Genome-Wide Characterization and Haplotype Module Stacking Analysis of the KTI Gene Family in Soybean (Glycine max L. Merr.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of KTI Gene Family Members
2.2. Analysis of Protein Characteristics and Physicochemical Properties of GmKTIs
2.3. Construction of Phylogenetic Tree
2.4. Gene Structure, Conserved Motifs, Promoter Cis-Acting Elements, and Transcriptional Profiling ANALYSIS
2.5. Collinearity and Phylogenetic Analysis of Paralogous Genes
2.6. Haplotype and Haplotype Module Stacking Analysis
3. Results
3.1. Identification of GmKTI Family
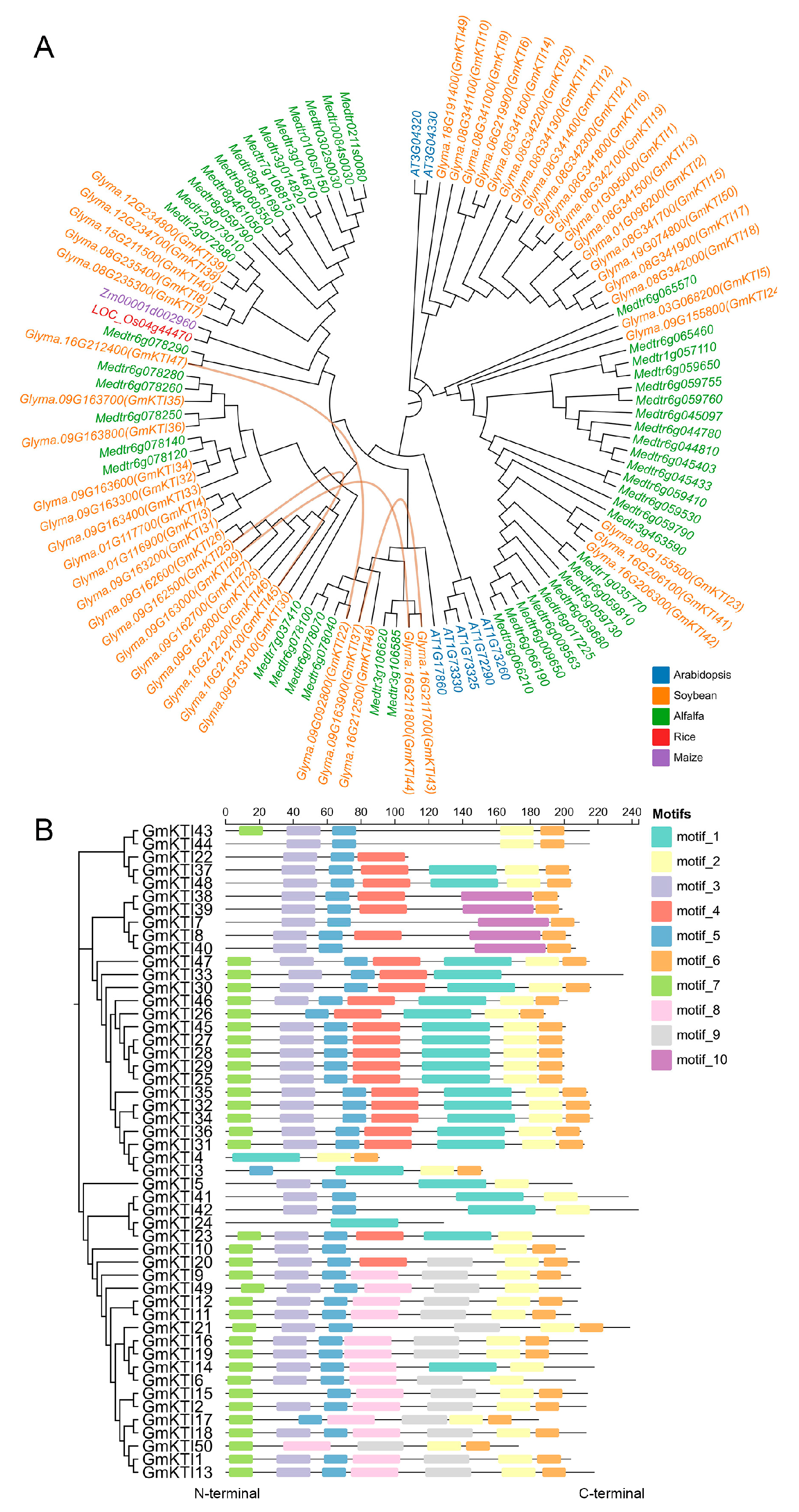
3.2. Phylogenetic Analysis of the GmKTI Family
3.3. Conservation Motif Analysis of GmKTIs
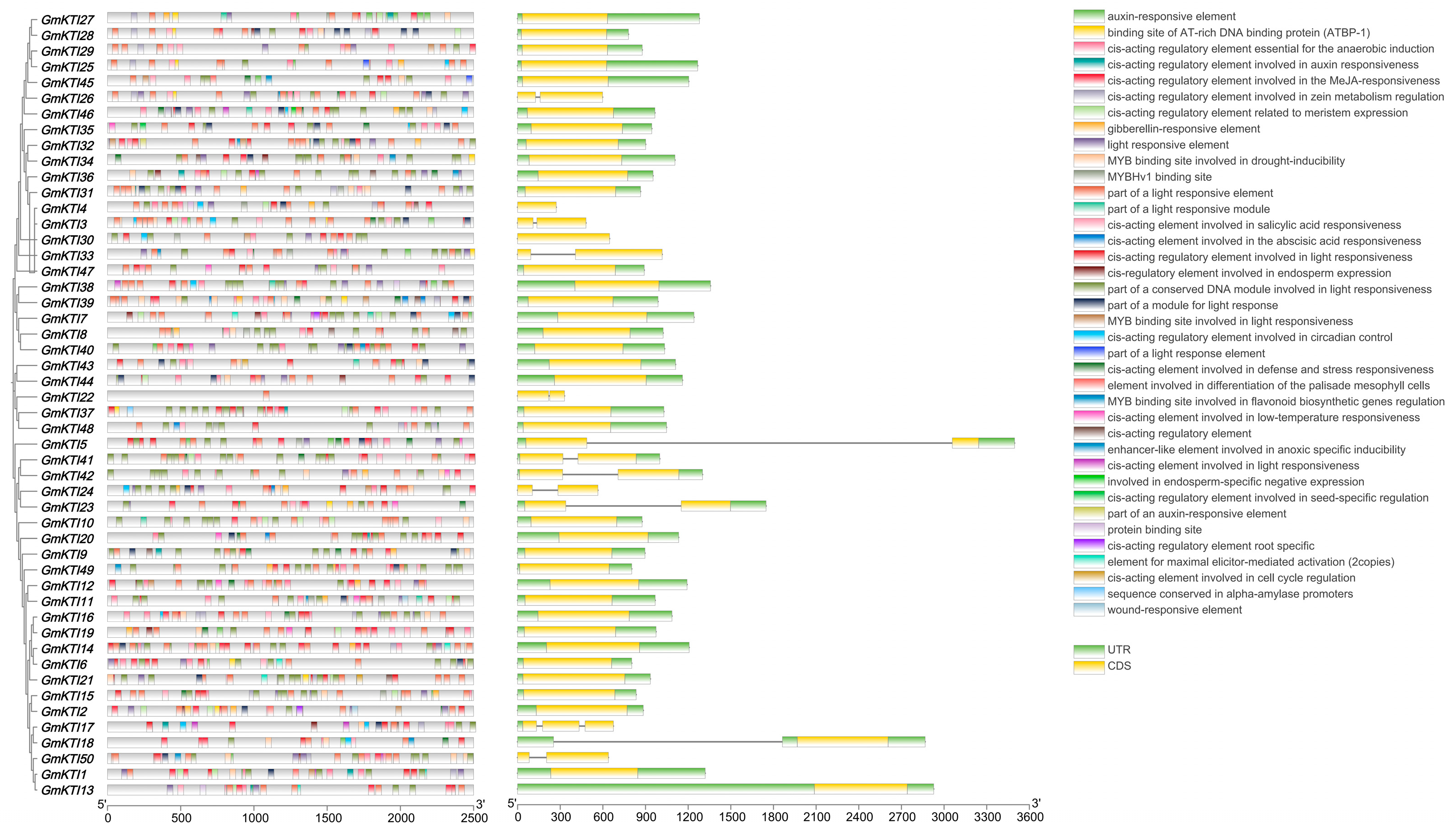
3.4. Analysis of Gene Structure and Promoter Cis-Acting Elements
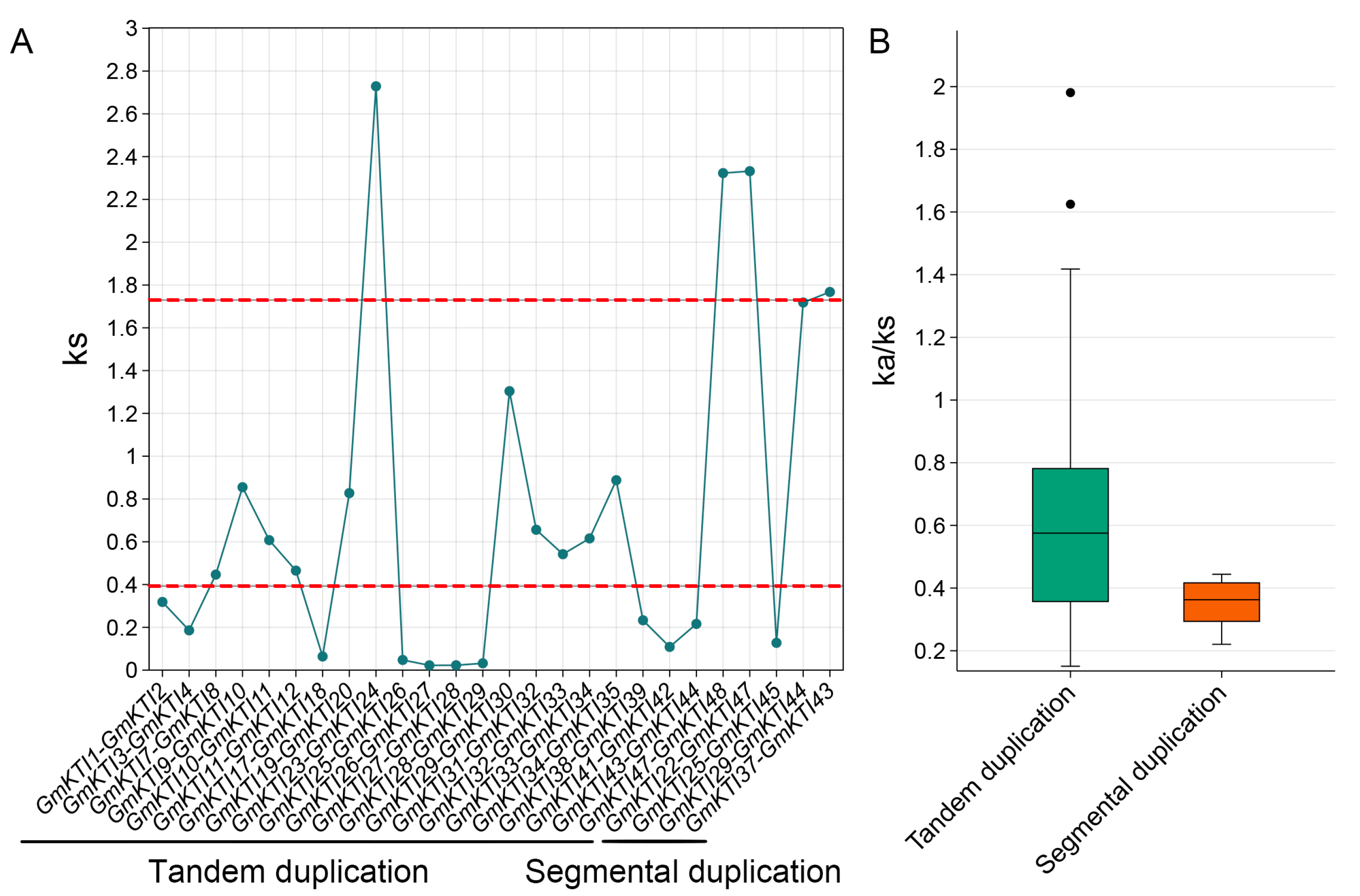
3.5. Phylogenetic Analysis of Paralogous Genes in the GmKTIs
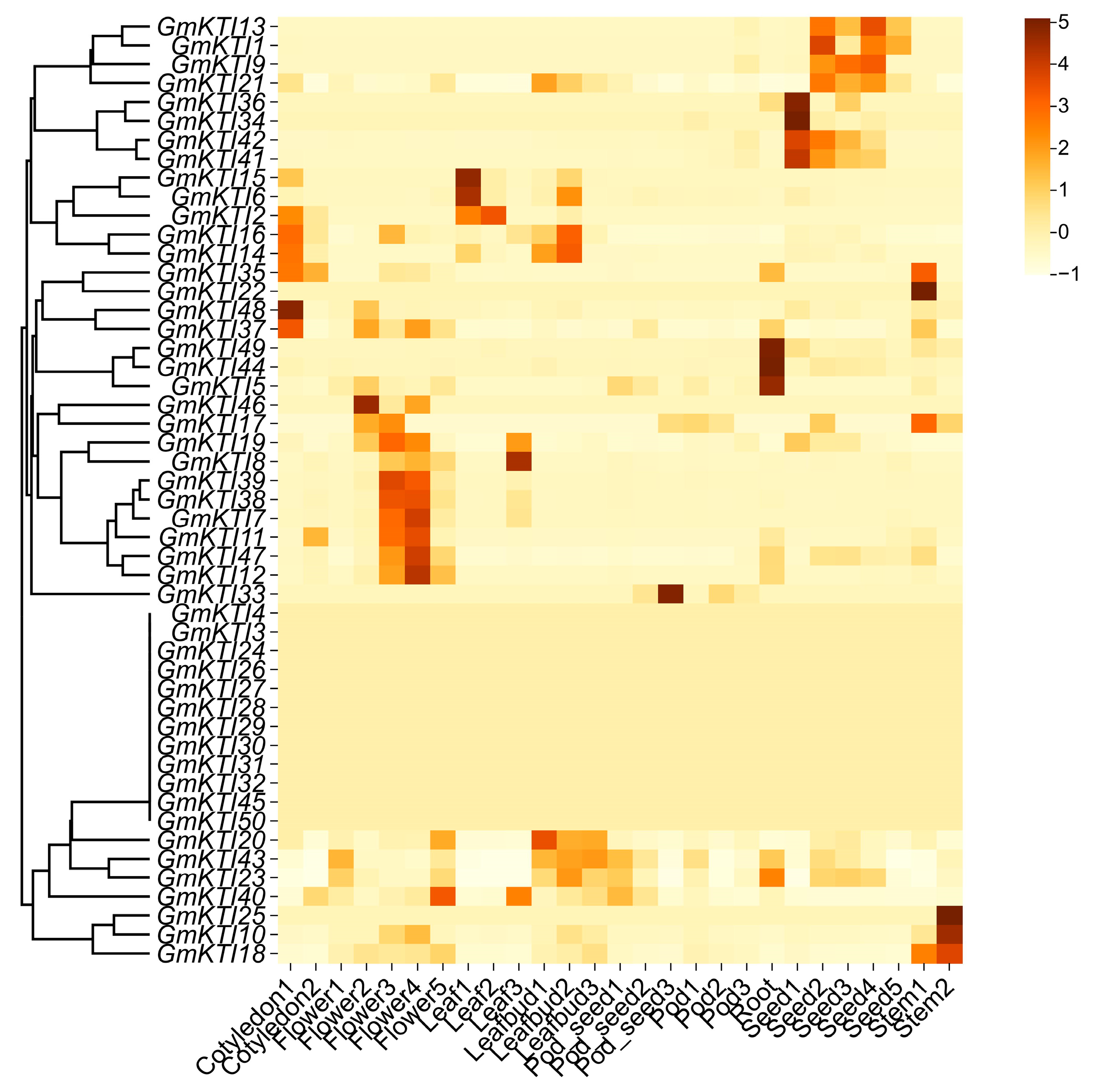
3.6. Transcriptional Profiling of GmKTIs
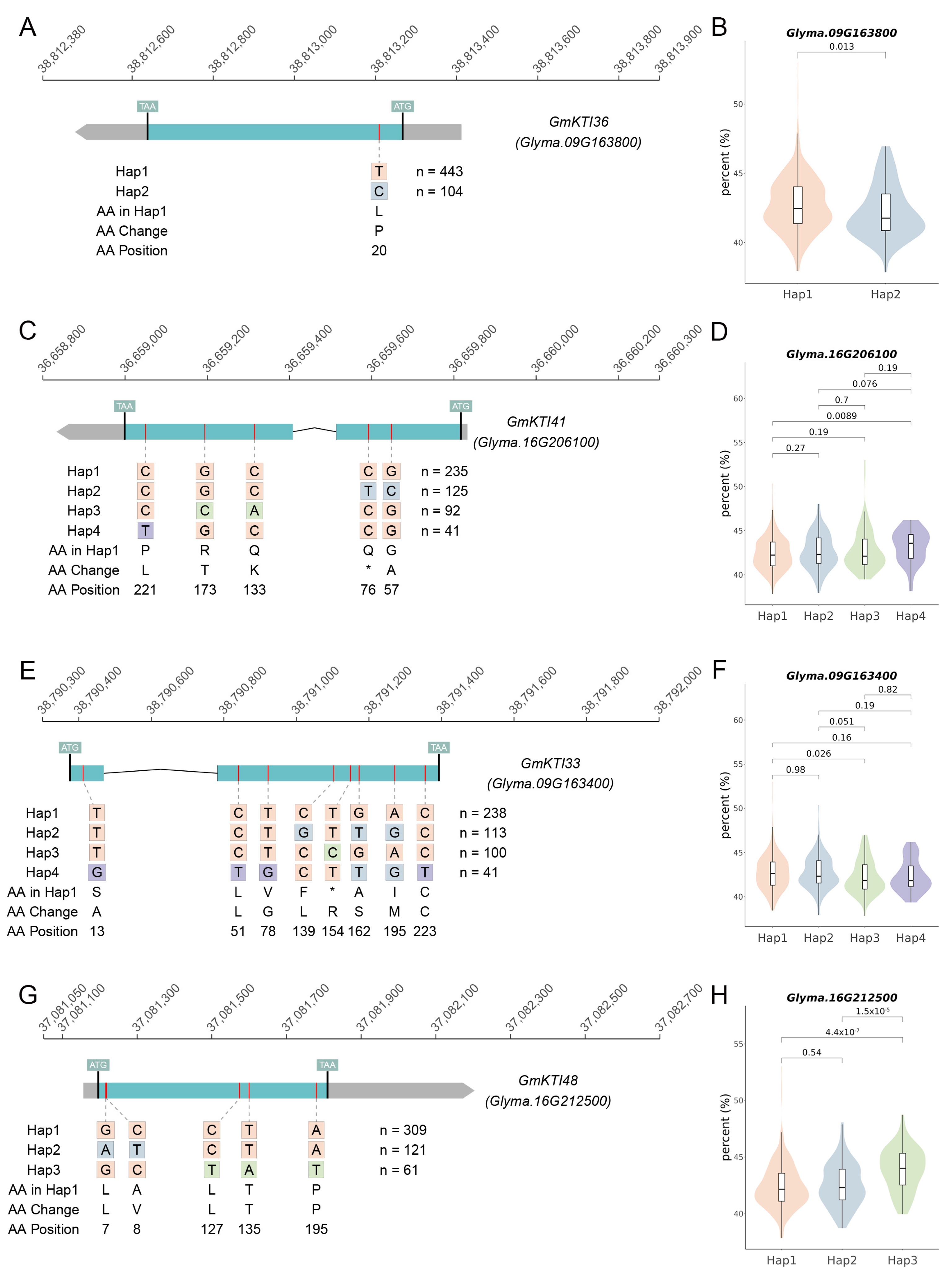
3.7. Haplotype Analysis of GmKTIs
3.8. Haplotype Module Stacking Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, E.H.; Ro, H.M.; Kim, S.L.; Kim, H.S.; Chung, I.M. Analysis of isoflavone, phenolic, soyasapogenol, and tocopherol compounds in soybean [Glycine max (L.) Merrill] germplasms of different seed weights and origins. J. Agric. Food Chem. 2012, 60, 6045–6055. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, A.; Ueda, Y.; Takase, H.; Yazaki, K. Do soybeans select specific species of Bradyrhizobium during growth? Commun. Integr. Biol. 2015, 8, e992734. [Google Scholar] [CrossRef] [PubMed]
