Multiple Transcriptomic Networks Regulate the Callus Development Process in Panax ginseng
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Gamma Irradiation, and Embryo Induction
2.2. Total RNA Isolation and RNA-Seq Data Production
2.3. DEG Identification and Functional Annotation
2.4. Network Analysis
3. Results and Discussion
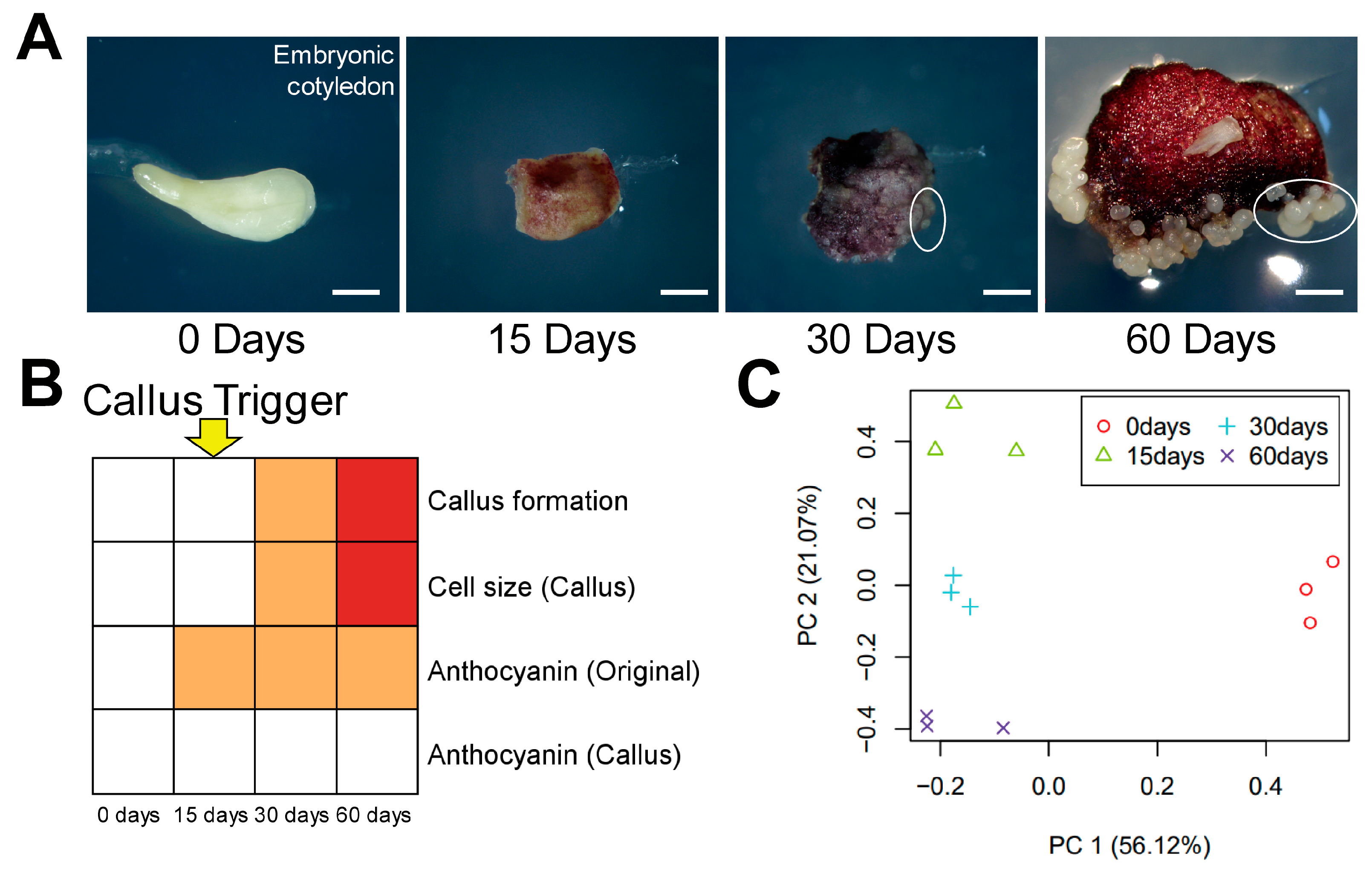
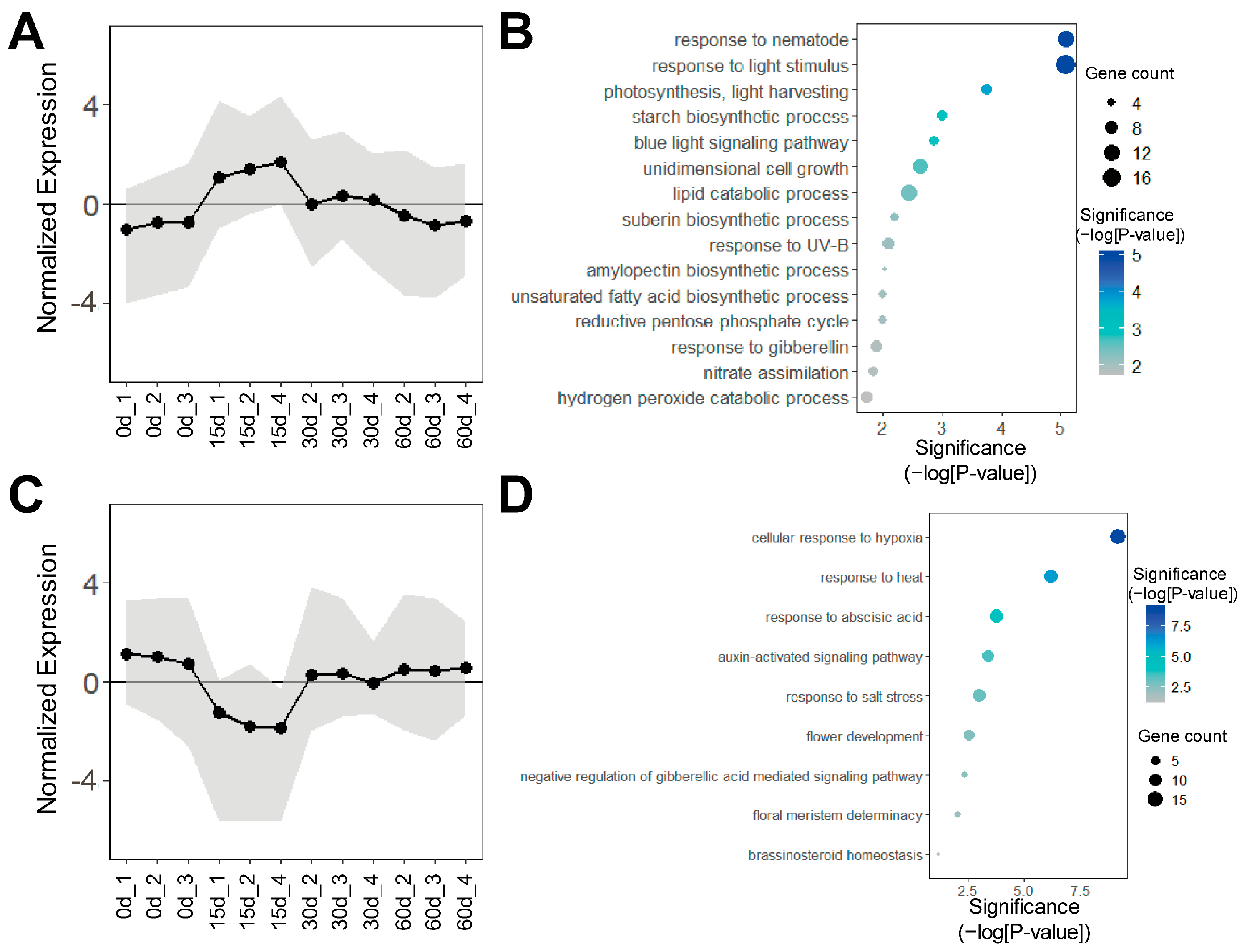
3.1. Time-Course Transcriptome Analysis of the Callus Induction Process
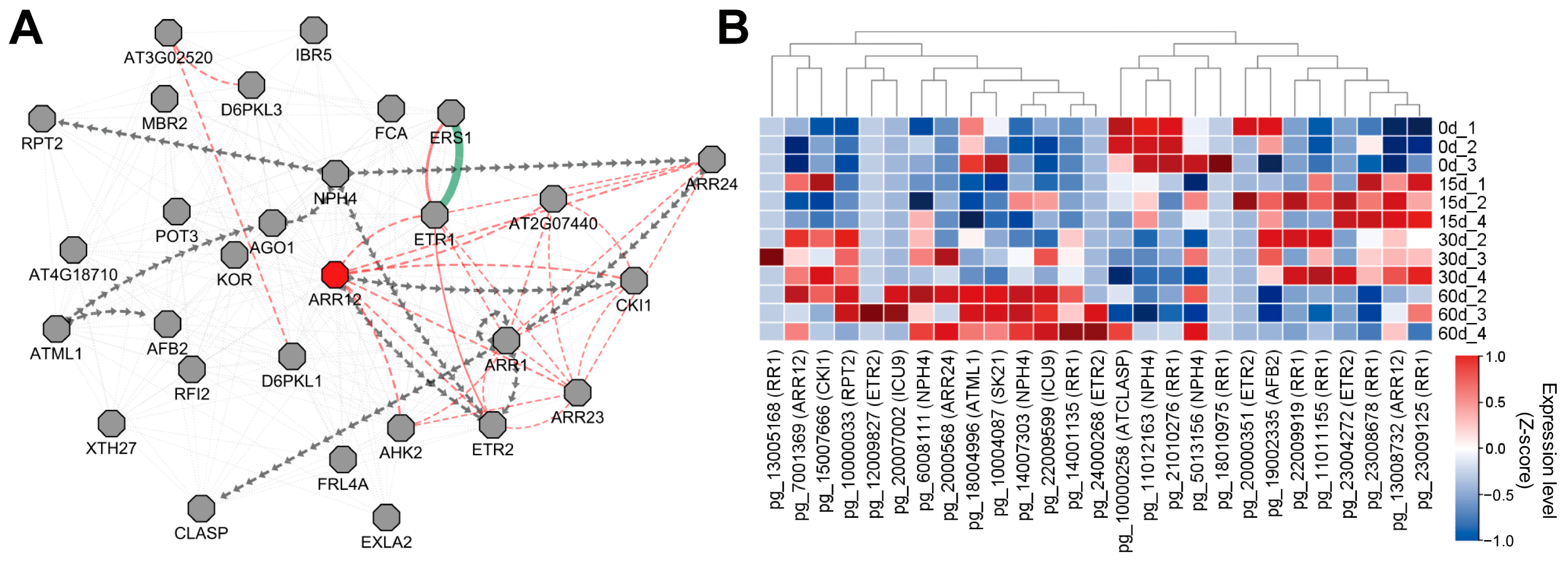
3.2. ARR12-Correlated Gene Networks Were Identified to Be Associated with Callus Induction
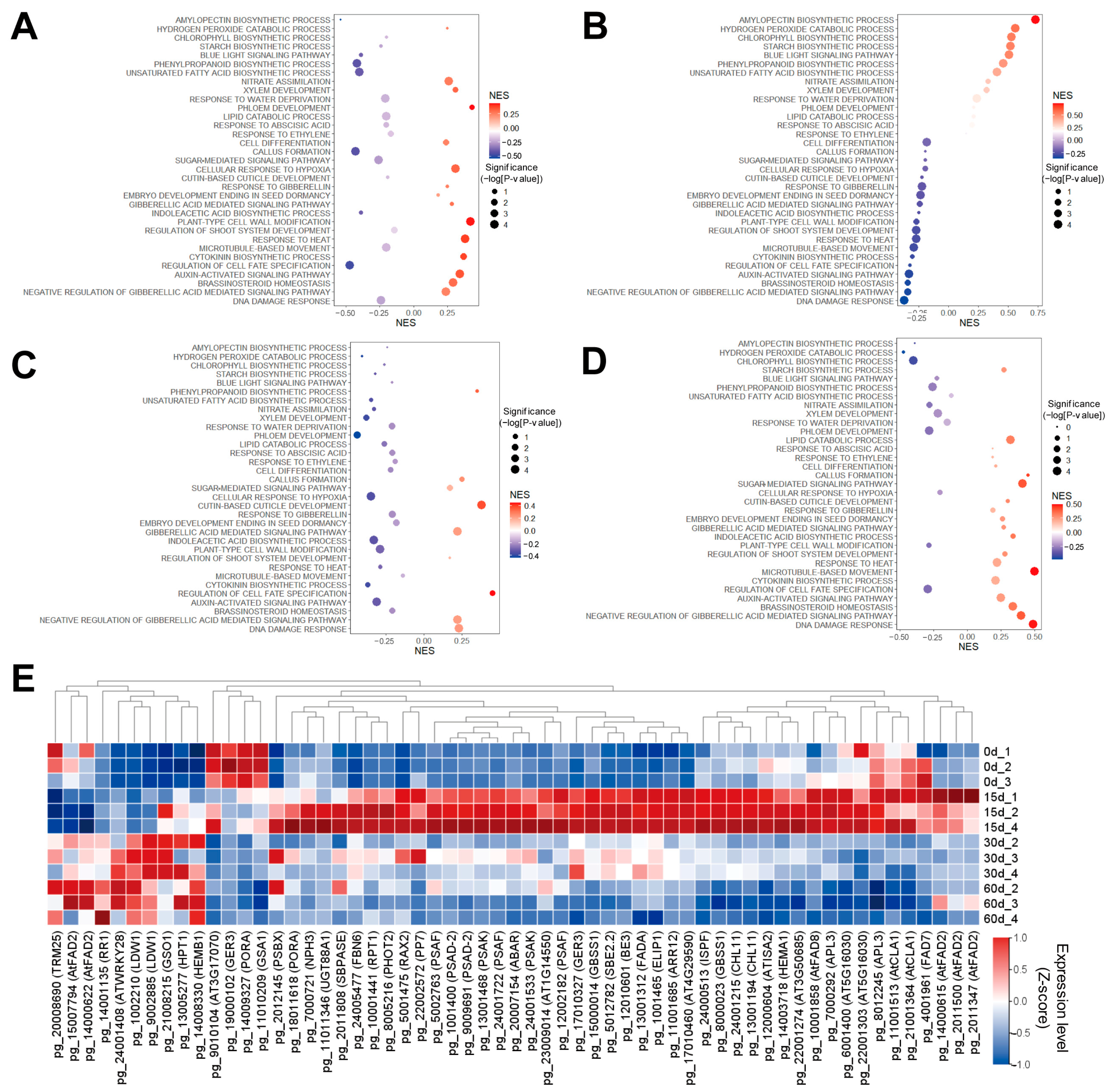
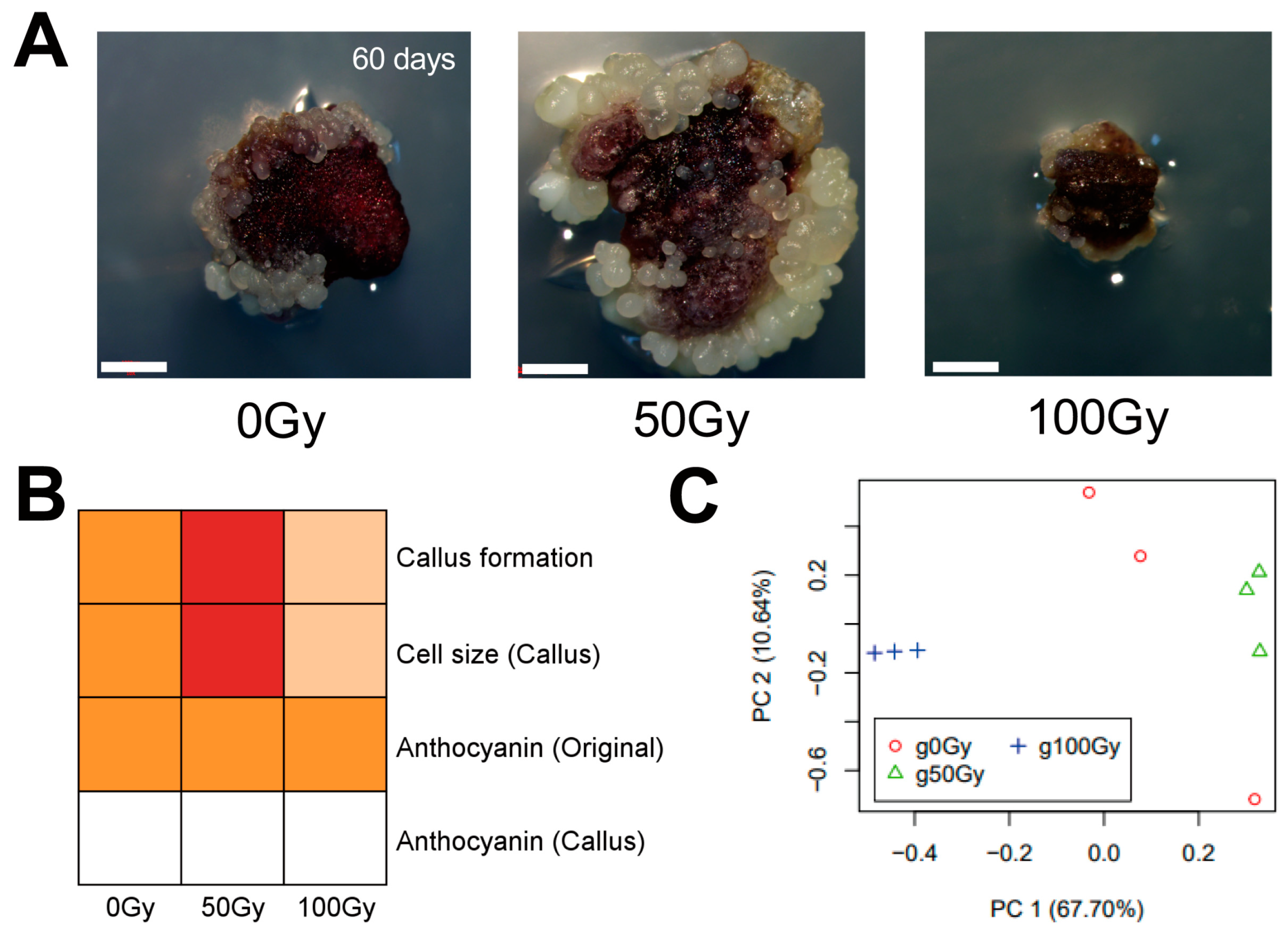
3.3. Gamma Radiation Treatment Modulates Cell Growth of Panax ginseng Callus
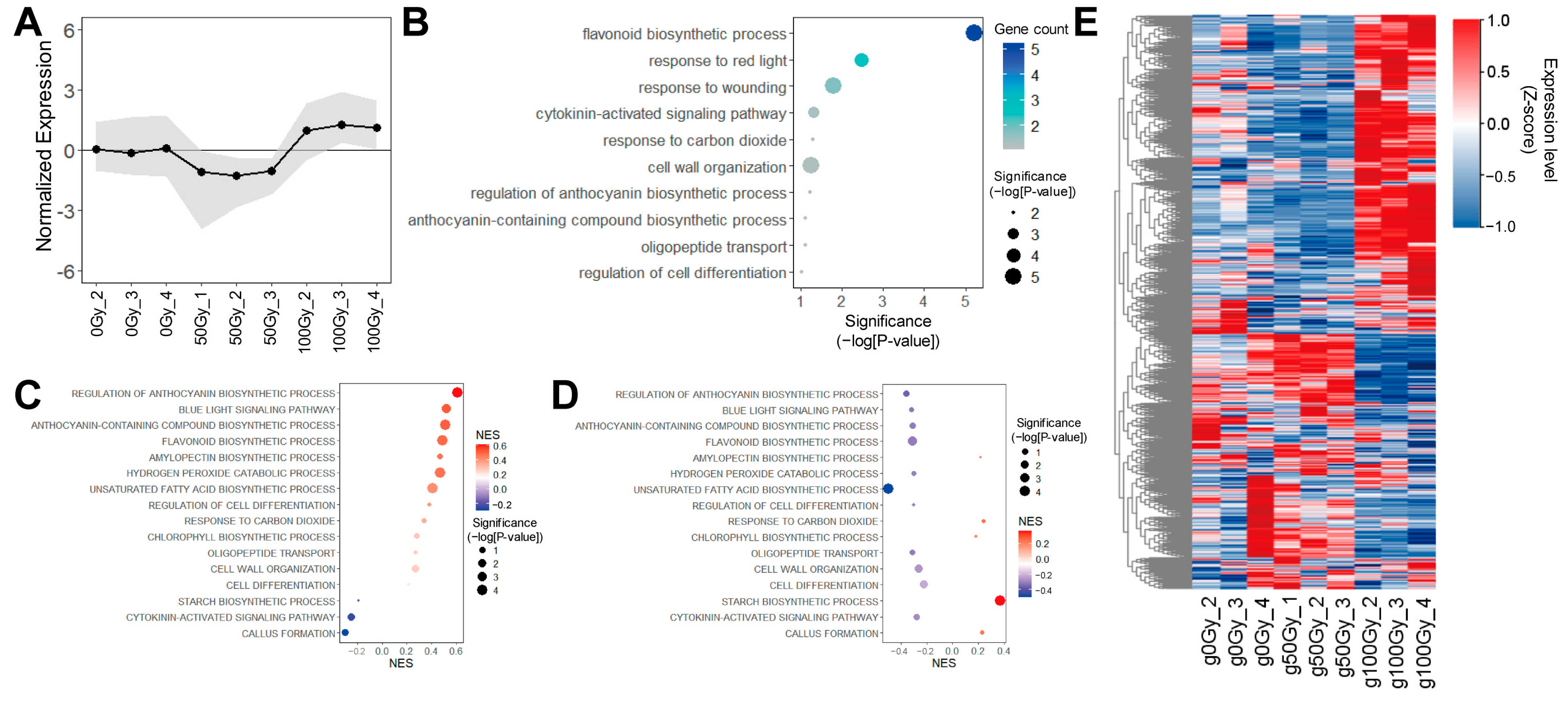
3.4. The WIND1-Associated Gene Network Was Associated with a Dose-Dependent Growth Pattern of Gamma-Ray Irradiation
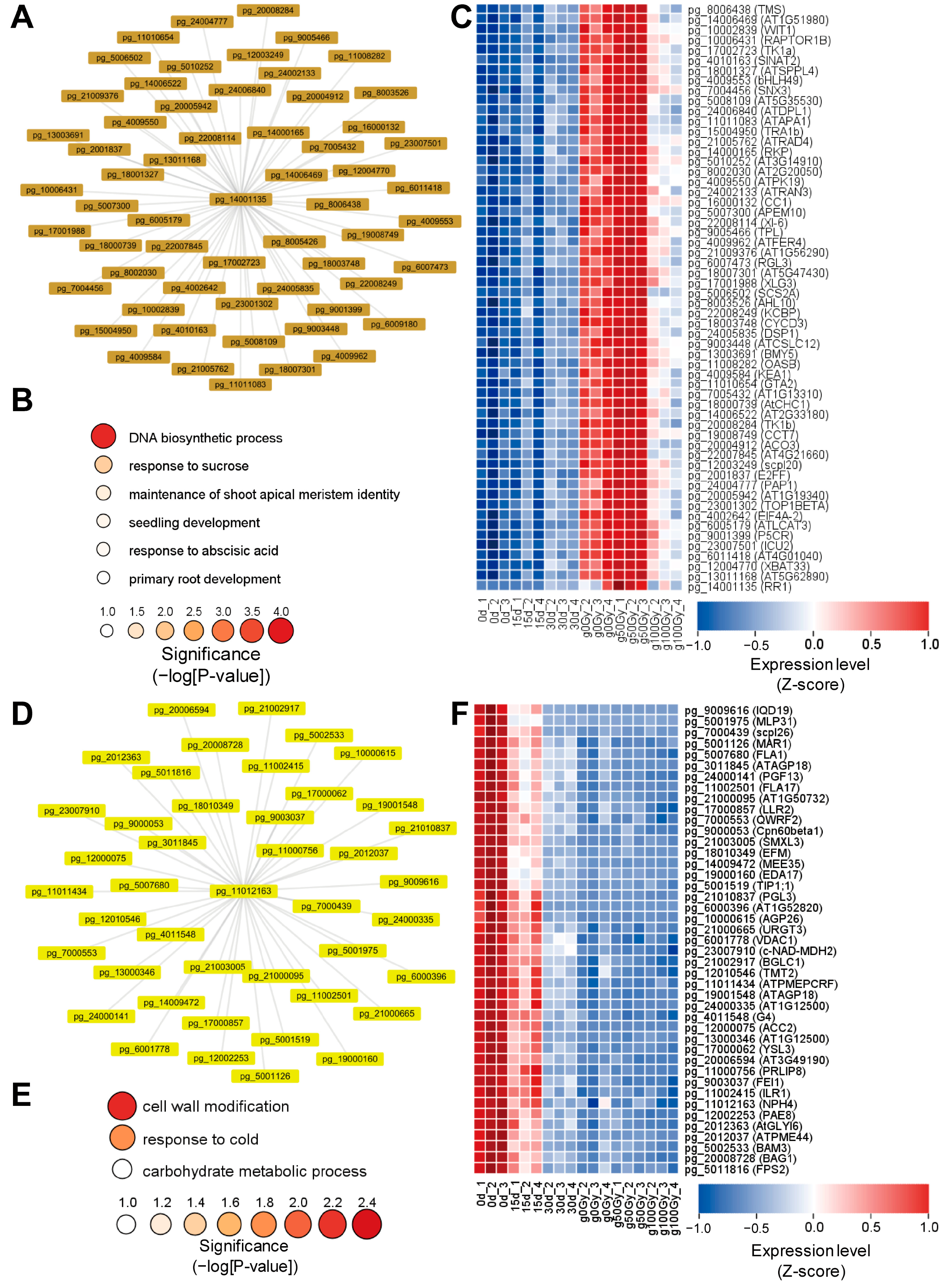
3.5. Global Co-Expression Network Analysis Reveals the RR1- and NPH4-Associated Networks to Be Associated with Callus Development
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, J.; Zhong, J.-J. Production of ginseng and its bioactive components in plant cell culture: Current technological and applied aspects. J. Biotechnol. 1999, 68, 89–99. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cao, H.; Xu, E.; Zhang, S.; Hu, Y. Genome-Wide Identification of Arabidopsis LBD29 Target Genes Reveals the Molecular Events behind Auxin-Induced Cell Reprogramming during Callus Formation. Plant Cell Physiol. 2018, 59, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dai, X.; Li, J.; Liu, N.; Liu, X.; Li, S.; Xiang, F. The Type-B Cytokinin Response Regulator ARR1 Inhibits Shoot Regeneration in an ARR12-Dependent Manner in Arabidopsis. Plant Cell 2020, 32, 2271–2291. [Google Scholar] [CrossRef]
