumulation in the seed coat of differently colored mung bean (Vigna radiata L.). Plant Physiol. Biochem. 2023, 200, 107739. [Google Scholar] [CrossRef]
- Wang, Q.; Cao, H.; Wang, J.; Gu, Z.; Lin, Q.; Zhang, Z.; Zhao, X.; Gao, W.; Zhu, H.; Yan, H. Fine-mapping and primary analysis of candidate genes associated with seed coat color in mung bean (Vigna radiata L.). J. Integr. Agric. 2024, 23, 2571–2588. [Google Scholar] [CrossRef]
- Song, J.; Xu, R.; Guo, Q.; Wu, C.; Li, Y.; Wang, X.; Wang, J.; Qiu, L.J. An omics strategy increasingly improves the discovery of genetic loci and genes for seed-coat color formation in soybean. Mol. Breed. 2023, 43, 71. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Ouyang, L.; Yao, R.; He, D.; Han, Z.; Li, W.; Ding, Y.; Wang, Z.; Kang, Y.; et al. Metabolomics combined with transcriptomics analyses of mechanism regulating testa pigmentation in peanut. Front. Plant Sci. 2022, 13, 1065049. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Lei, Y.; Yan, L.; Liu, Y.; Pandey, M.K.; Wan, X.; Varshney, R.K.; Fang, J.; Liao, B. Transcriptome and metabolome reveal redirection of flavonoids in a white testa peanut mutant. BMC Plant Biol. 2020, 20, 161. [Google Scholar] [CrossRef] [PubMed]
- Miernyk, J.A.; Johnston, M.L. Proteomic analysis of the testa from developing soybean seeds. J. Proteom. 2013, 89, 265–272. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, S.-J.; Lee, H.M.; Lee, B.W.; Ha, T.J.; Bae, D.W.; Son, B.-Y.; Kim, Y.H.; Baek, S.-B.; Kim, Y.C. Comparative proteomics analysis of seed coat from two black colored soybean cultivars during seed development. Plant Omics 2013, 6, 456–463. [Google Scholar]
- Gupta, R.; Min, C.W.; Kim, S.W.; Wang, Y.; Agrawal, G.K.; Rakwal, R.; Kim, S.G.; Lee, B.W.; Ko, J.M.; Baek, I.Y.; et al. Comparative investigation of seed coats of brown- versus yellow-colored soybean seeds using an integrated proteomics and metabolomics approach. Proteomics 2015, 15, 1706–1716. [Google Scholar] [CrossRef] [PubMed]
- Smykal, P.; Vernoud, V.; Blair, M.W.; Soukup, A.; Thompson, R.D. The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 2014, 5, 351. [Google Scholar] [CrossRef]
- Paauw, M.; Koes, R.; Quattrocchio, F.M. Alteration of flavonoid pigmentation patterns during domestication of food crops. J. Exp. Bot. 2019, 70, 3719–3735. [Google Scholar] [CrossRef] [PubMed]
- von Wettberg, E.J.B.; Chang, P.L.; Basdemir, F.; Carrasquila-Garcia, N.; Korbu, L.B.; Moenga, S.M.; Bedada, G.; Greenlon, A.; Moriuchi, K.S.; Singh, V.; et al. Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nat. Commun. 2018, 9, 649. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Klčová, B.; Balarynová, J.; Trněný, O.; Krejčí, P.; Cechová, M.Z.; Leonova, T.; Gorbach, D.; Frolova, N.; Kysil, E.; Orlova, A. Domestication has altered gene expression and secondary metabolites in pea seed coat. Plant J. 2024, 118, 2269–2295. [Google Scholar] [CrossRef]
- Hradilova, I.; Duchoslav, M.; Brus, J.; Pechanec, V.; Hybl, M.; Kopecky, P.; Smrzova, L.; Stefelova, N.; Vaclavek, T.; Bariotakis, M.; et al. Variation in wild pea (Pisum sativum subsp. elatius) seed dormancy and its relationship to the environment and seed coat traits. PeerJ 2019, 7, e6263. [Google Scholar] [CrossRef]
- Wang, M.; Li, W.; Fang, C.; Xu, F.; Liu, Y.; Wang, Z.; Yang, R.; Zhang, M.; Liu, S.; Lu, S.; et al. Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat. Genet. 2018, 50, 1435–1441. [Google Scholar] [CrossRef]
- Ku, Y.S.; Contador, C.A.; Ng, M.S.; Yu, J.; Chung, G.; Lam, H.M. The Effects of Domestication on Secondary Metabolite Composition in Legumes. Front. Genet. 2020, 11, 581357. [Google Scholar] [CrossRef]
- Puozaa, D.K.; Jaiswal, S.K.; Dakora, F.D. Black Seedcoat Pigmentation Is a Marker for Enhanced Nodulation and N2 Fixation in Bambara Groundnut (Vigna subterranea L. Verdc.) Landraces. Front. Agron. 2021, 3, 692238. [Google Scholar] [CrossRef]
- Hungria, M.; Phillips, D.A. Effects of a Seed Color Mutation on Rhizobial Nod-Gene-Inducing Flavonoids and Nodulation in Common Bean. Mol. Plant Microbe 1993, 6, 418–422. [Google Scholar] [CrossRef]
- Mishra, G.P.; Ankita; Aski, M.S.; Tontang, M.T.; Choudhary, P.; Tripathi, K.; Singh, A.; Kumar, R.R.; Thimmegowda, V.; Stobdan, T.; et al. Morphological, Molecular, and Biochemical Characterization of a Unique Lentil (Lens culinaris Medik.) Genotype Showing Seed-Coat Color Anomalies Due to Altered Anthocyanin Pathway. Plants 2022, 11, 1815. [Google Scholar] [CrossRef]
- Shumilina, J.; Soboleva, A.; Abakumov, E.; Shtark, O.Y.; Zhukov, V.A.; Frolov, A. Signaling in Legume-Rhizobia Symbiosis. Int. J. Mol. Sci. 2023, 24, 17397. [Google Scholar] [CrossRef]
- Compton, K.K.; Scharf, B.E. Rhizobial Chemoattractants, the Taste and Preferences of Legume Symbionts. Front. Plant Sci. 2021, 12, 686465. [Google Scholar] [CrossRef] [PubMed]
- Samac, D.A.; Graham, M.A. Recent advances in legume-microbe interactions: Recognition, defense response, and symbiosis from a genomic perspective. Plant Physiol. 2007, 144, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.E. Early interactions between legumes and rhizobia: Disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 2007, 103, 1355–1365. [Google Scholar] [CrossRef]
- Zhang, Y.; Bhat, J.A.; Zhang, Y.; Yang, S. Understanding the Molecular Regulatory Networks of Seed Size in Soybean. Int. J. Mol. Sci. 2024, 25, 1441. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, M.; Viana, V.E.; Banfi, C.; Cassina, V.; Corti, R.; Herrera-Ubaldo, H.; Babolin, N.; Guazzotti, A.; Kiegle, E.; Gregis, V.; et al. Cell wall modifications by alpha-XYLOSIDASE1 are required for control of seed and fruit size in Arabidopsis. J. Exp. Bot. 2022, 73, 1499–1515. [Google Scholar] [CrossRef]
- Boccaccini, A.; Cimini, S.; Kazmi, H.; Lepri, A.; Longo, C.; Lorrai, R.; Vittorioso, P. When Size Matters: New Insights on How Seed Size Can Contribute to the Early Stages of Plant Development. Plants 2024, 13, 1793. [Google Scholar] [CrossRef]
- Noguero, M.; Le Signor, C.; Vernoud, V.; Bandyopadhyay, K.; Sanchez, M.; Fu, C.; Torres-Jerez, I.; Wen, J.; Mysore, K.S.; Gallardo, K.; et al. DASH transcription factor impacts Medicago truncatula seed size by its action on embryo morphogenesis and auxin homeostasis. Plant J. 2015, 81, 453–466. [Google Scholar] [CrossRef]
- Chen, Z.; Lancon-Verdier, V.; Le Signor, C.; She, Y.M.; Kang, Y.; Verdier, J. Genome-wide association study identified candidate genes for seed size and seed composition improvement in M. truncatula. Sci. Rep. 2021, 11, 4224. [Google Scholar] [CrossRef]
- Sita, K.; Sehgal, A.; HanumanthaRao, B.; Nair, R.M.; Vara Prasad, P.V.; Kumar, S.; Gaur, P.M.; Farooq, M.; Siddique, K.H.M.; Varshney, R.K.; et al. Food Legumes and Rising Temperatures: Effects, Adaptive Functional Mechanisms Specific to Reproductive Growth Stage and Strategies to Improve Heat Tolerance. Front. Plant Sci. 2017, 8, 1658. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, S.P.; Araújo, M.D.; de Aragao, W.F.L.; Damasceno-Silva, K.J.; Rocha, M.D. Genetic analysis of yield component traits in cowpea [Vigna unguiculata (L.) Walp.]. Crop Breed. Appl. Biot. 2024, 24, e46432413. [Google Scholar] [CrossRef]