- Loveday, S.M. Food Proteins: Technological, Nutritional, and Sustainability Attributes of Traditional and Emerging Proteins. Annu. Rev. Food Sci. Technol. 2019, 10, 311–339. [Google Scholar] [CrossRef] [PubMed]
- Osborne, T.B. Our present knowledge of plant proteins. Science 1908, 28, 417–427. [Google Scholar] [CrossRef]
- Guan, X.; Yao, H.; Chen, Z.; Shan, L.; Zhang, M. Some functional properties of oat bran protein concentrate modified by trypsin. Food Chem. 2007, 101, 163–170. [Google Scholar] [CrossRef]
- Naismith, W.E.F. Ultracentrifuge studies on soybean protein. Biochim. Et Biophys. Acta 1955, 16, 203–210. [Google Scholar] [CrossRef]
- Wolf, W.J. Ultracentrifugal investigation of the effect of neutral salts on the extraction of soybean proteins. Arch. Biochem. Biophys. 1956, 63, 40–49. [Google Scholar] [CrossRef]
- Wolf, W.; Briggs, D. Purification and characterization of the 11S component of soybean proteins. Arch. Biochem. Biophys. 1959, 85, 186–199. [Google Scholar] [CrossRef]
- Liwang, C.; Wolf, W.J. Soybean protein aggregation by sonication: Ultracentrifugal analysis. J. Food Sci. 1983, 48, 1260–1264. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, T.; Jiang, L. Soy Protein: Molecular Structure Revisited and Recent Advances in Processing Technologies. Annu. Rev. Food Sci. Technol. 2021, 12, 119–147. [Google Scholar] [CrossRef]
- Vagadia, B.H.; Vanga, S.K.; Raghavan, V. Inactivation methods of soybean trypsin inhibitor–A review. Trends Food Sci. Technol. 2017, 64, 115–125. [Google Scholar] [CrossRef]
