Identification of Loci and Candidate Genes Associated with Arginine Content in Soybean
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotypic Identification
2.3. Genotyping and Data Quality Control
2.4. Genetic Diversity and Population Structure Analysis
2.5. Genome-Wide Association Study
2.6. Candidate Gene Annotation
2.7. Genomic Prediction
2.7.1. Genomic Prediction Using Different SNP Sets
2.7.2. Genomic Prediction Using GAPIT Version 3 for Whole Panel
2.7.3. Genomic Prediction Using GWAS-Derived SNP Markers
GWAS-Derived SNP Markers from the Whole Panel and Self-Prediction
GWAS-Derived SNP Markers from 80% of the Whole Panel
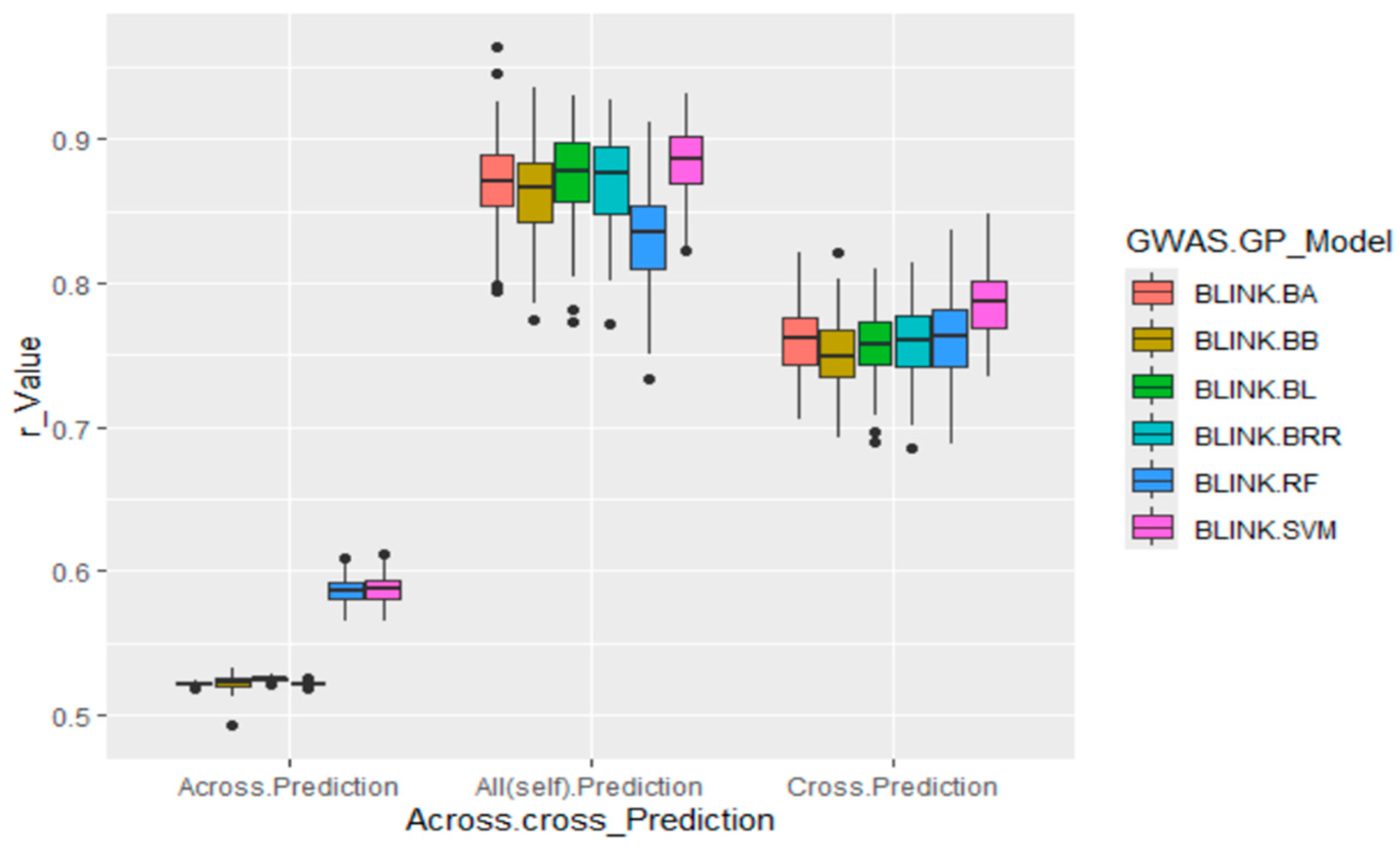
- Across. Prediction uses GWAS-derived SNP markers from the training set (80% of the population, 232 accessions) to predict the validation set (20–58% accessions).
- Cross. Prediction uses GWAS-derived SNP markers from the training set (80% of the population, 232 accessions) to predict itself.
- All(self). Prediction uses all associated SNP markers from the five repeats to predict the entire population (290 accessions).