- Ge, L.; Yu, J.; Wang, H.; Luth, D.; Bai, G.; Wang, K.; Chen, R. Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. USA 2016, 113, 12414–12419. [Google Scholar] [CrossRef]
- D’Erfurth, I.; Le Signor, C.; Aubert, G.; Sanchez, M.; Vernoud, V.; Darchy, B.; Lherminier, J.; Bourion, V.; Bouteiller, N.; Bendahmane, A.; et al. A role for an endosperm-localized subtilase in the control of seed size in legumes. New Phytol. 2012, 196, 738–751. [Google Scholar] [CrossRef]
- Confalonieri, M.; Carelli, M.; Galimberti, V.; Macovei, A.; Panara, F.; Biggiogera, M.; Scotti, C.; Calderini, O. Seed-Specific Expression of AINTEGUMENTA in Medicago truncatula Led to the Production of Larger Seeds and Improved Seed Germination. Plant Mol. Biol. Rep. 2014, 32, 957–970. [Google Scholar] [CrossRef]
- Basu, U.; Upadhyaya, H.D.; Srivastava, R.; Daware, A.; Malik, N.; Sharma, A.; Bajaj, D.; Narnoliya, L.; Thakro, V.; Kujur, A.; et al. ABC Transporter-Mediated Transport of Glutathione Conjugates Enhances Seed Yield and Quality in Chickpea. Plant Physiol. 2019, 180, 253–275. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.; Hayes, J.E.; Harris, J.; Sutton, T. Fine Mapping of a Vigor QTL in Chickpea (Cicer arietinum L.) Reveals a Potential Role for Ca4_TIFY4B in Regulating Leaf and Seed Size. Front. Plant Sci. 2022, 13, 829566. [Google Scholar] [CrossRef]
- Assefa, T.; Otyama, P.I.; Brown, A.V.; Kalberer, S.R.; Kulkarni, R.S.; Cannon, S.B. Genome-wide associations and epistatic interactions for internode number, plant height, seed weight and seed yield in soybean. BMC Genom. 2019, 20, 527. [Google Scholar] [CrossRef]
- Lu, X.; Xiong, Q.; Cheng, T.; Li, Q.T.; Liu, X.L.; Bi, Y.D.; Li, W.; Zhang, W.K.; Ma, B.; Lai, Y.C.; et al. A PP2C-1 Allele Underlying a Quantitative Trait Locus Enhances Soybean 100-Seed Weight. Mol. Plant 2017, 10, 670–684. [Google Scholar] [CrossRef]
- Lu, X.; Li, Q.T.; Xiong, Q.; Li, W.; Bi, Y.D.; Lai, Y.C.; Liu, X.L.; Man, W.Q.; Zhang, W.K.; Ma, B.; et al. The transcriptomic signature of developing soybean seeds reveals the genetic basis of seed trait adaptation during domestication. Plant J. 2016, 86, 530–544. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, K.; Ulucay, O.; Sakiroglu, M.; Udvardi, M.K.; Verdier, J. Analysis of Large Seeds from Three Different Medicago truncatula Ecotypes Reveals a Potential Role of Hormonal Balance in Final Size Determination of Legume Grains. Int. J. Mol. Sci. 2016, 17, 1472. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, Y.; Tao, J.J.; Lu, L.; Jiang, Z.H.; Wei, J.J.; Wu, C.M.; Yin, C.C.; Li, W.; Bi, Y.D.; et al. GmJAZ3 interacts with GmRR18a and GmMYC2a to regulate seed traits in soybean. J. Integr. Plant Biol. 2023, 65, 1983–2000. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhang, X.; Song, X.; Yang, J.; Pang, Y. Genome-Wide Identification and Characterization of APETALA2/Ethylene-Responsive Element Binding Factor Superfamily Genes in Soybean Seed Development. Front. Plant Sci. 2020, 11, 566647. [Google Scholar] [CrossRef]
- Tayade, R.; Imran, M.; Ghimire, A.; Khan, W.; Nabi, R.B.S.; Kim, Y. Molecular, genetic, and genomic basis of seed size and yield characteristics in soybean. Front. Plant Sci. 2023, 14, 1195210. [Google Scholar] [CrossRef]
- Wang, Z.; Lei, Y.; Liao, B. Omics-driven advances in the understanding of regulatory landscape of peanut seed development. Front. Plant Sci. 2024, 15, 1393438. [Google Scholar] [CrossRef]
- Sivakumar, K.B.; Gautam, A.; Singh, S.; Panwar, R.K.; Arora, A.; Verma, S.K. A Pragmatic Study on Seed Shape Classification and its Association among Seed Quality Attributes in Chickpea (Cicer arietinum L.). Legume Res. 2024, 47, 597–602. [Google Scholar] [CrossRef]
- Thompson, R.D.; Verdier, J. Networks of Seed Storage Protein Regulation in Cereals and Legumes at the Dawn of the Omics Era. In Seed Development: OMICS Technologies Toward Improvement of Seed Quality and Crop Yield: OMICS in Seed Biology; Agrawal, G.K., Rakwal, R., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 187–210. [Google Scholar]
- Bhattacharyya, M.; Martin, C.; Smith, A. The importance of starch biosynthesis in the wrinkled seed shape character of peas studied by Mendel. Plant Mol. Biol. 1993, 22, 525–531. [Google Scholar] [CrossRef]
- Khajuria, B.; Rajput, P.; Chowdhary, R.; Urfan, M.; Sharma, S.; Hakla, H.R.; Choudhary, S.P. Exploring novel SNPs and candidate genes associated with seed allometry in Pisum sativum L. Physiol. Mol. Biol. Plants 2024, 30, 1449–1462. [Google Scholar] [CrossRef]
- Sankaran, R.P.; Huguet, T.; Grusak, M.A. Identification of QTL affecting seed mineral concentrations and content in the model legume Medicago truncatula. Theor. Appl. Genet. 2009, 119, 241–253. [Google Scholar] [CrossRef]
- Alam, A.M.; Somta, P.; Muktadir, M.; Srinives, P. Quantitative trait loci associated with seed weight in mungbean (Vigna radiata (L.) Wilczek). Agric. Nat. Resour. 2014, 48, 197–204. [Google Scholar]
- Egbadzor, K.; Dadoza, M.; Danquah, E.; Yeboah, M.; Offei, S.; Ofori, K. Genetic control of seed size in cowpea (Vigna unguiculata (L.) Walp). Int. J. Agric. Sci. 2013, 5, 367–371. [Google Scholar] [CrossRef]
- Bohra, A.; Jha, R.; Lamichaney, A.; Singh, D.; Jha, U.C.; Naik, S.S.; Datta, D.; Maurya, A.K.; Tiwari, A.; Yadav, V. Mapping QTL for important seed traits in an interspecific F2 population of pigeonpea. 3 Biotech. 2020, 10, 434. [Google Scholar] [CrossRef]
- Giordani, W.; Gama, H.C.; Chiorato, A.F.; Garcia, A.A.F.; Vieira, M.L.C. Genome-wide association studies dissect the genetic architecture of seed shape and size in common bean. G3 (Bethesda) 2022, 12, jkac048. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhang, H.; Kan, G.; Ma, D.; Zhang, D.; Shi, G.; Hong, D.; Zhang, G.; Yu, D. Determination of the genetic architecture of seed size and shape via linkage and association analysis in soybean (Glycine max L. Merr.). Genetica 2013, 141, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Shao, J.; Huo, X.; Li, W.; Kong, Y.; Du, H.; Li, X.; Zhang, C. Identification of closely associated SNPs and candidate genes with seed size and shape via deep re-sequencing GWAS in soybean. Theor. Appl. Genet. 2022, 135, 2341–2351. [Google Scholar] [CrossRef] [PubMed]
- Gali, K.K.; Sackville, A.; Tafesse, E.G.; Lachagari, V.B.R.; McPhee, K.; Hybl, M.; Mikic, A.; Smykal, P.; McGee, R.; Burstin, J.; et al. Genome-Wide Association Mapping for Agronomic and Seed Quality Traits of Field Pea (Pisum sativum L.). Front. Plant Sci. 2019, 10, 1538. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, X.; Miao, H.; Chu, Y.; Cui, F.; Yang, W.; Wang, C.; Shen, Y.; Xu, T.; Zhao, L.; et al. QTL identification for seed weight and size based on a high-density SLAF-seq genetic map in peanut (Arachis hypogaea L.). BMC Plant Biol. 2019, 19, 537. [Google Scholar] [CrossRef]
- Lucas, M.R.; Huynh, B.L.; da Silva Vinholes, P.; Cisse, N.; Drabo, I.; Ehlers, J.D.; Roberts, P.A.; Close, T.J. Association Studies and Legume Synteny Reveal Haplotypes Determining Seed Size in Vigna unguiculata. Front. Plant Sci. 2013, 4, 95. [Google Scholar] [CrossRef]