- Singh, L.; Wilson, C.M.; Hadley, H.H. Genetic Differences in Soybean Trypsin Inhibitors Separated by Disc Electrophoresis 1. Crop Sci. 1969, 9, 489–491. [Google Scholar] [CrossRef]
- Hymowitz, T. Electrophoretic Analysis of SBTI-A2 in the USDA Soybean Germplasm Collection1. Crop Sci. 1973, 13, 420–421. [Google Scholar] [CrossRef]
- Zhao, S. A new electrophoretic variant of SBTi-A_2 in soybean seed protein. Soybean Genet. News Lett. 1992, 19, 22–24. [Google Scholar]
- Wang, K.-j.; Kaizuma, N.; Takahata, Y.; Hatakeyama, S. Detection of Two New Variants of Soybean Kunitz Trypsin Inhibitor through Electrophoresis. Breed. Sci. 1996, 46, 39–44. [Google Scholar] [CrossRef][Green Version]
- Orf, J.H.; Hymowitz, T. Inheritance of the absence of the kunitz trypsin inhibitor in seed protein of soybeans 1. Crop Sci. 1979, 19, 107–109. [Google Scholar] [CrossRef]
- Wang, K.J.; Yamashita, T.; Watanabe, M.; Takahata, Y. Genetic characterization of a novel Tib-derived variant of soybean Kunitz trypsin inhibitor detected in wild soybean (Glycine soja). Genome 2004, 47, 9–14. [Google Scholar] [CrossRef]
- Wang, K.; Li, X. Tif type of soybean Kunitz trypsin inhibitor exists in wild soybean of northern China. In Proceedings of the 8th National Soybean Research Conference of China, Beijing, China, 10–15 August 2009; pp. 167–168. [Google Scholar]
- Wang, K.; Takahata, Y.; Kono, Y.; Kaizuma, N. Allelic differentiation of Kunitz trypsin inhibitor in wild soybean (Glycine soja). Theor. Appl. Genet. 2008, 117, 565–573. [Google Scholar] [CrossRef]