- Ye, B.B.; Shang, G.D.; Pan, Y.; Xu, Z.G.; Zhou, C.M.; Mao, Y.B.; Bao, N.; Sun, L.; Xu, T.; Wang, J.W. AP2/ERF Transcription Factors Integrate Age and Wound Signals for Root Regeneration. Plant Cell 2020, 32, 226–241. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.; Yoshikawa, T.; Ishii, T.; Kajii, K. Regulation of saponin production in callus cultures of Panax ginseng [1]. Planta Med. 1983, 47, 200–204. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Furuya, T. Saponin production by cultures of Panax ginseng transformed with Agrobacterium rhizogenes. Plant Cell Rep. 1987, 6, 449–453. [Google Scholar] [CrossRef]
- Shoyama, Y.; Kamura, K.; Nishioka, I. Somatic Embryogenesis and Clonal Multiplication of Panax ginseng. Planta Med. 1988, 54, 155–156. [Google Scholar] [CrossRef]
- Arya, S.; Liu, J.R.; Eriksson, T. Plant regeneration from protoplasts of Panax ginseng (C.A. Meyer) through somatic embryogenesis. Plant Cell Rep. 1991, 10, 277–281. [Google Scholar] [CrossRef]
- Chang, W.C.; Hsing, Y.I. Plant regeneration through somatic embryogenesis in root-derived callus of ginseng (Panax ginseng C. A. Meyer). Theor. Appl. Genet. 1980, 57, 133–135. [Google Scholar] [CrossRef]
- Du, L.G.; Hou, Y.H.; Chang, W.C.; Li, A.S.; Fu, Z.M.; Shao, Q.Q.; Chen, Z.G.; Yang, Z.T. In vitro pollen plant induction and embryoid clone establishment of Panax ginseng. Sci. Sin. B 1987, 30, 941–945. [Google Scholar]