- Wang, R.; Gangola, M.P.; Irvine, C.; Gaur, P.M.; Baga, M.; Chibbar, R.N. Co-localization of genomic regions associated with seed morphology and composition in a desi chickpea (Cicer arietinum L.) population varying in seed protein concentration. Theor. Appl. Genet. 2019, 132, 1263–1281. [Google Scholar] [CrossRef]
- Yadav, P.; Saxena, K.B.; Hingane, A.; Kumar, C.V.S.; Kandalkar, V.S.; Varshney, R.K.; Saxena, R.K. An “Axiom Cajanus SNP Array” based high density genetic map and QTL mapping for high-selfing flower and seed quality traits in pigeonpea. BMC Genom. 2019, 20, 235. [Google Scholar] [CrossRef]
- Hina, A.; Cao, Y.; Song, S.; Li, S.; Sharmin, R.A.; Elattar, M.A.; Bhat, J.A.; Zhao, T. High-Resolution Mapping in Two RIL Populations Refines Major “QTL Hotspot” Regions for Seed Size and Shape in Soybean (Glycine max L.). Int. J. Mol. Sci. 2020, 21, 1040. [Google Scholar] [CrossRef] [PubMed]
- Jurado, M.; Garcia-Fernandez, C.; Campa, A.; Ferreira, J.J. Identification of consistent QTL and candidate genes associated with seed traits in common bean by combining GWAS and RNA-Seq. Theor. Appl. Genet. 2024, 137, 143. [Google Scholar] [CrossRef]
- Jiang, A.; Liu, J.; Gao, W.; Ma, R.; Tan, P.; Liu, F.; Zhang, J. Construction of a genetic map and QTL mapping of seed size traits in soybean. Front. Genet. 2023, 14, 1248315. [Google Scholar] [CrossRef]
- Gao, W.; Ma, R.; Li, X.; Liu, J.; Jiang, A.; Tan, P.; Xiong, G.; Du, C.; Zhang, J.; Zhang, X.; et al. Construction of Genetic Map and QTL Mapping for Seed Size and Quality Traits in Soybean (Glycine max L.). Int. J. Mol. Sci. 2024, 25, 2857. [Google Scholar] [CrossRef] [PubMed]
- Geravandi, M.; Cheghamirza, K.; Farshadfar, E.; Gepts, P. QTL analysis of seed size and yield-related traits in an inter-genepool population of common bean (Phaseolus vulgaris L.). Sci. Hortic. 2020, 274, 109678. [Google Scholar] [CrossRef]
- Kumar, R.; Saini, M.; Taku, M.; Debbarma, P.; Mahto, R.K.; Ramlal, A.; Sharma, D.; Rajendran, A.; Pandey, R.; Gaikwad, K.; et al. Identification of quantitative trait loci (QTLs) and candidate genes for seed shape and 100-seed weight in soybean [Glycine max (L.) Merr.]. Front. Plant Sci. 2023, 13, 1074245. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, D.; Mao, Y.; Zhou, Y.; Zhao, L.; Zhang, C.; Liu, Y.; Chen, J. The Organ Size and Morphological Change During the Domestication Process of Soybean. Front. Plant Sci. 2022, 13, 913238. [Google Scholar] [CrossRef]
- Dutta, H.; Mishra, G.P.; Aski, M.S.; Bosamia, T.C.; Mishra, D.C.; Bhati, J.; Sinha, S.K.; Vijay, D.; Prasad, C.T.M.; Das, S.; et al. Comparative transcriptome analysis, unfolding the pathways regulating the seed-size trait in cultivated lentil (Lens culinaris Medik.). Front. Genet. 2022, 13, 942079. [Google Scholar] [CrossRef]
- Du, J.; Wang, S.; He, C.; Zhou, B.; Ruan, Y.L.; Shou, H. Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J. Exp. Bot. 2017, 68, 1955–1972. [Google Scholar] [CrossRef]
- Quiroz-Figueroa, F.R.; Monribot-Villanueva, J.L.; Bojorquez-Velazquez, E.; Gomez-Peraza, R.L.; Elizalde-Contreras, J.M.; Bautista-Valle, M.V.; Guerrero-Analco, J.A.; Valdez-Morales, M.; Singh, R.K.; Ruiz-May, E. Proteometabolomic Analysis Reveals Molecular Features Associated with Grain Size and Antioxidant Properties amongst Chickpea (Cicer arietinum L.) Seeds Genotypes. Antioxidants 2022, 11, 1850. [Google Scholar] [CrossRef] [PubMed]
- Roland, W.S.U.; Pouvreau, L.; Curran, J.; van de Velde, F.; de Kok, P.M.T. Flavor Aspects of Pulse Ingredients. Cereal Chem. 2017, 94, 58–65. [Google Scholar] [CrossRef]
- Xiang, L.; Jiang, B.; Xiong, Y.L.; Zhou, L.; Zhong, F.; Zhang, R.; Tahir, A.B.; Xiao, Z. Beany flavor in pea protein: Recent advances in formation mechanism, analytical techniques and microbial fermentation mitigation strategies. Food Biosci. 2023, 56, 103166. [Google Scholar] [CrossRef]
- Maoz, I.; Lewinsohn, E.; Gonda, I. Amino acids metabolism as a source for aroma volatiles biosynthesis. Curr. Opin. Plant Biol. 2022, 67, 102221. [Google Scholar] [CrossRef]
- Akanni, G.B.; Adebo, O.A. Metabolite perturbations in fermented legumes as elucidated using metabolomics: A review. Int. J. Food Sci. Technol. 2024, 59, 4234–4250. [Google Scholar] [CrossRef]
- Adebo, J.A.; Njobeh, P.B.; Gbashi, S.; Oyedeji, A.B.; Ogundele, O.M.; Oyeyinka, S.A.; Adebo, O.A. Fermentation of Cereals and Legumes: Impact on Nutritional Constituents and Nutrient Bioavailability. Fermentation 2022, 8, 63. [Google Scholar] [CrossRef]
- Ritter, S.W.; Gastl, M.I.; Becker, T.M. The modification of volatile and nonvolatile compounds in lupines and faba beans by substrate modulation and lactic acid fermentation to facilitate their use for legume-based beverages—A review. Compr. Rev. Food Sci. Food Saf. 2022, 21, 4018–4055. [Google Scholar] [CrossRef] [PubMed]
- Karolkowski, A.; Guichard, E.; Briand, L.; Salles, C. Volatile Compounds in Pulses: A Review. Foods 2021, 10, 3140. [Google Scholar] [CrossRef] [PubMed]
- Oomah, B.D.; Razafindrainibe, M.; Drover, J.C. Headspace volatile components of Canadian grown low-tannin faba bean (Vicia faba L.) genotypes. J. Sci. Food Agric. 2014, 94, 473–481. [Google Scholar] [CrossRef]
- Trindler, C.; Kopf-Bolanz, K.A.; Denkel, C. Aroma of peas, its constituents and reduction strategies-Effects from breeding to processing. Food Chem. 2022, 376, 131892. [Google Scholar] [CrossRef] [PubMed]
- Bott, L.; Chambers, E., IV. Sensory characteristics of combinations of chemicals potentially associated with beany aroma in foods. J. Sens. Stud. 2006, 21, 308–321. [Google Scholar] [CrossRef]
- Ang, E.; Boatright, W. Olfactory perception of major odorants found in the headspace of aqueous soy protein isolate slurries. J. Food Sci. 2003, 68, 388–393. [Google Scholar] [CrossRef]
- Szczygiel, E.J.; Harte, J.B.; Strasburg, G.M.; Cho, S. Consumer acceptance and aroma characterization of navy bean (Phaseolus vulgaris) powders prepared by extrusion and conventional processing methods. J. Sci. Food Agric. 2017, 97, 4142–4150. [Google Scholar] [CrossRef]
- Oomah, B.D.; Liang, L.S.; Balasubramanian, P. Volatile compounds of dry beans (Phaseolus vulgaris L.). Plant Foods Hum. Nutr. 2007, 62, 177–183. [Google Scholar] [CrossRef]
- Wang, X.; Ho, C.; Shahidi, F. Flavor Components of Fats and Oils. In Bailey’s Industrial Oil and Fat Products; John Wiley & Sons: Hoboken, NJ, USA, 2020; pp. 1–25. [Google Scholar] [CrossRef]
- Lampi, A.M.; Yang, Z.; Mustonen, O.; Piironen, V. Potential of faba bean lipase and lipoxygenase to promote formation of volatile lipid oxidation products in food models. Food Chem. 2020, 311, 125982. [Google Scholar] [CrossRef]
- Murat, C.; Bard, M.H.; Dhalleine, C.; Cayot, N. Characterisation of odour active compounds along extraction process from pea flour to pea protein extract. Food Res. Int. 2013, 53, 31–41. [Google Scholar] [CrossRef]