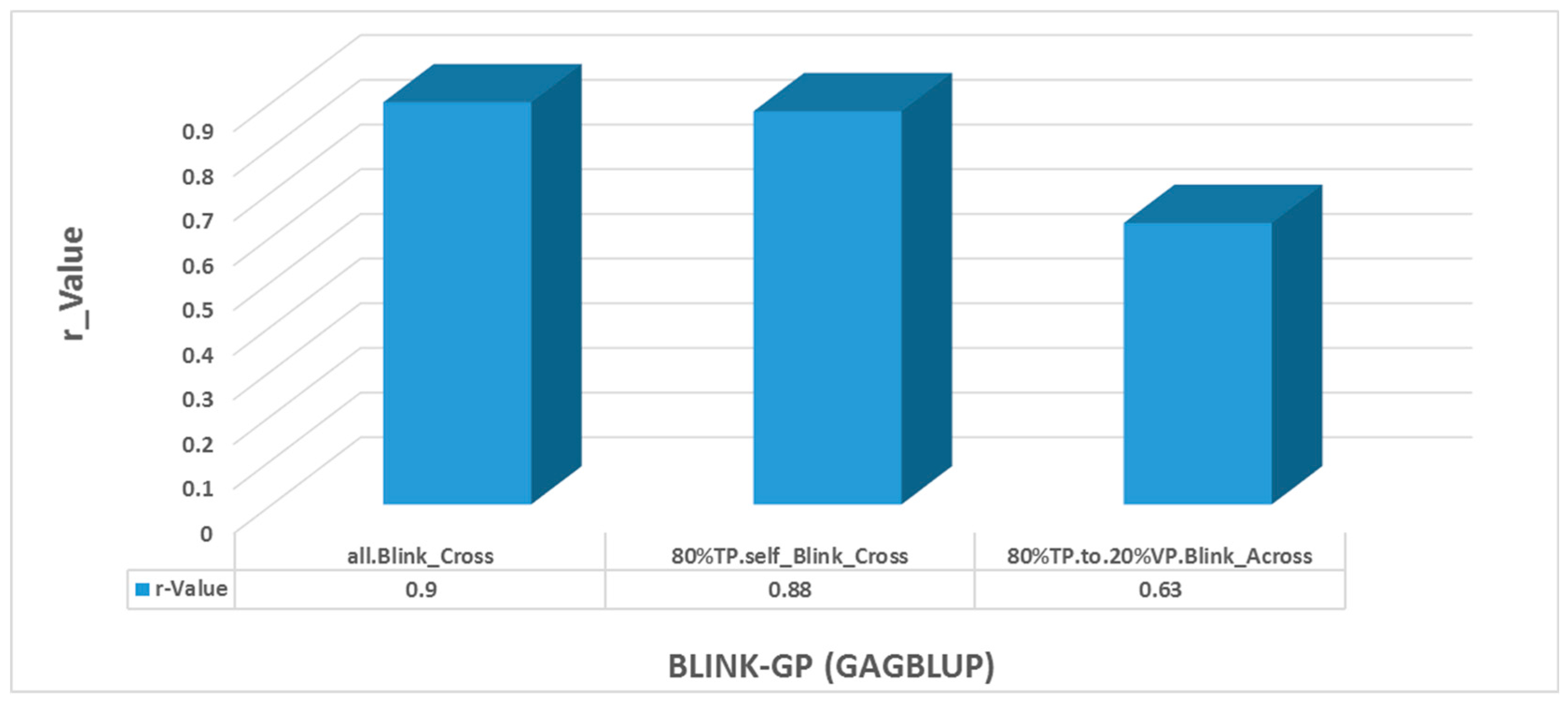
GWAS-Derived SNP Markers Using GAGBLUP in GAPIT Version 3
- Cross-population prediction for entire panel self (all. Blink_Cross)—GWAS-derived SNP markers identified by the BLINK model were used to predict Genomic Estimated Breeding Values (GEBVs) for the entire population of 290 accessions;
- Cross-population prediction for training population (80%TP.self_Blink_Cross)—SNP markers derived from the training population (TP; 232 accessions) were used for self-prediction within the same training set;
- Across-population prediction (80%TP.to.20%VP. Blink_Across)—SNP markers identified from the TP (80%; 232 accessions) were applied to predict GEBVs in the validation population (VP; 20%; 53 accessions).
3. Results
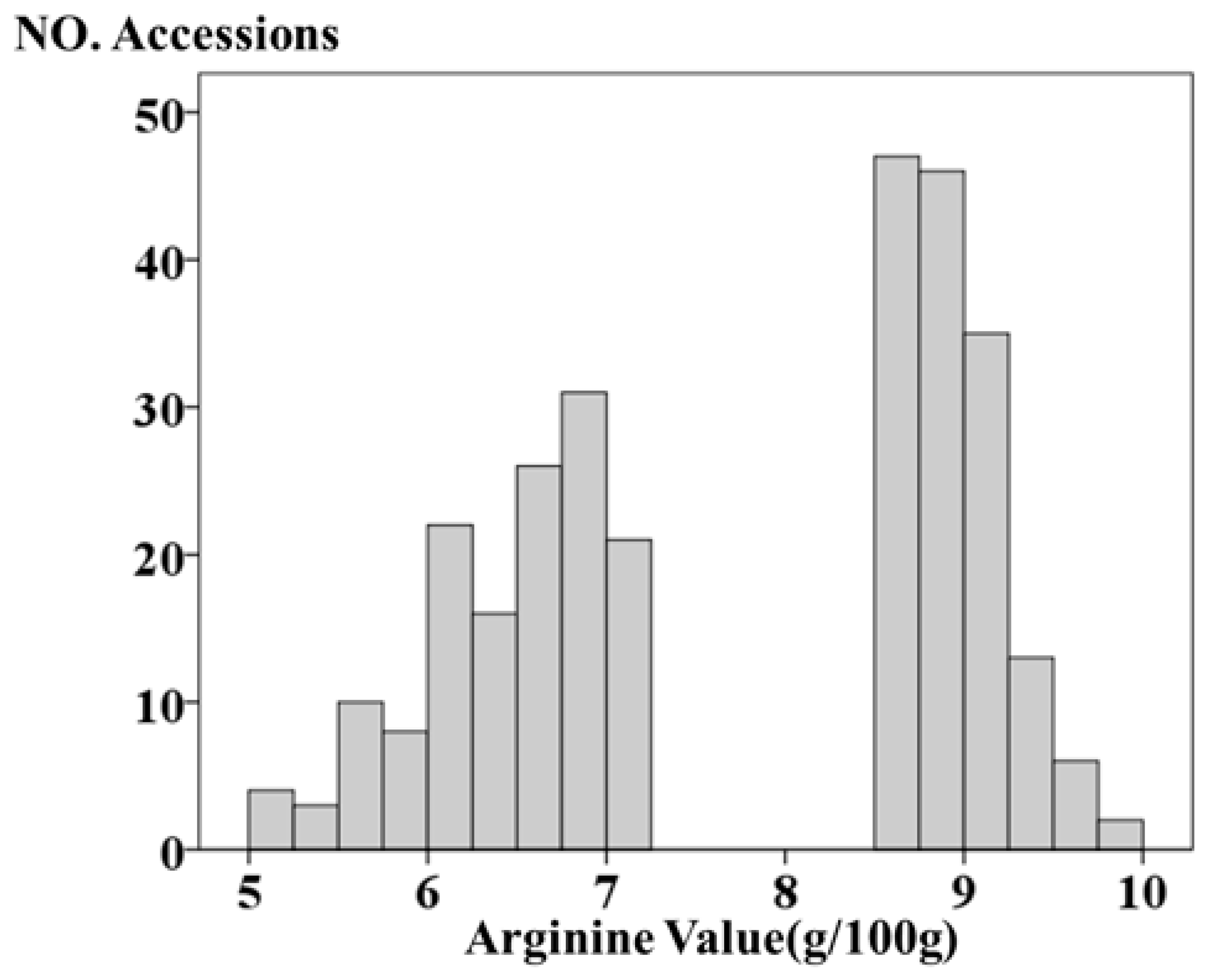
3.1. Phenotypic Analysis of Arginine Content
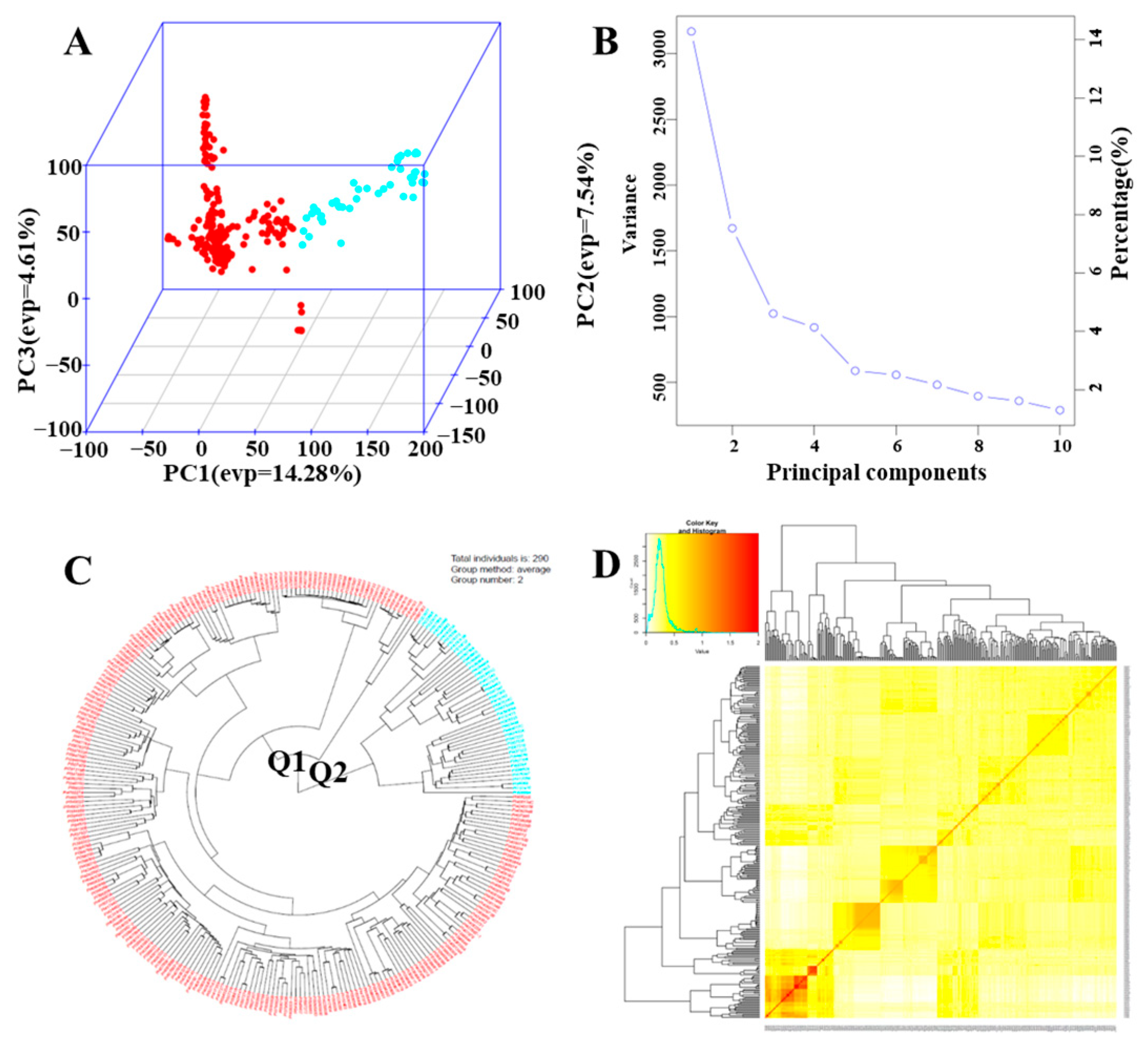
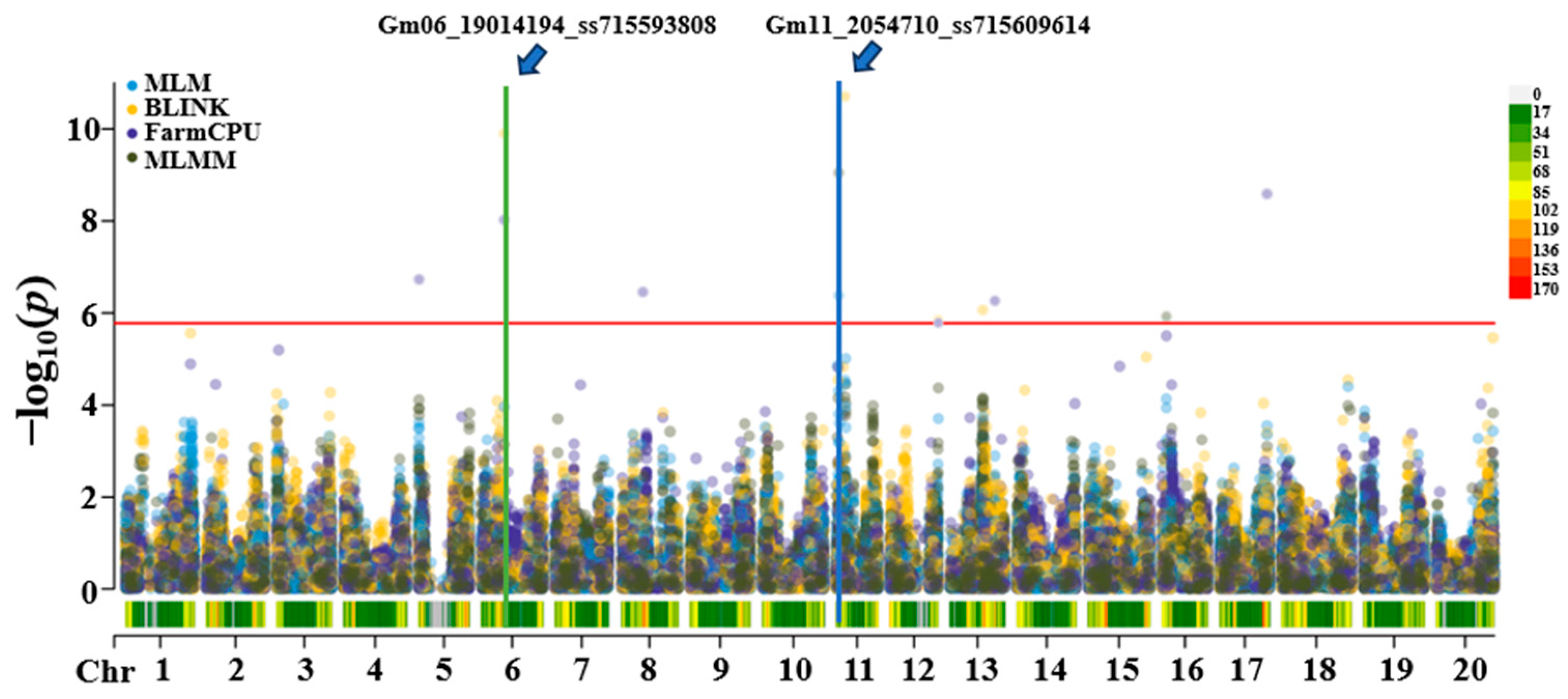
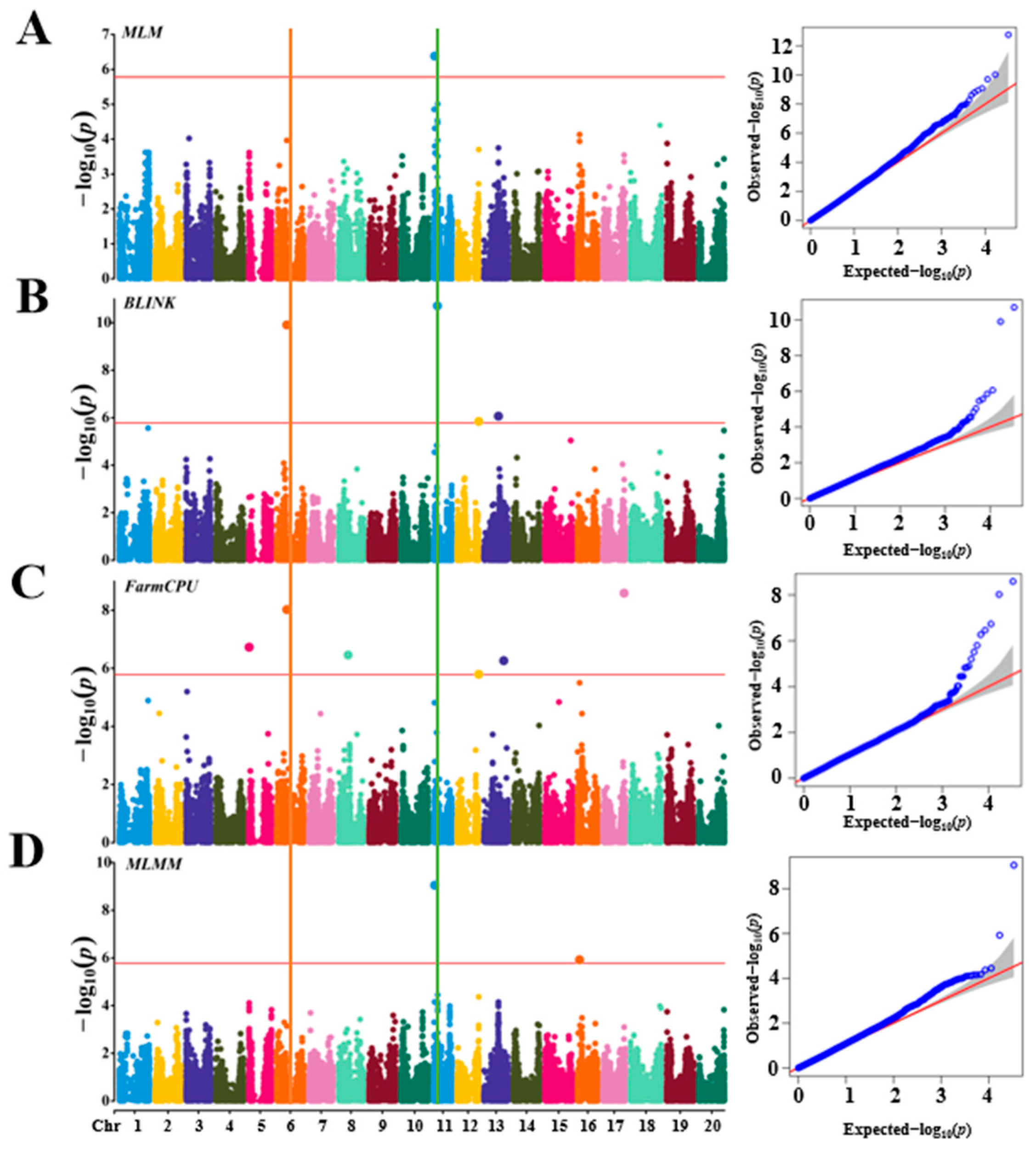
3.2. Population Structure and GWAS
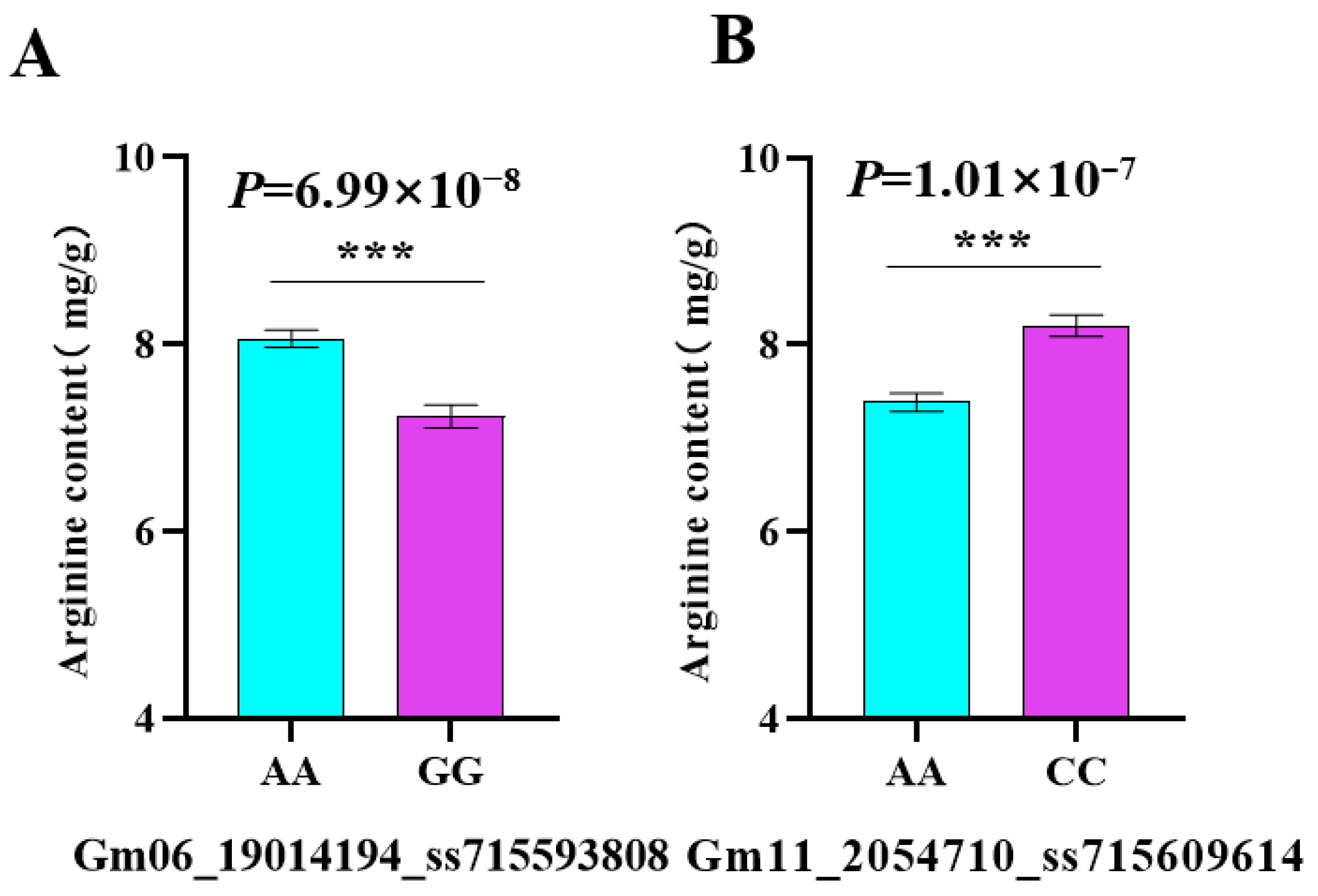
3.3. Haplotype Analysis
3.4. Candidate Gene Detection
3.5. Genomic Prediction Using Whole Panel to Predict Itself
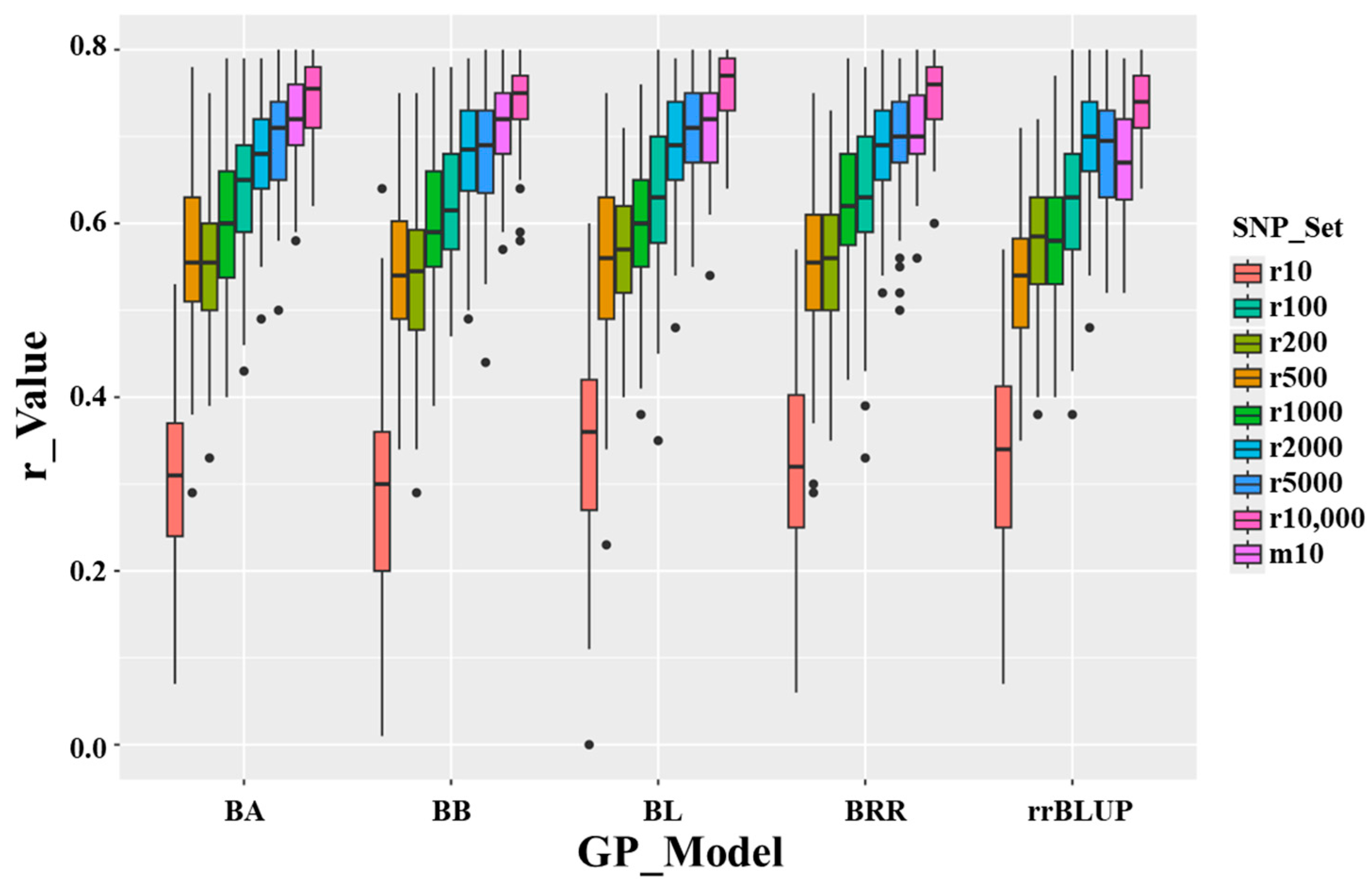
3.6. Genomic Prediction Using Randomly Selected SNPs for Cross-Prediction
3.7. Genomic Prediction by GWAS-Derived SNP Markers
3.7.1. GWAS-Derived SNP Markers from the Whole Panel and Self-Prediction
3.7.2. GWAS-Derived SNP Markers from 80% of the Whole Panel
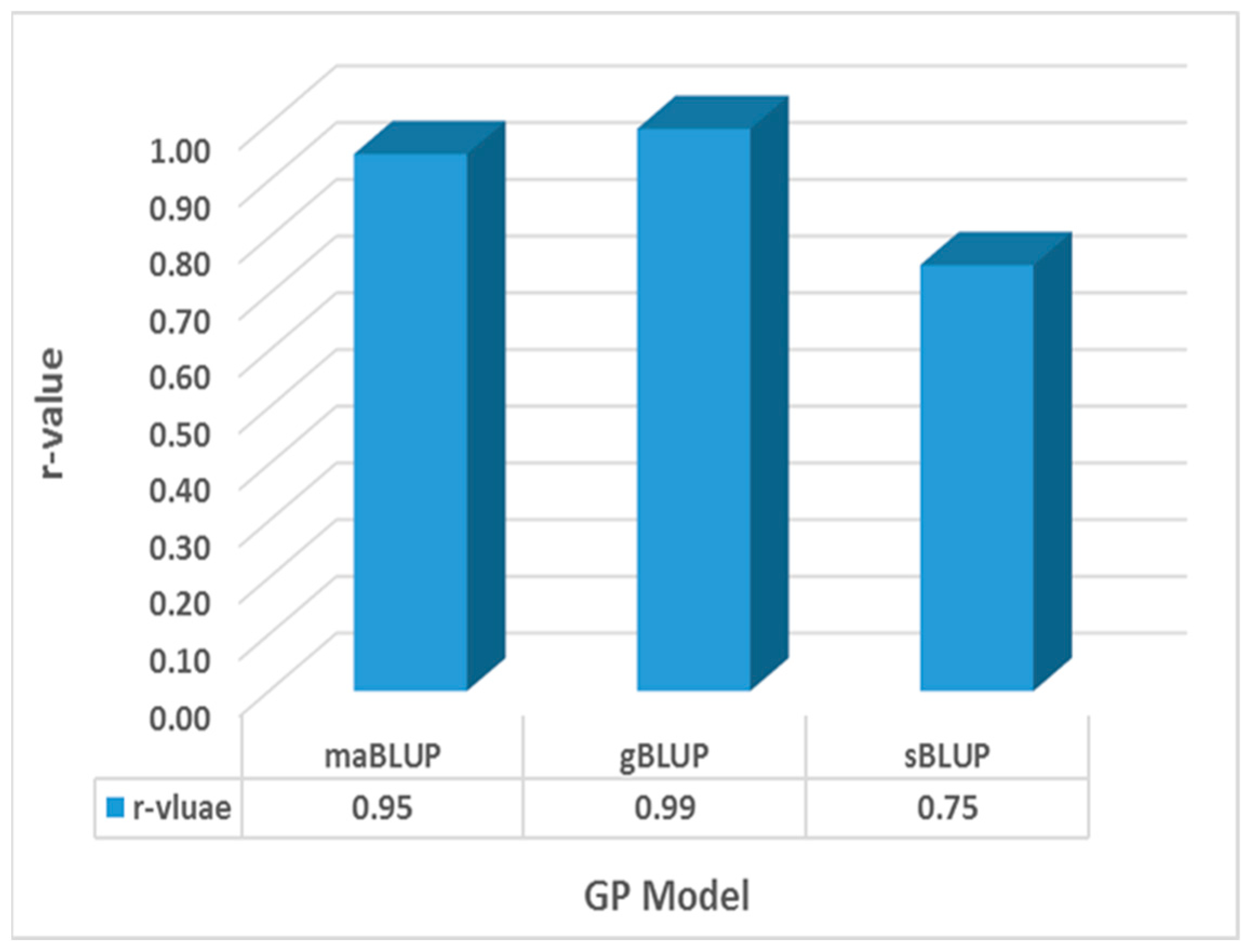
3.8. Genomic Prediction by GAGBLUP from 80% of the Whole Panel
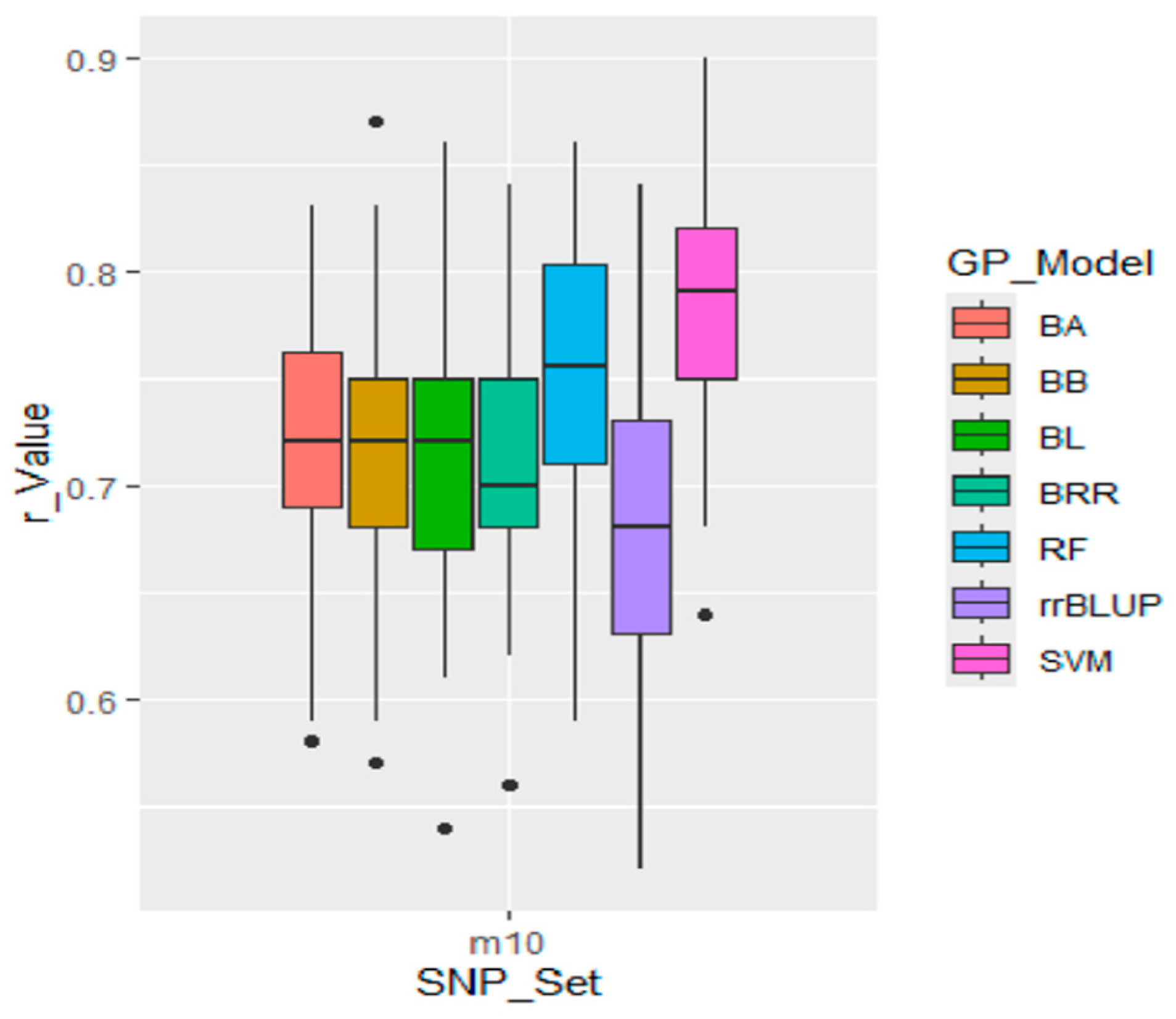
3.9. Genomic Prediction Using Difference Genomic Models
4. Discussion
4.1. Importance of Studying Arginine Content