- Song, S.I.; Kim, C.H.; Baek, S.J.; Choi, Y.D. Nucleotide sequences of cDNAs encoding the precursors for soybean (Glycine max) trypsin inhibitors (Kunitz type). Plant Physiol. 1993, 101, 1401–1402. [Google Scholar] [CrossRef]
- Kim, S.H.; Hara, S.; Hase, S.; Ikenaka, T.; Toda, H.; Kitamura, K.; Kaizuma, N. Comparative Study on Amino Acid Sequences of Kunitz-Type Soybean Trypsin Inhibitors, Tia, Tib, and Tic. J. Biochem. 1985, 98, 435–448. [Google Scholar] [CrossRef]
- Wang, K.-j.; Takahata, Y.; Ito, K.; Zhao, Y.; Tsutsumi, K.-i.; Kaizuma, N. Genetic Characterization of a Novel Soybean Kunitz Trypsin Inhibitor. Breed. Sci. 2001, 51, 185–190. [Google Scholar] [CrossRef]
- Kaizuma, N.; Oikawa, K.; Miura, M. Consideration on the cause of the differential t i alleles frequency distributions found among some regional populations of soybean glycine max land varieties. J. Fac. Agric. Iwate Univ. 1980, 15, 81–96. [Google Scholar]
- Hymowitz, T.; Kaizuma, N. Soybean Seed Protein Electrophoresis Profiles from 15 Asian Countries or Regions: Hypotheses on Paths of Dissemination of Soybeans from China. Econ. Bot. 1981, 35, 10–23. [Google Scholar] [CrossRef]
- Fushan, L.J. Studies on the ecological and geographical distribution ofthe Chinese resources of wild soybean (G. Soja). Sci. Agric. Sin. 1993, 26, 47–55. [Google Scholar]
- Xin, H.; Xie, K.; Dong, A.W.; Uan, Q.Y.; Gu, O.M. The amino acid sequence determination of a new variant of Kunitz soybean trypsin inhibitor (SBTi-A2). Soybean Genet. Newslett. 1999. [Google Scholar]