- Bi, S.; Xu, X.; Luo, D.; Lao, F.; Pang, X.; Shen, Q.; Hu, X.; Wu, J. Characterization of Key Aroma Compounds in Raw and Roasted Peas (Pisum sativum L.) by Application of Instrumental and Sensory Techniques. J. Agric. Food Chem. 2020, 68, 2718–2727. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, V.G.; Kraus, S.; Schott, M.; Muranyi, I.; Schweiggert-Weisz, U.; Eisner, P. Screening of Twelve Pea (Pisum sativum L.) Cultivars and Their Isolates Focusing on the Protein Characterization, Functionality, and Sensory Profiles. Foods 2021, 10, 758. [Google Scholar] [CrossRef]
- Trikusuma, M.; Paravisini, L.; Peterson, D.G. Identification of aroma compounds in pea protein UHT beverages. Food Chem. 2020, 312, 126082. [Google Scholar] [CrossRef]
- Khrisanapant, P.; Leong, S.Y.; Kebede, B.; Oey, I. Effects of Hydrothermal Processing Duration on the Texture, Starch and Protein In Vitro Digestibility of Cowpeas, Chickpeas and Kidney Beans. Foods 2021, 10, 1415. [Google Scholar] [CrossRef]
- Mishra, P.K.; Tripathi, J.; Gupta, S.; Variyar, P.S. Effect of cooking on aroma profile of red kidney beans (Phaseolus vulgaris) and correlation with sensory quality. Food Chem. 2017, 215, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.K.; Tripathi, J.; Gupta, S.; Variyar, P.S. GC-MS olfactometric characterization of odor active compounds in cooked red kidney beans (Phaseolus vulgaris). Heliyon 2019, 5, e02459. [Google Scholar] [CrossRef]
- Khrisanapant, P.; Kebede, B.; Leong, S.Y.; Oey, I. A Comprehensive Characterisation of Volatile and Fatty Acid Profiles of Legume Seeds. Foods 2019, 8, 651. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Chapman, M.A.; Abberton, M.T.; Akpojotor, U.L.; Ortiz, R. Exploiting genetic and genomic resources to enhance productivity and abiotic stress adaptation of underutilized pulses. Front. Genet. 2023, 14, 1193780. [Google Scholar] [CrossRef]
- Wakte, K.; Zanan, R.; Hinge, V.; Khandagale, K.; Nadaf, A.; Henry, R. Thirty-three years of 2-acetyl-1-pyrroline, a principal basmati aroma compound in scented rice (Oryza sativa L.): A status review. J. Sci. Food Agric. 2017, 97, 384–395. [Google Scholar] [CrossRef]
- Arikit, S.; Yoshihashi, T.; Wanchana, S.; Uyen, T.T.; Huong, N.T.; Wongpornchai, S.; Vanavichit, A. Deficiency in the amino aldehyde dehydrogenase encoded by GmAMADH2, the homologue of rice Os2AP, enhances 2-acetyl-1-pyrroline biosynthesis in soybeans (Glycine max L.). Plant Biotechnol. J. 2011, 9, 75–87. [Google Scholar] [CrossRef]
- Arikit, S.; Yoshihashi, T.; Wanchana, S.; Tanya, P.; Juwattanasomran, R.; Srinives, P.; Vanavichit, A. A PCR-based marker for a locus conferring aroma in vegetable soybean (Glycine max L.). Theor. Appl. Genet. 2011, 122, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.L.; Jin, H.X.; Yang, Q.H.; Zhu, L.M.; Yu, X.M.; Fu, X.J.; Zhao, M.; Yuan, F.J. A Sequence Variation in GmBADH2 Enhances Soybean Aroma and Is a Functional Marker for Improving Soybean Flavor. Int. J. Mol. Sci. 2022, 23, 4116. [Google Scholar] [CrossRef]
- Attar, U.; Hinge, V.; Zanan, R.; Adhav, R.; Nadaf, A. Identification of aroma volatiles and understanding 2-acetyl-1-pyrroline biosynthetic mechanism in aromatic mung bean (Vigna radiata (L.) Wilczek). Physiol. Mol. Biol. Plants 2017, 23, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Basanagouda, G.; Ramesh, S.; Siddu, C.B.; Chandana, B.R.; Kalpana, M.P.; Rotti, K.; Sathish, H. A non-synonymous SNP in homolog of BADH2 gene is associated with fresh pod fragrance in dolichos bean (Lablab purpureus var. lignosus (Prain) Kumari). Genet. Resour. Crop Evol. 2023, 70, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Qian, L.; Chi, Z.; Jia, X.; Qi, F.; Yuan, F.; Liu, Z.; Zheng, Y. Combined Metabolomic and Quantitative RT-PCR Analyses Revealed the Synthetic Differences of 2-Acetyl-1-pyrroline in Aromatic and Non-Aromatic Vegetable Soybeans. Int. J. Mol. Sci. 2022, 23, 14529. [Google Scholar] [CrossRef]
- Turquetti-Moraes, D.K.; Cardoso-Silva, C.B.; Almeida-Silva, F.; Venancio, T.M. Multiomic analysis of genes related to oil traits in legumes provide insights into lipid metabolism and oil richness in soybean. Plant Physiol. Bioch. 2025, 218, 109180. [Google Scholar] [CrossRef]
- Mehle, H.; Paravisini, L.; Peterson, D.G. Impact of temperature and water activity on the aroma composition and flavor stability of pea (Pisum sativum) protein isolates during storage. Food Funct. 2020, 11, 8309–8319. [Google Scholar] [CrossRef]
- Karolkowski, A.; Belloir, C.; Briand, L.; Salles, C. Non-Volatile Compounds Involved in Bitterness and Astringency of Pulses: A Review. Molecules 2023, 28, 3298. [Google Scholar] [CrossRef]
- Heng, L.; Vincken, J.P.; van Koningsveld, G.; Legger, A.; Gruppen, H.; van Boekel, T.; Roozen, J.; Voragen, F. Bitterness of saponins and their content in dry peas. J. Sci. Food Agric. 2006, 86, 1225–1231. [Google Scholar] [CrossRef]
- Glaser, P.; Dawid, C.; Meister, S.; Bader-Mittermaier, S.; Schott, M.; Eisner, P.; Hofmann, T. Molecularization of Bitter Off-Taste Compounds in Pea-Protein Isolates (Pisum sativum L.). J. Agric. Food Chem. 2020, 68, 10374–10387. [Google Scholar] [CrossRef] [PubMed]
- Temussi, P.A. The good taste of peptides. J. Pept. Sci. 2012, 18, 73–82. [Google Scholar] [CrossRef]
- Meinlschmidt, P.; Schweiggert-Weisz, U.; Brode, V.; Eisner, P. Enzyme assisted degradation of potential soy protein allergens with special emphasis on the technofunctionality and the avoidance of a bitter taste formation. LWT-Food Sci. Technol. 2016, 68, 707–716. [Google Scholar] [CrossRef]
- Keast, R.S.; Breslin, P.A. An overview of binary taste–taste interactions. Food Qual. Prefer. 2003, 14, 111–124. [Google Scholar] [CrossRef]
- Ma, Z.; Boye, J.I.; Azarnia, S.; Simpson, B.K. Volatile flavor profile of Saskatchewan grown pulses as affected by different thermal processing treatments. Int. J. Food Prop. 2016, 19, 2251–2271. [Google Scholar] [CrossRef]
- Vurro, F.; De Angelis, D.; Squeo, G.; Caponio, F.; Summo, C.; Pasqualone, A. Exploring Volatile Profiles and De-Flavoring Strategies for Enhanced Acceptance of Lentil-Based Foods: A Review. Foods 2024, 13, 2608. [Google Scholar] [CrossRef]
- Leonard, W.; Zhang, P.; Ying, D.; Fang, Z. Surmounting the off-flavor challenge in plant-based foods. Crit. Rev. Food Sci. Nutr. 2023, 63, 10585–10606. [Google Scholar] [CrossRef]
- Vara-Ubol, S.; Chambers, E.; Chambers, D.H. Sensory Characteristics of Chemical Compounds Potentially Associated with Beany Aroma in Foods. J. Sens. Stud. 2004, 19, 15–26. [Google Scholar] [CrossRef]
- Wang, Y.; Tuccillo, F.; Lampi, A.M.; Knaapila, A.; Pulkkinen, M.; Kariluoto, S.; Coda, R.; Edelmann, M.; Jouppila, K.; Sandell, M.; et al. Flavor challenges in extruded plant-based meat alternatives: A review. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2898–2929. [Google Scholar] [CrossRef]
- Troszynska, A.; Estrella, I.; Lamparski, G.; Hernández, T.; Amarowicz, R.; Pegg, R.B. Relationship between the sensory quality of lentil (Lens culinaris) sprouts and their phenolic constituents. Food Res. Int. 2011, 44, 3195–3201. [Google Scholar] [CrossRef]
- Simons, C.W. Characterization of Edible Bean Flours: Properties and Functionality; North Dakota State University: Fargo, ND, USA, 2013. [Google Scholar]