- Furuya, T.; Yoshikawa, T.; Ishii, T.; Kajii, K. Effects of auxins on growth and saponin production in callus cultures of Panax ginseng. Planta Med. 1983, 47, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Le, K.-C.; Ho, T.-T.; Paek, K.-Y.; Park, S.-Y. Low dose gamma radiation increases the biomass and ginsenoside content of callus and adventitious root cultures of wild ginseng (Panax ginseng Mayer). Ind. Crops Prod. 2019, 130, 16–24. [Google Scholar] [CrossRef]
- Kim, D.S.; Song, M.; Kim, S.H.; Jang, D.S.; Kim, J.B.; Ha, B.K.; Kim, S.H.; Lee, K.J.; Kang, S.Y.; Jeong, I.Y. The improvement of ginsenoside accumulation in Panax ginseng as a result of γ-irradiation. J. Ginseng Res. 2013, 37, 332–340. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Bae, T.W.; Boo, K.H.; Sun, H.J.; Song, I.J.; Pham, C.H.; Ganesan, M.; Yang, D.H.; Kang, H.G.; Ko, S.M.; et al. Ginsenoside Production and Morphological Characterization of Wild Ginseng (Panax ginseng Meyer) Mutant Lines Induced by γ-irradiation (60Co) of Adventitious Roots. J. Ginseng. Res. 2011, 35, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Sun, H.J.; Song, I.J.; Bae, T.W.; Kang, H.G.; Ko, S.M.; Kwon, Y.I.; Kim, I.W.; Lee, J.; Park, S.Y.; et al. Plant regeneration of Korean wild ginseng (Panax ginseng Meyer) mutant lines induced by γ-irradiation (60Co) of adventitious roots. J. Ginseng. Res. 2014, 38, 220–225. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Tang, M.; Qu, Z.; Shao, C.; Zheng, P.; Hou, W. Transcriptomic and Metabolomic Research on the Germination Process of Panax ginseng Overwintering Buds. Plants 2024, 13, 1041. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-X.; Niu, Y.-Q.; Wang, X.-F.; Wang, Z.-H.; Wang, M.-L.; Yang, J.; Wang, Y.-G.; Zhang, W.-J.; Song, Z.-P.; Li, L.-F. Phenotypic and transcriptomic responses of the shade-grown species Panax ginseng to variable light conditions. Ann. Bot. 2022, 130, 749–762. [Google Scholar] [CrossRef]