- Birk, Y.; Gertler, A. Khalef S A pure trypsin inhibitor from soya beans. J. Biochem. 1963, 87, 281. [Google Scholar] [CrossRef]
- Frattali, V.P.; Steiner, R.F. Soybean inhibitors. I. Separation and some properties of three inhibitors from commercial crude soybean trypsin inhibitor. Biochemistry 1968, 7, 521–530. [Google Scholar] [CrossRef]
- Rachis, I.J.; Anderson, R. Isolation of four soybean trypsin inhibitors by DEAE-cellulose chromatography. Biochem. Biophys. Res. Commun. 1964, 15, 230–235. [Google Scholar] [CrossRef]
- Yamamoto, M.; Ikenaka, T. Studies on soybean trypsin inhibitorPurification and characterization of two soybean trypsin inhibitors. J. Biochem. 1967, 62, 141–149. [Google Scholar] [CrossRef]
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.; Liu, Z.; Frommer, W.; Ma, J.; Chen, L. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]
- Tian, H.; Yin, Y.; Li, X.; Zhang, Z.; Feng, S.; Jin, S.; Han, X.; Yang, M.; Xu, C.; Hu, L. Identification of HSSP1 as a regulator of soybean protein content through QTLanalysis Soy-SPCCnetwork. Plant Biotechnol. J. 2025. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Guo, C.; Li, H.; Qiu, H.; Li, H.; Jong, C.; Yu, G.; Zhang, Y.; Hu, L.; Wu, X. Natural variation in Fatty Acid 9 is a determinant of fatty acid protein content. Plant Biotechnol. J. 2024, 22, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L. The TIGR rice genome annotation resource: Improvements and new features. Nucleic Acids Res. 2007, 35 (Suppl. 1), D883–D887. [Google Scholar] [CrossRef]