- Han, J.J.; Janz, J.A.; Gerlat, M. Development of gluten-free cracker snacks using pulse flours and fractions. Food Res. Int. 2010, 43, 627–633. [Google Scholar] [CrossRef]
- Mkanda, A.V.; Minnaar, A.; de Kock, H.L. Relating consumer preferences to sensory and physicochemical properties of dry beans (Phaseolus vulgaris). J. Sci. Food Agric. 2007, 87, 2868–2879. [Google Scholar] [CrossRef]
- Subuola, F.; Widodo, Y.; Kehinde, T. Processing and utilization of legumes in the tropics. Trends Vital Food Control Eng. 2012, 77, 71–84. [Google Scholar]
- Nyombaire, G.; Siddiq, M.; Dolan, K. Physico-chemical and sensory quality of extruded light red kidney bean (Phaseolus vulgaris L.) porridge. LWT-Food Sci. Technol. 2011, 44, 1597–1602. [Google Scholar] [CrossRef]
- Ulrich, D.; Hoberg, E.; Neugebauer, W.; Tiemann, H.; Darsow, U. Investigation of the boiled potato flavor by human sensory and instrumental methods. Am. J. Potato Res. 2000, 77, 111–117. [Google Scholar] [CrossRef]
- Dhull, S.B.; Kinabo, J.; Uebersax, M.A. Nutrient profile and effect of processing methods on the composition and functional properties of lentils (Lens culinaris Medik): A review. Legume Sci. 2023, 5, e156. [Google Scholar] [CrossRef]
- Rudra, S.G.; Singh, A.; Pal, P.; Thakur, R.K. Antinutritional factors in lentils: Their effect on bioavailability of nutrients and significance in human health. In Lentils: Production, Processing Technologies, Products, and Nutritional Profile; John Wiley & Sons: Hoboken, NJ, USA, 2023; pp. 339–364. [Google Scholar] [CrossRef]
- Elango, D.; Rajendran, K.; Van der Laan, L.; Sebastiar, S.; Raigne, J.; Thaiparambil, N.A.; El Haddad, N.; Raja, B.; Wang, W.; Ferela, A.; et al. Raffinose Family Oligosaccharides: Friend or Foe for Human and Plant Health? Front. Plant Sci. 2022, 13, 829118. [Google Scholar] [CrossRef]
- Kubicka, E.; Troszyńska, A. Antioxidative potential of lentil seed coat towards lipoxygenase activity and β-carotene oxidation. Pol. J. Food Nutr. Sci. 2003, 53, 147–150. [Google Scholar]
- Gepts, P.; Beavis, W.D.; Brummer, E.C.; Shoemaker, R.C.; Stalker, H.T.; Weeden, N.F.; Young, N.D. Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol 2005, 137, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, D.; Pasqualone, A.; Costantini, M.; Ricciardi, L.; Lotti, C.; Pavan, S.; Summo, C. Data on the proximate composition, bioactive compounds, physicochemical and functional properties of a collection of faba beans (Vicia faba L.) and lentils (Lens culinaris Medik.). Data Brief. 2021, 34, 106660. [Google Scholar] [CrossRef]
- Liburdi, K.; Esti, M.; Petroselli, V.; Mendler-Drienyovszki, N.; Radicetti, E.; Mancinelli, R. Catalytic properties of lipoxygenase extracted from different varieties of Pisum sativum and Lens culinaris. J. Food Biochem. 2021, 45, e13617. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.M.; Erskine, W.; Berger, J.D.; Nelson, M.N. Phenotypic characterisation and linkage mapping of domestication syndrome traits in yellow lupin (Lupinus luteus L.). Theor. Appl. Genet. 2020, 133, 2975–2987. [Google Scholar] [CrossRef]
- Frick, K.M.; Foley, R.C.; Kamphuis, L.G.; Siddique, K.H.M.; Garg, G.; Singh, K.B. Characterization of the genetic factors affecting quinolizidine alkaloid biosynthesis and its response to abiotic stress in narrow-leafed lupin (Lupinus angustifolius L.). Plant Cell Environ. 2018, 41, 2155–2168. [Google Scholar] [CrossRef]
- Martineau-Cote, D.; Achouri, A.; Karboune, S.; L’Hocine, L. Faba Bean: An Untapped Source of Quality Plant Proteins and Bioactives. Nutrients 2022, 14, 1541. [Google Scholar] [CrossRef]
- Wang, J.; Kuang, H.Q.; Zhang, Z.H.; Yang, Y.Q.; Yan, L.; Zhang, M.C.; Song, S.K.; Guan, Y.F. Generation of seed lipoxygenase-free soybean using CRISPR-Cas9. Crop J. 2020, 8, 432–439. [Google Scholar] [CrossRef]
- Bhowmik, P.; Yan, W.; Hodgins, C.; Polley, B.; Warkentin, T.; Nickerson, M.; Ro, D.K.; Marsolais, F.; Domoney, C.; Shariati-Ievari, S.; et al. CRISPR/Cas9-mediated lipoxygenase gene-editing in yellow pea leads to major changes in fatty acid and flavor profiles. Front. Plant Sci. 2023, 14, 1246905. [Google Scholar] [CrossRef] [PubMed]
- Dierking, E.C.; Bilyeu, K.D. Association of a soybean raffinose synthase gene with low raffinose and stachyose seed phenotype. Plant Genome 2008, 1, 135–145. [Google Scholar] [CrossRef]
- Lippolis, A.; Roland, W.S.U.; Bocova, O.; Pouvreau, L.; Trindade, L.M. The challenge of breeding for reduced off-flavor in faba bean ingredients. Front. Plant Sci. 2023, 14, 1286803. [Google Scholar] [CrossRef] [PubMed]
- Lippolis, A.; Hollebrands, B.; Acierno, V.; de Jong, C.; Pouvreau, L.; Paulo, J.; Gezan, S.A.; Trindade, L.M. GWAS Identifies SNP Markers and Candidate Genes for Off-Flavours and Protein Content in Faba Bean (Vicia faba L.). Plants 2025, 14, 193. [Google Scholar] [CrossRef]
- Bassett, A.; Kamfwa, K.; Ambachew, D.; Cichy, K. Genetic variability and genome-wide association analysis of flavor and texture in cooked beans (Phaseolus vulgaris L.). Theor. Appl. Genet. 2021, 134, 959–978. [Google Scholar] [CrossRef] [PubMed]
- Dhungana, S.K.; Kulkarni, K.P.; Park, C.W.; Jo, H.; Song, J.T.; Shin, D.H.; Lee, J.D. Mapping quantitative trait loci controlling soybean seed starch content in an interspecific cross of ‘Williams 82’ (Glycine max) and ‘PI 366121’ (Glycine soja). Plant Breed. 2017, 136, 379–385. [Google Scholar] [CrossRef]
- Zhao, N.; Xue, D.; Miao, Y.; Wang, Y.; Zhou, E.; Zhou, Y.; Yao, M.; Gu, C.; Wang, K.; Li, B.; et al. Construction of a high-density genetic map for faba bean (Vicia faba L.) and quantitative trait loci mapping of seed-related traits. Front. Plant Sci. 2023, 14, 1201103. [Google Scholar] [CrossRef]
- Ohm, H.; Astrand, J.; Ceplitis, A.; Bengtsson, D.; Hammenhag, C.; Chawade, A.; Grimberg, A. Novel SNP markers for flowering and seed quality traits in faba bean (Vicia faba L.): Characterization and GWAS of a diversity panel. Front. Plant Sci. 2024, 15, 1348014. [Google Scholar] [CrossRef] [PubMed]
- Elango, D.; Wang, W.; Thudi, M.; Sebastiar, S.; Ramadoss, B.R.; Varshney, R.K. Genome-wide association mapping of seed oligosaccharides in chickpea. Front. Plant Sci. 2022, 13, 1024543. [Google Scholar] [CrossRef]
- Luzzatto, L.; Arese, P. Favism and Glucose-6-Phosphate Dehydrogenase Deficiency. N. Engl. J. Med. 2018, 378, 60–71. [Google Scholar] [CrossRef]
- Bjornsdotter, E.; Nadzieja, M.; Chang, W.; Escobar-Herrera, L.; Mancinotti, D.; Angra, D.; Xia, X.; Tacke, R.; Khazaei, H.; Crocoll, C.; et al. VC1 catalyses a key step in the biosynthesis of vicine in faba bean. Nat. Plants 2021, 7, 923–931. [Google Scholar] [CrossRef]
- Sun, J.; Li, Y.; Cheng, X.; Zhang, H.; Yu, J.; Zhang, L.; Qiu, Y.; Diao, J.; Wang, C. Metabolomic Analysis of Flavour Development in Mung Bean Foods: Impact of Thermal Processing and Storage on Precursor and Volatile Compounds. Foods 2025, 14, 797. [Google Scholar] [CrossRef]
- Wallace, L.T. Sensory Analysis and Genetic Mapping of Green Bean Flavor. Ph.D. Thesis, Oregon State University, Corvallis, OR, USA, 2018. Available online: https://ir.library.oregonstate.edu/concern/graduate_thesis_or_dissertations/kh04dv69b (accessed on 24 July 2025).