4.2. Research Background of the Identified Loci
4.3. Rationale for Selecting These Two Candidate Genes
4.4. Harnessing GP for the Efficient Selection of Arginine Content in Soybean Breeding Programs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GWAS | Genome-Wide Association Study |
| SNP | Single Nucleotide Polymorphism |
| GP | Genomic Prediction |
| GS | Genomic Selection |
| PA | Predictive Accuracy |
| BLUP | Best Linear Unbiased Prediction |
| BA | Bayesian A |
| BB | Bayesian B |
| BRR | Bayesian Ridge Regression |
| SVM | Support Vector Machine |
| LOD | Logarithm of the Odds |
References
- Wang, Y.; Wang, X.; Zhang, R.; Chen, T.; Xiao, J.; Li, Q.; Ding, X.; Sun, X. Genome-Scale Identification of Wild Soybean Serine/Arginine-Rich Protein Family Genes and Their Responses to Abiotic Stresses. Int. J. Mol. Sci. 2024, 25, 11175. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Jia, Q.; Zhou, M.; Zhang, H.; Chen, H. Identification of excellent germplasm with high content of four free amino acids in soybean and GWAS analysis. J. Plant Genet. Resour. 2024, 25, 957–966. [Google Scholar]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- King, J.E.; Gifford, D.J. Amino Acid Utilization in Seeds of Loblolly Pine during Germination and Early Seedling Growth (I. Arginine and Arginase Activity). Plant Physiol. 1997, 113, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Siddappa, S.; Marathe, G.K. What we know about plant arginases? Plant Physiol. Biochem. 2020, 156, 600–610. [Google Scholar] [CrossRef]
- Pál, M.; Szalai, G.; Gondor, O.K.; Janda, T. Unfinished story of polyamines: Role of conjugation, transport and light-related regulation in the polyamine metabolism in plants. Plant Sci. 2021, 308, 110923. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Davis, T.A.; Kim, S.W.; Li, P.; Rhoads, J.M.; Yin, Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids 2009, 37, 153–168. [Google Scholar] [CrossRef]