- Hufford, M.B.; Seetharam, A.S.; Woodhouse, M.R.; Chougule, K.M.; Ou, S.; Liu, J.; Ricci, W.A.; Guo, T.; Olson, A.; Qiu, Y. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 2021, 373, 655–662. [Google Scholar] [CrossRef]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.; Gouzy, J.; Schoof, H. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Yang, Z.; Luo, C.; Pei, X.; Wang, S.; Huang, Y.; Li, J.; Liu, B.; Kong, F.; Yang, Q.-Y.; Fang, C. SoyMD: A platform combining multi-omics data with various tools for soybean research and breeding. Nucleic Acids Res. 2024, 52, D1639–D1650. [Google Scholar] [CrossRef]
- Madden, T. The BLAST sequence analysis tool. In The NCBI Handbook; National Center for Biotechnology Information: Bethesda, MD, USA, 2013; Volume 2, pp. 425–436. [Google Scholar]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef]
- Van, K.; Kim, D.H.; Cai, C.M.; Kim, M.Y.; Shin, J.H.; Graham, M.A.; Shoemaker, R.C.; Choi, B.-S.; Yang, T.-J.; Lee, S.-H. Sequence level analysis of recently duplicated regions in soybean [Glycine max (L.) Merr.] genome. DNA Res. 2008, 15, 93–102. [Google Scholar] [CrossRef]
- Severin, A.J.; Cannon, S.B.; Graham, M.M.; Grant, D.; Shoemaker, R.C. Changes in twelve homoeologous genomic regions in soybean following three rounds of polyploidy. Plant Cell 2011, 23, 3129–3136. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Panzade, S.; Chimote, V.; Shinde, C.; Aher, A. Identification of Soybean (Glycine max L.) Segregants for Kunitz Trypsin Inhibitor and Lipoxygenase Free Gene Using Linked Molecular Markers. Plant Cell Biotechnol. Mol. Biol. 2025, 26, 33–34. [Google Scholar]