- Hong, C.P.; Kim, J.; Lee, J.; Yoo, S.I.; Bae, W.; Geem, K.R.; Yu, J.; Jang, I.; Jo, I.H.; Cho, H.; et al. Gibberellin Signaling Promotes the Secondary Growth of Storage Roots in Panax ginseng. Int. J. Mol. Sci. 2021, 22, 8694. [Google Scholar] [CrossRef]
- Geem, K.R.; Kim, J.; Bae, W.; Jee, M.-G.; Yu, J.; Jang, I.; Lee, D.-Y.; Hong, C.P.; Shim, D.; Ryu, H. Nitrate enhances the secondary growth of storage roots in Panax ginseng. J. Ginseng Res. 2023, 47, 469–478. [Google Scholar] [CrossRef]
- Hong, J.; Han, S.; Geem, K.R.; Bae, W.; Kim, J.; Jee, M.-G.; Lee, J.-W.; Kim, J.-U.; Lee, G.; Joo, Y.; et al. Identification of a key signaling network regulating perennating bud dormancy in Panax ginseng. J. Ginseng Res. 2024, 48, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yun, Y.; Huh, J.; Um, Y.; Shim, D. Comparative transcriptome analysis on wild-simulated ginseng of different age revealed possible mechanism of ginsenoside accumulation. Plant Physiol. Biochem. 2023, 201, 107870. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhao, J.; Liu, Y.; Li, N.; Wang, Z.; Wang, X.; Liu, X.; Jiang, L.; Liu, B.; Fu, X.; et al. High Chromosomal Stability and Immortalized Totipotency Characterize Long-Term Tissue Cultures of Chinese Ginseng (Panax ginseng). Genes 2021, 12, 514. [Google Scholar] [CrossRef]
- Vardhan, P.V.; Shukla, L.I. Gamma irradiation of medicinally important plants and the enhancement of secondary metabolite production. Int. J. Radiat. Biol. 2017, 93, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-W.; Jo, I.-H.; Kim, J.-U.; Hong, C.-E.; Kim, Y.-C.; Kim, D.-H.; Park, Y.-D. Improvement of seed dehiscence and germination in ginseng by stratification, gibberellin, and/or kinetin treatments. Hortic. Environ. Biotechnol. 2018, 59, 293–301. [Google Scholar] [CrossRef]