- Shi, S.H.; Lee, S.S.; Zhu, Y.M.; Jin, Z.Q.; Wu, F.B.; Qiu, C.W. Comparative Metabolomic Profiling Reveals Key Secondary Metabolites Associated with High Quality and Nutritional Value in Broad Bean (Vicia faba L.). Molecules 2022, 27, 8995. [Google Scholar] [CrossRef]
- Correia, B.S.B.; Sørensen, E.B.; Aaslyng, M.A.D.; Bertram, H.C. Metabolome of different cultivars of peas, lentils, faba beans and lupins–An 1H NMR spectroscopic exploration of their sensory attributes and potential biofunctionality. Food Chem. 2025, 477, 143579. [Google Scholar] [CrossRef]
- Karolkowski, A.; Meudec, E.; Bruguière, A.; Mitaine-Offer, A.-C.; Bouzidi, E.; Levavasseur, L.; Sommerer, N.; Briand, L.; Salles, C. Faba Bean (Vicia faba L. minor) bitterness: An untargeted Metabolomic Approach to highlight the impact of the non-volatile fraction. Metabolites 2023, 13, 964. [Google Scholar] [CrossRef]
- Karolkowski, A.; Belloir, C.; Martin, C.; Lucchi, G.; Meudec, E.; Sommerer, N.; Bouzidi, E.; Levavasseur, L.; Briand, L.; Salles, C. Combining sensory profiling and metabolomic approach to better understand the origins of bitter perception in faba bean (Vicia faba L. minor) fractions. Sci. Talks 2024, 11, 100379. [Google Scholar] [CrossRef]
- Zha, F.; Rao, J.; Chen, B. Modification of pulse proteins for improved functionality and flavor profile: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3036–3060. [Google Scholar] [CrossRef] [PubMed]
- Cosson, A. Pea Protein Based Foods for the Future: From Chemical Composition to Statistical Models to Understand the Mechanisms Behind Perceptions of Beany, Bitterness and Astringency. Ph.D. Thesis, Université Paris-Saclay, Orsay, France, 2021. [Google Scholar]
- Ashogbon, A.O.; Akintayo, E.T.; Oladebeye, A.O.; Oluwafemi, A.D.; Akinsola, A.F.; Imanah, O.E. Developments in the isolation, composition, and physicochemical properties of legume starches. Crit. Rev. Food Sci. Nutr. 2021, 61, 2938–2959. [Google Scholar] [CrossRef] [PubMed]
- Hedley, C.L. Carbohydrates in Grain Legume Seeds: Improving Nutritional Quality and Agronomic Characteristics; CABI Publishing: Oxfordshire, UK, 2000. [Google Scholar]
- Tayade, R.; Kulkarni, K.P.; Jo, H.; Song, J.T.; Lee, J.D. Insight Into the Prospects for the Improvement of Seed Starch in Legume-A Review. Front. Plant Sci. 2019, 10, 1213. [Google Scholar] [CrossRef]
- Bhatty, R.S. Cooking Quality of Lentils—The Role of Structure and Composition of Cell-Walls. J. Agric. Food Chem. 1990, 38, 376–383. [Google Scholar] [CrossRef]
- Reyes-Moreno, C.; Paredes-Lopez, O. Hard-to-cook phenomenon in common beans—A review. Crit. Rev. Food Sci. Nutr. 1993, 33, 227–286. [Google Scholar] [CrossRef]
- Gu, Y.; Rasmussen, C.G. Cell biology of primary cell wall synthesis in plants. Plant Cell 2022, 34, 103–128. [Google Scholar] [CrossRef]
- Geitmann, A.; Bacic, A.T. Focus on cell walls. Plant Physiol. 2023, 194, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, J.C.; Clemente, A.; Ochatt, S.J.; Vaz Patto, M.C.; Von Wettberg, E.; Smykal, P. Editorial: Biological and genetic basis of agronomical and seed quality traits in legumes. Front. Plant Sci. 2022, 13, 1009980. [Google Scholar] [CrossRef] [PubMed]
- Mendu, V.; Griffiths, J.S.; Persson, S.; Stork, J.; Downie, A.B.; Voiniciuc, C.; Haughn, G.W.; DeBolt, S. Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol. 2011, 157, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Bashline, L.; Lei, L.; Gu, Y. Cellulose synthesis and its regulation. Arab. Book. 2014, 12, e0169. [Google Scholar] [CrossRef]
- Pauly, M.; Gille, S.; Liu, L.; Mansoori, N.; de Souza, A.; Schultink, A.; Xiong, G. Hemicellulose biosynthesis. Planta 2013, 238, 627–642. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 2016, 67, 463–476. [Google Scholar] [CrossRef]
- Zhu, X.; Xin, X.; Gu, Y. Cellulose and hemicellulose synthesis and their regulation in plant cells. Extracell. Sugar-Based Biopolym. Matrices 2019, 12, 303–353. [Google Scholar]
- Ma, Y.; Chen, F. Plant protein heat-induced gels: Formation mechanisms and regulatory strategies. Coatings 2023, 13, 1899. [Google Scholar] [CrossRef]
- Tang, Q.; Roos, Y.H.; Miao, S. Structure, gelation mechanism of plant proteins versus dairy proteins and evolving modification strategies. Trends Food Sci. Technol. 2024, 147, 104464. [Google Scholar] [CrossRef]
- Yiu, C.C.-Y.; Liang, S.W.; Mukhtar, K.; Kim, W.; Wang, Y.; Selomulya, C. Food emulsion gels from plant-based ingredients: Formulation, processing, and potential applications. Gels 2023, 9, 366. [Google Scholar] [CrossRef]
- El-Sohaimy, S.A.; Brennan, M.; Darwish, A.M.; Brennan, C. Physicochemical, texture and sensorial evaluation of pasta enriched with chickpea flour and protein isolate. Ann. Agric. Sci. 2020, 65, 28–34. [Google Scholar] [CrossRef]
- Singh, B.B.; Ishiyaku, M.F. Genetics of rough seed coat texture in cowpea. J. Hered. 2000, 91, 170–174. [Google Scholar] [CrossRef]
- Mashi, D.S. Genetic Studies on Seed Coat Texture and Cooking Time in some Varieties of Cowpea (Vigna unguiculata (L.) Walp). Ph.D. Thesis, University of Jos, Jos, Nigeria, 2006. [Google Scholar]
- Oladejo, A.S.; Bolaji, A.O.; Olawuyi, O. Inheritance pattern of seed coat texture in cowpea (Vigna unguiculata (L.) WALP. Ife J. Agric. 2020, 32, 46–51. [Google Scholar]
- Ohto, M.A.; Fischer, R.L.; Goldberg, R.B.; Nakamura, K.; Harada, J.J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3123–3128. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Lu, X.; Wen, J.; Wang, Z.; Chai, M. Physical Seed Dormancy in Legumes: Molecular Advances and Perspectives. Plants 2024, 13, 1473. [Google Scholar] [CrossRef]
- Yu, A.M.; Wang, Z.Q.; Zhang, Y.; Li, F.; Liu, A.Z. Global Gene Expression of Seed Coat Tissues Reveals a Potential Mechanism of Regulating Seed Size Formation in Castor Bean. Int. J. Mol. Sci. 2019, 20, 1282. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Garcia, A.; Balarynova, J.; Smykal, P.; von Wettberg, E.J.; Noble, S.D.; Bett, K.E. Genetic and transcriptomic analysis of lentil seed imbibition and dormancy in relation to its domestication. Plant Genome 2025, 18, e70021. [Google Scholar] [CrossRef]
- Matilla, A.J. Auxin: Hormonal Signal Required for Seed Development and Dormancy. Plants 2020, 9, 705. [Google Scholar] [CrossRef]
- Miransari, M.; Smith, D.L. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Soltani, A.; Walter, K.A.; Wiersma, A.T.; Santiago, J.P.; Quiqley, M.; Chitwood, D.; Porch, T.G.; Miklas, P.; McClean, P.E.; Osorno, J.M.; et al. The genetics and physiology of seed dormancy, a crucial trait in common bean domestication. BMC Plant Biol. 2021, 21, 58. [Google Scholar] [CrossRef]
- Chandra, S.; Taak, Y.; Rathod, D.R.; Yadav, R.R.; Poonia, S.; Sreenivasa, V.; Talukdar, A. Genetics and mapping of seed coat impermeability in soybean using inter-specific populations. Physiol. Mol. Biol. Plants 2020, 26, 2291–2299. [Google Scholar] [CrossRef] [PubMed]
- Le, B.H.; Wagmaister, J.A.; Kawashima, T.; Bui, A.Q.; Harada, J.J.; Goldberg, R.B. Using genomics to study legume seed development. Plant Physiol. 2007, 144, 562–574. [Google Scholar] [CrossRef]