- Liu, K.; Wang, Z.; An, Y.-Q. Developing evaluating CRISPR-Cas9 edited transgene-free soybeans with dramatic reduction in trypsin chymotrypsin inhibition based on selfing phenotyping T1, T3 Seeds. J. Agric. Food Res. 2025, 101811, in press. [Google Scholar] [CrossRef]
- Bukan, M.; Andrijanić, Z.; Pejić, I.; Ključarić, M.; Čižmek, L.; Tomaz, I.; Buljević, N.; Šarčević, H. Validation of Molecular Markers for Low Kunitz Trypsin Inhibitor Content in European Soybean (Glycine max, L. Merr.) Germplasm. Genes 2024, 15, 1028. [Google Scholar] [CrossRef]
- Rani, A.; Kumar, V.; Shukla, S.; Jha, P.; Tayalkar, T.; Mittal, P. Changes in storage protein composition on genetic removal of Kunitz trypsin inhibitor maintain protein content in soybean (Glycine max). J. Agric. Food Res. 2020, 2, 100065. [Google Scholar] [CrossRef]
- Khan, R.; Alkharouf, N.; Beard, H.; MacDonald, M.; Chouikha, I.; Meyer, S.; Grefenstette, J.; Knap, H.; Matthews, B. Resistance mechanisms in soybean: Gene expression profile at an early stage of soybean cyst nematode invasion. J. Nematol. 2004, 36, 241–248. [Google Scholar]
- Rashed, N.A.; MacDonald, M.H.; Matthews, B.F. Protease inhibitor expression in soybean roots exhibiting susceptible and resistant interactions with soybean cyst nematode. J. Nematol. 2008, 40, 138. [Google Scholar] [PubMed]
- Hilder, V.A.; Gatehouse, A.M.; Sheerman, S.E.; Barker, R.F.; Boulter, D. A novel mechanism of insect resistance engineered into tobacco. Nature 1987, 330, 160–163. [Google Scholar] [CrossRef]