- Lee, J.-W.; Do, G.-R.; Jo, I.-H.; Hong, C.-E.; Bang, K.-H.; Kim, J.-U.; Park, Y.-D. Zygotic embryo culture is an efficient way to optimize in vitro growth in Panax ginseng. Ind. Crops Prod. 2021, 167, 113497. [Google Scholar] [CrossRef]
- Lee, J.-W.; Kim, J.-U.; Bang, K.-H.; Kim, D.-H.; Jo, I.-H.; Park, Y.-D. Efficient Micropropagation of Genetically Stable Panax ginseng Meyer by Somatic Embryogenesis. Agronomy 2023, 13, 1139. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, Y.; Wang, X.; Yu, X.; Liao, Y.; Zhang, H.; Li, L.; Wang, Y.; Liu, B.; Li, W. Telomere-to-telomere reference genome for Panax ginseng highlights the evolution of saponin biosynthesis. Hortic. Res. 2024, 11, uhae107. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis, 2nd edition. Meas.-Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kakimoto, T. CKI1, a Histidine Kinase Homolog Implicated in Cytokinin Signal Transduction. Science 1996, 274, 982–985. [Google Scholar] [CrossRef]
- Zhao, X.-C.; Qu, X.; Mathews, D.E.; Schaller, G.E. Effect of Ethylene Pathway Mutations upon Expression of the Ethylene Receptor ETR1 from Arabidopsis. Plant Physiol. 2002, 130, 1983–1991. [Google Scholar] [CrossRef]
- Ambrose, J.C.; Shoji, T.; Kotzer, A.M.; Pighin, J.A.; Wasteneys, G.O. The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 2007, 19, 2763–2775. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.J.; Estelle, M. Mechanism of Auxin-Regulated Gene Expression in Plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef]
- Morel, A.; Teyssier, C.; Trontin, J.-F.; Eliášová, K.; Pešek, B.; Beaufour, M.; Morabito, D.; Boizot, N.; Le Metté, C.; Belal-Bessai, L.; et al. Early molecular events involved in Pinus pinaster Ait. somatic embryo development under reduced water availability: Transcriptomic and proteomic analyses. Physiol. Plant. 2014, 152, 184–201. [Google Scholar] [CrossRef]