- Xu, Y.Q.; Guo, Y.; Shi, B.; Yan, S.; Guo, X. Dietary arginine supplementation enhances the growth performance and immune status of broiler chickens. Livest. Sci. 2018, 209, 8–13. [Google Scholar] [CrossRef]
- Böger, R.H. The emerging role of asymmetric dimethylarginine as a novel cardiovascular risk factor. Cardiovasc. Res. 2003, 59, 824–833. [Google Scholar] [CrossRef]
- Bronte, V.; Zanovello, P. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 2005, 5, 641–654. [Google Scholar] [CrossRef]
- Qin, J.; Shi, A.; Song, Q.; Li, S.; Wang, F.; Cao, Y.; Ravelombola, W.; Song, Q.; Yang, C.; Zhang, M. Genome Wide Association Study and Genomic Selection of Amino Acid Concentrations in Soybean Seeds. Front. Plant Sci. 2019, 10, 1445. [Google Scholar] [CrossRef] [PubMed]
- Fallen, B.; Hatcher, C.; Allen, F.; Kopsell, D.; Saxton, A.; Chen, P.; Kantartzi, S.; Cregan, P.; Hyten, D.; Pantalone, V. Soybean Seed Amino Acid Content QTL Detected Using the Universal Soy Linkage Panel 1.0 with 1,536 SNPs. J. Plant Genome Sci. 2013, 1, 68–79. [Google Scholar] [CrossRef]
- Warrington, C.V.; Abdel-Haleem, H.; Hyten, D.L.; Cregan, P.B.; Orf, J.H.; Killam, A.S.; Bajjalieh, N.; Li, Z.; Boerma, H.R. QTL for seed protein and amino acids in the Benning × Danbaekkong soybean population. Theor. Appl. Genet. Theor. Angew. Genet. 2015, 128, 839–850. [Google Scholar] [CrossRef]