- Wang, K.; Henry, R.J.; Gilbert, R.G. Causal relations among starch biosynthesis, structure, and properties. Springer Sci. Rev. 2014, 2, 15–33. [Google Scholar] [CrossRef]
- Masari, A.; Kaewwongwal, A.; Somta, P.; Srinives, P. Inheritance and a major quantitative trait locus of seed starch content in mungbean (Vigna radiata (L.) Wilczek). Euphytica 2017, 213, 166. [Google Scholar] [CrossRef]
- Casanas, F.; Pérez-Vega, E.; Almirall, A.; Plans, M.; Sabaté, J.; Ferreira, J.J. Mapping of QTL associated with seed chemical content in a RIL population of common bean (Phaseolus vulgaris L.). Euphytica 2013, 192, 279–288. [Google Scholar] [CrossRef]
- Jha, A.B.; Tar’an, B.; Diapari, M.; Warkentin, T.D. SNP variation within genes associated with amylose, total starch and crude protein concentration in field pea. Euphytica 2015, 206, 459–471. [Google Scholar] [CrossRef]
- Bogracheva, T.Y.; Cairns, P.; Noel, T.R.; Hulleman, S.; Wang, T.L.; Morris, V.J.; Ring, S.G.; Hedley, C.L. The effect of mutant genes at the r, rb, rug3, rug4, rug5 and lam loci on the granular structure and physico-chemical properties of pea seed starch. Carbohyd. Polym. 1999, 39, 303–314. [Google Scholar] [CrossRef]
- Lloyd, J.R.; Hedley, C.L.; Bull, V.J.; Ring, S.G. Determination of the effect of r and rb mutations on the structure of amylose and amylopectin in pea (Pisum sativum L). Carbohyd. Polym. 1996, 29, 45–49. [Google Scholar] [CrossRef]
- Wang, T.L.; Domoney, C.; Hedley, C.L.; Casey, R.; Grusak, M.A. Can we improve the nutritional quality of legume seeds? Plant Physiol. 2003, 131, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, F.; Jiang, X.; Yang, X.; He, F.; Wang, Z.; Long, R.; Chen, L.; Yang, T.; Wang, C.; et al. Identification of Genetic Loci Associated With Crude Protein Content and Fiber Composition in Alfalfa (Medicago sativa L.) Using QTL Mapping. Front. Plant Sci. 2021, 12, 608940. [Google Scholar] [CrossRef]
- Suanum, W.; Somta, P.; Kongjaimun, A.; Yimram, T.; Kaga, A.; Tomooka, N.; Takahashi, Y.; Srinives, P. Co-localization of QTLs for pod fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea. Mol. Breed. 2016, 36, 80. [Google Scholar] [CrossRef]
- Wang, J.; Mao, L.; Zeng, Z.; Yu, X.; Lian, J.; Feng, J.; Yang, W.; An, J.; Wu, H.; Zhang, M.; et al. Genetic mapping high protein content QTL from soybean ‘Nanxiadou 25’ and candidate gene analysis. BMC Plant Biol. 2021, 21, 388. [Google Scholar] [CrossRef]
- Chakraborty, A.; Junaid, A.; Parida, S.K.; Bhatia, S. Integrated genomic approaches delineate a novel role of ROP1 ENHANCER1 in controlling seed protein content of chickpea. J. Exp. Bot. 2023, 74, 817–834. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Wobus, U. Seed-Development Programs: A Systems Biology-Based Comparison Between Dicots and Monocots. Annu. Rev. Plant Biol. 2013, 64, 189–217. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, G.; Xu, S.; Mao, W.; Hu, Q.; Gong, Y. Comparative Transcriptomic Analyses of Vegetable and Grain Pea (Pisum sativum L.) Seed Development. Front. Plant Sci. 2015, 6, 1039. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Bandhiwal, N.; Shah, N.; Kant, C.; Gaur, R.; Bhatia, S. Global transcriptome analysis of developing chickpea (Cicer arietinum L.) seeds. Front. Plant Sci. 2014, 5, 698. [Google Scholar] [CrossRef] [PubMed]
- Umnajkitikorn, K.; Boonchuen, P.; Senavongse, R.; Tongta, S.; Tian, Y.; Hu, Y.; Petersen, B.L.; Blennow, A. Transcriptomics and starch biosynthesis analysis in leaves and developing seeds of mung bean provide a basis for genetic engineering of starch composition and seed quality. Front. Plant Sci. 2024, 15, 1332150. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.F.; Zheng, A.; Li, G.Q.; Zhang, Y.M. Integrated Bioinformatics and Multi-Omics Analyses Reveal Possible Molecular Mechanisms for Seed Starch Content Differences between Glycine max and Cicer arietinum. Agronomy 2024, 14, 328. [Google Scholar] [CrossRef]
- Gupta, K.; Gupta, S.; Faigenboim-Doron, A.; Patil, A.S.; Levy, Y.; Carrus, S.C.; Hovav, R. Deep transcriptomic study reveals the role of cell wall biosynthesis and organization networks in the developing shell of peanut pod. BMC Plant Biol. 2021, 21, 509. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.H.; Chen, F.; Torres-Jerez, I.; Tang, Y.; Wang, M.; Du, Q.; Cheng, X.; Wen, J.; Dixon, R. Transcriptome analysis of secondary cell wall development in Medicago truncatula. BMC Genom. 2016, 17, 23. [Google Scholar] [CrossRef]
- Yuan, X.; Jiang, X.; Zhang, M.; Wang, L.; Jiao, W.; Chen, H.; Mao, J.; Ye, W.; Song, Q. Integrative omics analysis elucidates the genetic basis underlying seed weight and oil content in soybean. Plant Cell 2024, 36, 2160–2175. [Google Scholar] [CrossRef]
- Vigeolas, H.; Chinoy, C.; Zuther, E.; Blessington, B.; Geigenberger, P.; Domoney, C. Combined metabolomic and genetic approaches reveal a link between the polyamine pathway and albumin 2 in developing pea seeds. Plant Physiol. 2008, 146, 74–82. [Google Scholar] [CrossRef]
- Garcia-Villalba, R.; Leon, C.; Dinelli, G.; Segura-Carretero, A.; Fernandez-Gutierrez, A.; Garcia-Canas, V.; Cifuentes, A. Comparative metabolomic study of transgenic versus conventional soybean using capillary electrophoresis-time-of-flight mass spectrometry. J. Chromatogr. A 2008, 1195, 164–173. [Google Scholar] [CrossRef]
- Sedlakova, V.; Zeljkovic, S.C.; Stefelova, N.; Smykal, P.; Hanacek, P. Phenylpropanoid Content of Chickpea Seed Coats in Relation to Seed Dormancy. Plants 2023, 12, 2687. [Google Scholar] [CrossRef]
- Sheaffer, C.C.; Wyse, D.L.; Ehlke, N.J. Palatability and nutritive value of native legumes. Nativ. Plants J. 2009, 10, 224–231. [Google Scholar] [CrossRef]
- Kirilov, A.; Vasileva, V. Palatability of subterranean clover and some perennial grasses and legume forage crops. J. Glob. Innov. Agric. Soc. Social. Sci. 2016, 4, 152–155. [Google Scholar] [CrossRef]
- Akibode, C.; Mywish, K. Global and Regional Trends in Production, Trade and Consumption of Food Legume; Crops Report Submitted to SPIA; AgEcon Search: St. Paul, MN, USA, 2011. [Google Scholar] [CrossRef]
- Serventi, L.; Brennan, C.; Mustafa, R. Physicochemical and Sensory Evaluation of Grain-Based Food. Foods 2022, 11, 1237. [Google Scholar] [CrossRef]
- Pang, Z.; Cao, J.; Li, H.; Chen, C.; Liu, X. Rheology and tribology properties of cereal and legume flour paste from different botanical origins. J. Food Sci. 2020, 85, 4130–4140. [Google Scholar] [CrossRef] [PubMed]
- Kohyama, K. Food texture–sensory evaluation and instrumental measurement. In Textural Characteristics of World Foods; John Wiley & Sons: Hoboken, NJ, USA, 2020; pp. 1–13. [Google Scholar] [CrossRef]
- Pevicharova, G.; Sofkova-Bobcheva, S.; Zsivanovits, G. Sensory and instrumental texture of snap bean (Phaseolus vulgaris L.). Int. J. Food Prop. 2015, 18, 1169–1180. [Google Scholar] [CrossRef]
- Bassett, A.; Katuuramu, D.N.; Song, Q.J.; Cichy, K. QTL Mapping of Seed Quality Traits Including Cooking Time, Flavor, and Texture in a Yellow Dry Bean (Phaseolus vulgaris L.) Population. Front. Plant Sci. 2021, 12, 670284. [Google Scholar] [CrossRef] [PubMed]
- Holland, C.; Ryden, P.; Edwards, C.H.; Grundy, M.M. Plant Cell Walls: Impact on Nutrient Bioaccessibility and Digestibility. Foods 2020, 9, 201. [Google Scholar] [CrossRef]