- Xavier, L.R.; Corrêa, C.C.G.; da Paschoa, R.P.; Vieira, K.d.S.; Pacheco, D.D.R.; Gomes, L.d.E.S.; Duncan, B.C.; da Conceição, L.d.S.; Pinto, V.B.; Santa-Catarina, C.; et al. Time-Dependent Proteomic Signatures Associated with Embryogenic Callus Induction in Carica papaya L. Plants 2023, 12, 3891. [Google Scholar] [CrossRef]
- Iida, H.; Mähönen, A.P.; Jürgens, G.; Takada, S. Epidermal injury-induced derepression of key regulator ATML1 in newly exposed cells elicits epidermis regeneration. Nat. Commun. 2023, 14, 1031. [Google Scholar] [CrossRef]
- Takada, S. Post-Embryonic Induction of ATML1-SRDX Alters the Morphology of Seedlings. PLoS ONE 2013, 8, e79312. [Google Scholar] [CrossRef]
- Zhu, X.; Xu, Z.; Wang, G.; Cong, Y.; Yu, L.; Jia, R.; Qin, Y.; Zhang, G.; Li, B.; Yuan, D.; et al. Single-cell resolution analysis reveals the preparation for reprogramming the fate of stem cell niche in cotton lateral meristem. Genome Biol. 2023, 24, 194. [Google Scholar]
- Iwase, A.; Kondo, Y.; Laohavisit, A.; Takebayashi, A.; Ikeuchi, M.; Matsuoka, K.; Asahina, M.; Mitsuda, N.; Shirasu, K.; Fukuda, H.; et al. WIND transcription factors orchestrate wound-induced callus formation, vascular reconnection and defense response in Arabidopsis. New Phytol. 2021, 232, 734–752. [Google Scholar] [CrossRef]
- Iwase, A.; Nobutaka, M.; Momoko, I.; Mariko, O.; Chie, K.; Koich, K.; Jun, I.; Hiroshi, E.; Sugimoto, K. Arabidopsis WIND1 induces callus formation in rapeseed, tomato, and tobacco. Plant Signal. Behav. 2013, 8, e27432. [Google Scholar] [CrossRef]