- Ravelombola, W.; Qin, J.; Shi, A.; Song, Q.; Yuan, J.; Wang, F.; Chen, P.; Yan, L.; Feng, Y.; Zhao, T.; et al. Genome-wide association study and genomic selection for yield and related traits in soybean. PLoS ONE 2021, 16, e0255761. [Google Scholar] [CrossRef]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J.; Meuwissen, T.H. Using the genomic relationship matrix to predict the accuracy of genomic selection. J. Anim. Breed. Genet. Z. Tierz. Zucht. 2011, 128, 409–421. [Google Scholar] [CrossRef]
- Zhang, J.; Song, Q.; Cregan, P.B.; Jiang, G.L. Genome-wide association study, genomic prediction and marker-assisted selection for seed weight in soybean (Glycine max). Theor. Appl. Genet. Theor. Angew. Genet. 2016, 129, 117–130. [Google Scholar] [CrossRef]
- Spindel, J.; Begum, H.; Akdemir, D.; Virk, P.; Collard, B.; Redoña, E.; Atlin, G.; Jannink, J.L.; McCouch, S.R. Correction: Genomic Selection and Association Mapping in Rice (Oryza sativa): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines. PLoS Genet. 2015, 11, e1005350. [Google Scholar]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Phung, N.T.; Mai, C.D.; Hoang, G.T.; Truong, H.T.; Lavarenne, J.; Gonin, M.; Nguyen, K.L.; Ha, T.T.; Do, V.N.; Gantet, P.; et al. Genome-wide association mapping for root traits in a panel of rice accessions from Vietnam. BMC Plant Biol. 2016, 16, 64. [Google Scholar] [CrossRef]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE 2013, 8, e54985. [Google Scholar] [CrossRef]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Fingerprinting Soybean Germplasm and Its Utility in Genomic Research. G3 Genes Genomes Genet. 2015, 5, 1999–2006. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Shi, A.; Gepts, P.; Song, Q.; Xiong, H.; Michaels, T.E.; Chen, S. Genome-Wide Association Study and Genomic Prediction for Soybean Cyst Nematode Resistance in USDA Common Bean (Phaseolus vulgaris) Core Collection. Front. Plant Sci. 2021, 12, 624156. [Google Scholar] [CrossRef]