- Mehmood, S.; Thirup, S.S.; Ahmed, S.; Bashir, N.; Saeed, A.; Rafiq, M.; Saeed, Q.; Najam-ul-Haq, M.; Khaliq, B.; Ibrahim, M. Crystal structure of Kunitz-type trypsin inhibitor: Entomotoxic effect of native and encapsulated protein targeting gut trypsin of Tribolium castaneum Herbst. Comput. Struct. Biotechnol. J. 2024, 23, 3132–3142. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, H.; Zhang, Z.; Feng, S.; Song, J.; Han, X.; Chen, X.; Li, C.; Liu, E.; Xu, L.; Yang, M.; et al. Genome-Wide Characterization and Haplotype Module Stacking Analysis of the KTI Gene Family in Soybean (Glycine max L. Merr.). Agronomy 2025, 15, 1210. https://doi.org/10.3390/agronomy15051210
Tian H, Zhang Z, Feng S, Song J, Han X, Chen X, Li C, Liu E, Xu L, Yang M, et al. Genome-Wide Characterization and Haplotype Module Stacking Analysis of the KTI Gene Family in Soybean (Glycine max L. Merr.). Agronomy. 2025; 15(5):1210. https://doi.org/10.3390/agronomy15051210
Chicago/Turabian StyleTian, Huilin, Zhanguo Zhang, Shaowei Feng, Jia Song, Xue Han, Xin Chen, Candong Li, Enliang Liu, Linli Xu, Mingliang Yang, and et al. 2025. "Genome-Wide Characterization and Haplotype Module Stacking Analysis of the KTI Gene Family in Soybean (Glycine max L. Merr.)" Agronomy 15, no. 5: 1210. https://doi.org/10.3390/agronomy15051210
APA StyleTian, H., Zhang, Z., Feng, S., Song, J., Han, X., Chen, X., Li, C., Liu, E., Xu, L., Yang, M., Chen, Q., Wu, X., & Qi, Z. (2025). Genome-Wide Characterization and Haplotype Module Stacking Analysis of the KTI Gene Family in Soybean (Glycine max L. Merr.). Agronomy, 15(5), 1210. https://doi.org/10.3390/agronomy15051210