- Suo, J.; Zhou, C.; Zeng, Z.; Li, X.; Bian, H.; Wang, J.; Zhu, M.; Han, N. Identification of regulatory factors promoting embryogenic callus formation in barley through transcriptome analysis. BMC Plant Biol. 2021, 21, 145. [Google Scholar] [CrossRef]
- Hao, J.; Xu, D.; Wang, C.; Cao, Q.; Zhao, Q.; Xie, M.; Zhang, H.; Zhang, L. Phylogeny and expression patterns of ERF genes that are potential reproductive inducers in hybrid larch. BMC Genom. 2024, 25, 288. [Google Scholar] [CrossRef]
- Ding, A.; Yang, X.; Yu, X.; Chen, Z.; Liu, Y.; Wang, W.; Sun, Y. A chimeric AtERF4 repressor modulates pleiotropic aspects of plant growth and abiotic stress tolerance in transgenic Arabidopsis. Plant Growth Regul. 2022, 96, 255–267. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Zhao, Q.; Li, T.; Wei, J.; Li, B.; Shen, W.; Yang, C.; Zeng, Y.; Rodriguez, P.L.; et al. The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat. Plants 2019, 5, 512–524. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Sánchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Hu, F.-X.; Zhong, J.-J. Jasmonic acid mediates gene transcription of ginsenoside biosynthesis in cell cultures of Panax notoginseng treated with chemically synthesized 2-hydroxyethyl jasmonate. Process Biochem. 2008, 43, 113–118. [Google Scholar] [CrossRef]
- Ali, M.B.; Yu, K.W.; Hahn, E.J.; Paek, K.Y. Methyl jasmonate and salicylic acid elicitation induces ginsenosides accumulation, enzymatic and non-enzymatic antioxidant in suspension culture Panax ginseng roots in bioreactors. Plant Cell Rep. 2006, 25, 613–620. [Google Scholar] [CrossRef]
- Gao, L.; Liu, J.; Liao, L.; Gao, A.; Njuguna, B.N.; Zhao, C.; Zheng, B.; Han, Y. Callus Induction and Adventitious Root Regeneration of Cotyledon Explants in Peach Trees. Horticulturae 2023, 9, 850. [Google Scholar] [CrossRef]