- Zhang, X.; Sallam, A.; Gao, L.; Kantarski, T.; Poland, J.; DeHaan, L.R.; Wyse, D.L.; Anderson, J.A. Establishment and Optimization of Genomic Selection to Accelerate the Domestication and Improvement of Intermediate Wheatgrass. Plant Genome 2016, 9, plantgenome2015.07.0059. [Google Scholar] [CrossRef] [PubMed]
- Meher, P.K.; Rustgi, S.; Kumar, A. Performance of Bayesian and BLUP alphabets for genomic prediction: Analysis, comparison and results. Heredity 2022, 128, 519–530. [Google Scholar] [CrossRef]
- Pérez, P.; de los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef]
- Jarquín, D.; Kocak, K.; Posadas, L.; Hyma, K.; Jedlicka, J.; Graef, G.; Lorenz, A. Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genom. 2014, 15, 740. [Google Scholar] [CrossRef]
- Krishnappa, G.; Savadi, S.; Tyagi, B.S.; Singh, S.K.; Mamrutha, H.M.; Kumar, S.; Mishra, C.N.; Khan, H.; Gangadhara, K.; Uday, G.; et al. Integrated genomic selection for rapid improvement of crops. Genomics 2021, 113, 1070–1086. [Google Scholar] [CrossRef]
- Qu, S.; Lu, S.; Liu, Y.; Li, M.; Chen, S. Accurate genomic selection using low-density SNP panels preselected by maximum likelihood estimation. Aquaculture 2024, 579, 740154. [Google Scholar] [CrossRef]
- Funck, D.; Eckard, S.; Müller, G. Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol. 2010, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, X.; Lu, Y.; Bhusal, S.J.; Song, Q.; Cregan, P.B.; Yen, Y.; Brown, M.; Jiang, G.L. Genome-wide Scan for Seed Composition Provides Insights into Soybean Quality Improvement and the Impacts of Domestication and Breeding. Mol. Plant 2018, 11, 460–472. [Google Scholar] [CrossRef] [PubMed]