- Ratanpaul, V.; Williams, B.A.; Black, J.L.; Gidley, M.J. Apparent amylase diffusion rates in milled cereal grains determined in vitro: Potential relevance to digestion in the small intestine of pigs. J. Cereal Sci. 2018, 82, 42–48. [Google Scholar] [CrossRef]
- Shi, D.; Stone, A.K.; Jafarian, Z.; Liu, E.; Xu, C.; Bhagwat, A.; Lu, Y.; Gao, P.; Polley, B.; Bhowmik, P.; et al. Submerged fermentation of lentil protein isolate and its impact on protein functionality, nutrition, and volatile profiles. J. Food Sci. 2024, 89, 3412–3429. [Google Scholar] [CrossRef] [PubMed]
- Khorsandi, A.; Shi, D.; Stone, A.K.; Bhagwat, A.; Lu, Y.; Xu, C.; Das, P.P.; Polley, B.; Akhov, L.; Gerein, J. Effect of solid-state fermentation on the protein quality and volatile profile of pea and navy bean protein isolates. Cereal Chem. 2024, 101, 131–143. [Google Scholar] [CrossRef]
- Massmann, C.M.; Berhow, M.; Gibbons, W.R.; Karki, B. The effects of fungal bioprocessing on air-classified pea protein concentrates. LWT 2022, 154, 112686. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Reynolds, M.P.; Ortiz, R. Mitigating tradeoffs in plant breeding. iScience 2021, 24, 102965. [Google Scholar] [CrossRef]
- Duraiswamy, A.; Sneha, A.N.; Jebakani, K.S.; Selvaraj, S.; Pramitha, J.L.; Selvaraj, R.; Petchiammal, K.I.; Kather Sheriff, S.; Thinakaran, J.; Rathinamoorthy, S.; et al. Genetic manipulation of anti-nutritional factors in major crops for a sustainable diet in future. Front. Plant Sci. 2022, 13, 1070398. [Google Scholar] [CrossRef]
- Perera, D.; Devkota, L.; Garnier, G.; Panozzo, J.; Dhital, S. Hard-to-cook phenomenon in common legumes: Chemistry, mechanisms and utilisation. Food Chem. 2023, 415, 135743. [Google Scholar] [CrossRef] [PubMed]
- Chigwedere, C.M.; Wanasundara, J.P.D.; Shand, P.J. Sensory descriptors for pulses and pulse-derived ingredients: Toward a standardized lexicon and sensory wheel. Compr. Rev. Food Sci. Food Saf. 2022, 21, 999–1023. [Google Scholar] [CrossRef]
- Nakitto, M.; Johanningsmeier, S.D.; Moyo, M.; Bugaud, C.; de Kock, H.; Dahdouh, L.; Forestier-Chiron, N.; Ricci, J.; Khakasa, E.; Ssali, R.T.; et al. Sensory guided selection criteria for breeding consumer-preferred sweetpotatoes in Uganda. Food Qual. Prefer. 2022, 101, 104628. [Google Scholar] [CrossRef]
- Martinez-Velasco, J.D.; Filomena-Ambrosio, A.; Garzon-Castro, C.L. Technological tools for the measurement of sensory characteristics in food: A review. F1000Research 2023, 12, 340. [Google Scholar] [CrossRef] [PubMed]
- Senarathne, C.; Thavarajah, D. Interdisciplinary approaches to enhance sensory properties and consumer acceptance in pulse crops. Plants People Planet 2025. [Google Scholar] [CrossRef]
- Sharma, N.; Raman, H.; Wheeler, D.; Kalenahalli, Y.; Sharma, R. Data-driven approaches to improve water-use efficiency and drought resistance in crop plants. Plant Sci. 2023, 336, 111852. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Sharma, L.; Onkarappa, D.; Yogendra, K.; Bose, J.; Sharma, R.A. Molecular Basis and Engineering Strategies for Transcription Factor-Mediated Reproductive-Stage Heat Tolerance in Crop Plants. Agronomy 2024, 14, 159. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; von Wettberg, E.J.B.; Naik, Y.D.; Thudi, M.; Siddique, K.H.M. Legume Pangenome: Status and Scope for Crop Improvement. Plants 2022, 11, 3041. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Prasad, M. Editorial: Dissecting antinutrient traits using omics approaches. Front. Plant Sci. 2023, 14, 1234245. [Google Scholar] [CrossRef]
- Fernandez, C.G.T.; Nestor, B.J.; Danilevicz, M.F.; Gill, M.; Petereit, J.; Bayer, P.E.; Finnegan, P.M.; Batley, J.; Edwards, D. Pangenomes as a Resource to Accelerate Breeding of Under-Utilised Crop Species. Int. J. Mol. Sci. 2022, 23, 2671. [Google Scholar] [CrossRef]
- Hu, H.; Yuan, X.; Saini, D.K.; Yang, T.; Wu, X.; Wu, R.; Liu, Z.; Jan, F.; Mir, R.R.; Liu, L.; et al. A panomics-driven framework for the improvement of major food legume crops: Advances, challenges, and future prospects. Hortic. Res. 2025, 12, uhaf091. [Google Scholar] [CrossRef]

| No. | Gene Name | Plant | Role | Reference |
|---|---|---|---|---|
| 1. | BIG SEEDS1 (BS1)—transcriptional regulator | Medicago truncatula, Soybean | Deletion/downregulation of BS1 in Medicago/Soybean significantly increases seed size, weight and amino acid content. | [85] |
| 2. | Subtilase gene (SBT1.1) | Medicago truncatula, Pisum sativum | Controls seed size in legumes through the regulation of embryo cell division. Co-located at a chromosomal position coinciding with a seed weight QTL. | [86] |
| 3. | USP (Unknown Seed Protein) and ANT (AINTEGUMENTA) | Medicago truncatula | Specific expression of ANT in seeds resulted in larger seeds. The gene driven by the seed-specific promoter USP leads to the expansion of storage parenchyma cells in the cotyledon and a significant increase in vacuole size, resulting in a large-seeded phenotype. | [87] |
| 4. | ABCC3-type transporter gene | Chickpea | Regulates seed weight by transcriptional regulation and modulation of the transport of glutathione conjugates in seeds. | [88] |
| 5. | Ca4_TIFY4B | Chickpea | Determines leaf and seed size. | [89] |
| 6. | Glyma.19G151900—gene encoding a histidine phosphor transfer protein | Soybean | Known to regulate seed weight. | [90] |
| 7. | PP2C-1 | Soybean | Regulates the brassinosteroid (BR) signalling pathway and controls the seed size. | [91] |
| 8. | GA20OX and NFYA | Soybean | Overexpression of genes enhanced seed size/weight and oil content in seeds of transgenic plants. | [92] |
| 9. | Isopentenyladenine (iPR) | Medicago truncatula | Associated with cell proliferation during seed development. | [93] |
| 10. | GmJAZ3 (JASMONATE-ZIM DOMAIN 3) | Soybean | Promotes increased cell proliferation and enhanced seed size/weight | [94] |
| 11. | GmAP2-1, GmAP2-4 and GmAP2-6 | Soybean | Play crucial roles in regulating seed size in soybeans by positively influencing seed weight and size. | [95] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, N.; Paul Mukhopadhyay, S.; Onkarappa, D.; Yogendra, K.; Ratanpaul, V. Bridging Genes and Sensory Characteristics in Legumes: Multi-Omics for Sensory Trait Improvement. Agronomy 2025, 15, 1849. https://doi.org/10.3390/agronomy15081849
Sharma N, Paul Mukhopadhyay S, Onkarappa D, Yogendra K, Ratanpaul V. Bridging Genes and Sensory Characteristics in Legumes: Multi-Omics for Sensory Trait Improvement. Agronomy. 2025; 15(8):1849. https://doi.org/10.3390/agronomy15081849
Chicago/Turabian StyleSharma, Niharika, Soumi Paul Mukhopadhyay, Dhanyakumar Onkarappa, Kalenahalli Yogendra, and Vishal Ratanpaul. 2025. "Bridging Genes and Sensory Characteristics in Legumes: Multi-Omics for Sensory Trait Improvement" Agronomy 15, no. 8: 1849. https://doi.org/10.3390/agronomy15081849
APA StyleSharma, N., Paul Mukhopadhyay, S., Onkarappa, D., Yogendra, K., & Ratanpaul, V. (2025). Bridging Genes and Sensory Characteristics in Legumes: Multi-Omics for Sensory Trait Improvement. Agronomy, 15(8), 1849. https://doi.org/10.3390/agronomy15081849