- Yasuda, T.; Yajima, Y.; Yamada, Y. Induction of DNA synthesis and callus formation from tuber tissue of Jerusalem artichoke by 2,4-dichlorophenoxyacetic acid. Plant Cell Physiol. 1974, 15, 321–329. [Google Scholar]
- Xu, X.; Legay, S.; Berni, R.; Hausman, J.-F.; Guerriero, G. Transcriptomic Changes in Internode Explants of Stinging Nettle during Callogenesis. Int. J. Mol. Sci. 2021, 22, 12319. [Google Scholar] [CrossRef]
- Lechon, T.; Kent, N.A.; Murray, J.A.H.; Scofield, S. Regulation of meristem and hormone function revealed through analysis of directly-regulated SHOOT MERISTEMLESS target genes. Sci. Rep. 2025, 15, 240. [Google Scholar] [CrossRef] [PubMed]
- Manzano, D.; Andrade, P.; Caudepón, D.; Altabella, T.; Arró, M.; Ferrer, A. Suppressing Farnesyl Diphosphate Synthase Alters Chloroplast Development and Triggers Sterol-Dependent Induction of Jasmonate- and Fe-Related Responses. Plant Physiol. 2016, 172, 93–117. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lee, J.-W.; Jo, I.-H. Multiple Transcriptomic Networks Regulate the Callus Development Process in Panax ginseng. Agronomy 2025, 15, 1244. https://doi.org/10.3390/agronomy15051244
Kim J, Lee J-W, Jo I-H. Multiple Transcriptomic Networks Regulate the Callus Development Process in Panax ginseng. Agronomy. 2025; 15(5):1244. https://doi.org/10.3390/agronomy15051244
Chicago/Turabian StyleKim, Jaewook, Jung-Woo Lee, and Ick-Hyun Jo. 2025. "Multiple Transcriptomic Networks Regulate the Callus Development Process in Panax ginseng" Agronomy 15, no. 5: 1244. https://doi.org/10.3390/agronomy15051244
APA StyleKim, J., Lee, J.-W., & Jo, I.-H. (2025). Multiple Transcriptomic Networks Regulate the Callus Development Process in Panax ginseng. Agronomy, 15(5), 1244. https://doi.org/10.3390/agronomy15051244