| SNP | Chr. | Position | LOD | Model | LOD | PVE | Beneficial _Allele | Unbeneficial _Allele | Link Gene |
|---|---|---|---|---|---|---|---|---|---|
| (t-test) | (%) | (0–5 k) | |||||||
| Gm05_464582_ss715592561 | 5 | 464582 | 6.73 | FarmCPU | 7.87 | 2.58 | C | T | Glyma.05G005300 |
| Gm06_19014194_ss715593808 | 6 | 19014194 | 9.91 8.02 | BLINK, FarmCPU | 7.23 | 3.91 | A | G | Glyma.06g203200 |
| Gm08_18566925_ss715600087 | 8 | 18566925 | 6.46 | FarmCPU | 1.50 | 1.16 | C | T | Glyma.08g227900 |
| Gm11_2054710_ss715609614 | 11 | 2054710 | 6.38 9.05 | MLM, MLMM | 21.24 | 19 | A | C | Glyma.11G028600 |
| Gm11_7143691_ss715611069 | 11 | 7143691 | 10.71 | BLINK | 10.40 | 10.19 | C | T | Glyma.11g094000 |
| Gm12_40011028_ss715613048 | 12 | 40011028 | 5.85 | BLINK | 1.78 | 3.81 | C | T | Glyma.12g241800 |
| Gm13_27198365_ss715614420 | 13 | 27198365 | 6.06 | BLINK | 5.69 | 7.61 | A | G | Glyma.13g156700 |
| Gm13_37091348_ss715615859 | 13 | 37091348 | 6.26 | FarmCPU | 5.37 | 0 | G | T | Glyma.13G268700 |
| Gm16_3557974_ss715624794 | 16 | 3557974 | 5.93 | MLMM | 4.93 | 5.8 | C | T | Glyma.16g037600 |
| Gm17_39308794_ss715627603 | 17 | 39308794 | 8.59 | FarmCPU | 3.05 | 0 | A | G | Glyma.17g237800 |
| SNP_Set | r-Value | SE of r-Value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rrBLUP | BA | BB | BL | BRR | SNP Set Mean | rrBLUP | BA | BB | BL | BRR | SNP Set Mean | |
| r10 | 0.34 | 0.30 | 0.28 | 0.34 | 0.32 | 0.32 | 0.11 | 0.10 | 0.13 | 0.11 | 0.11 | 0.12 |
| r100 | 0.63 | 0.64 | 0.62 | 0.63 | 0.64 | 0.63 | 0.08 | 0.08 | 0.07 | 0.08 | 0.09 | 0.08 |
| r200 | 0.58 | 0.55 | 0.54 | 0.57 | 0.56 | 0.56 | 0.07 | 0.08 | 0.09 | 0.07 | 0.08 | 0.08 |
| r500 | 0.32 | 0.32 | 0.32 | 0.33 | 0.32 | 0.32 | 0.08 | 0.09 | 0.08 | 0.10 | 0.09 | 0.09 |
| r1000 | 0.58 | 0.60 | 0.60 | 0.60 | 0.62 | 0.60 | 0.08 | 0.09 | 0.08 | 0.08 | 0.08 | 0.08 |
| r2000 | 0.70 | 0.68 | 0.68 | 0.69 | 0.69 | 0.69 | 0.06 | 0.07 | 0.07 | 0.06 | 0.07 | 0.07 |
| r5000 | 0.68 | 0.69 | 0.68 | 0.71 | 0.70 | 0.69 | 0.07 | 0.06 | 0.07 | 0.06 | 0.06 | 0.06 |
| r10,000 | 0.75 | 0.76 | 0.76 | 0.77 | 0.76 | 0.76 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 |
| m10 | 0.68 | 0.72 | 0.71 | 0.72 | 0.71 | 0.71 | 0.07 | 0.05 | 0.05 | 0.06 | 0.05 | 0.06 |
| GP Model Mean | 0.58 | 0.58 | 0.58 | 0.60 | 0.59 | 0.59 | 0.07 | 0.07 | 0.08 | 0.07 | 0.08 | 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Yang, Q.; Yu, C.; Liu, Z.; Shi, X.; Wu, X.; Xu, R.; Shen, P.; Zhang, Y.; Shi, A.; et al. Identification of Loci and Candidate Genes Associated with Arginine Content in Soybean. Agronomy 2025, 15, 1339. https://doi.org/10.3390/agronomy15061339
Ma J, Yang Q, Yu C, Liu Z, Shi X, Wu X, Xu R, Shen P, Zhang Y, Shi A, et al. Identification of Loci and Candidate Genes Associated with Arginine Content in Soybean. Agronomy. 2025; 15(6):1339. https://doi.org/10.3390/agronomy15061339
Chicago/Turabian StyleMa, Jiahao, Qing Yang, Cuihong Yu, Zhi Liu, Xiaolei Shi, Xintong Wu, Rongqing Xu, Pengshuo Shen, Yuechen Zhang, Ainong Shi, and et al. 2025. "Identification of Loci and Candidate Genes Associated with Arginine Content in Soybean" Agronomy 15, no. 6: 1339. https://doi.org/10.3390/agronomy15061339
APA StyleMa, J., Yang, Q., Yu, C., Liu, Z., Shi, X., Wu, X., Xu, R., Shen, P., Zhang, Y., Shi, A., & Yan, L. (2025). Identification of Loci and Candidate Genes Associated with Arginine Content in Soybean. Agronomy, 15(6), 1339. https://doi.org/10.3390/agronomy15061339







