Phylogenetic Diversity and Quantitative PCR Detection of Erwinia amylovora in Xinjiang, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Genetic Diversity Analysis of E. amylovora in Xinjiang
2.3. Determination of the CRR1 Genotype by PCR
2.4. Development of qPCR Detection System
2.4.1. Primer Design
2.4.2. Specificity of the Primers
2.4.3. Optimization of Primer Concentration
2.4.4. Optimization of Annealing Temperature
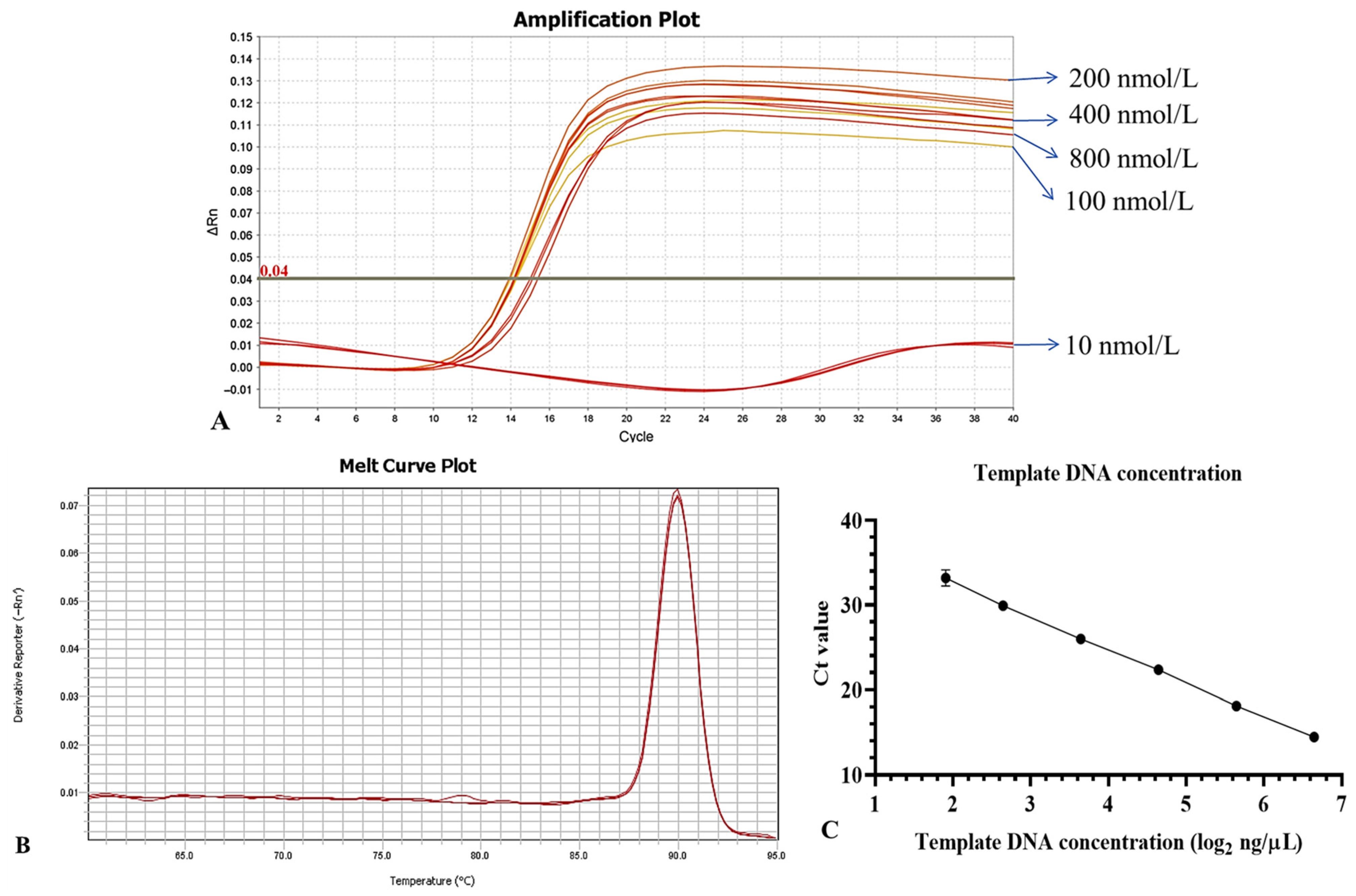
2.4.5. Standard Curve of Quantitative PCR
2.5. Evaluation of Primer Sensitivity
2.5.1. Sensitivity of the Primers in PCR
2.5.2. qPCR Sensitivity
2.6. qPCR Detection of Pear Tissues Infected by Fire Blight Pathogen
3. Results
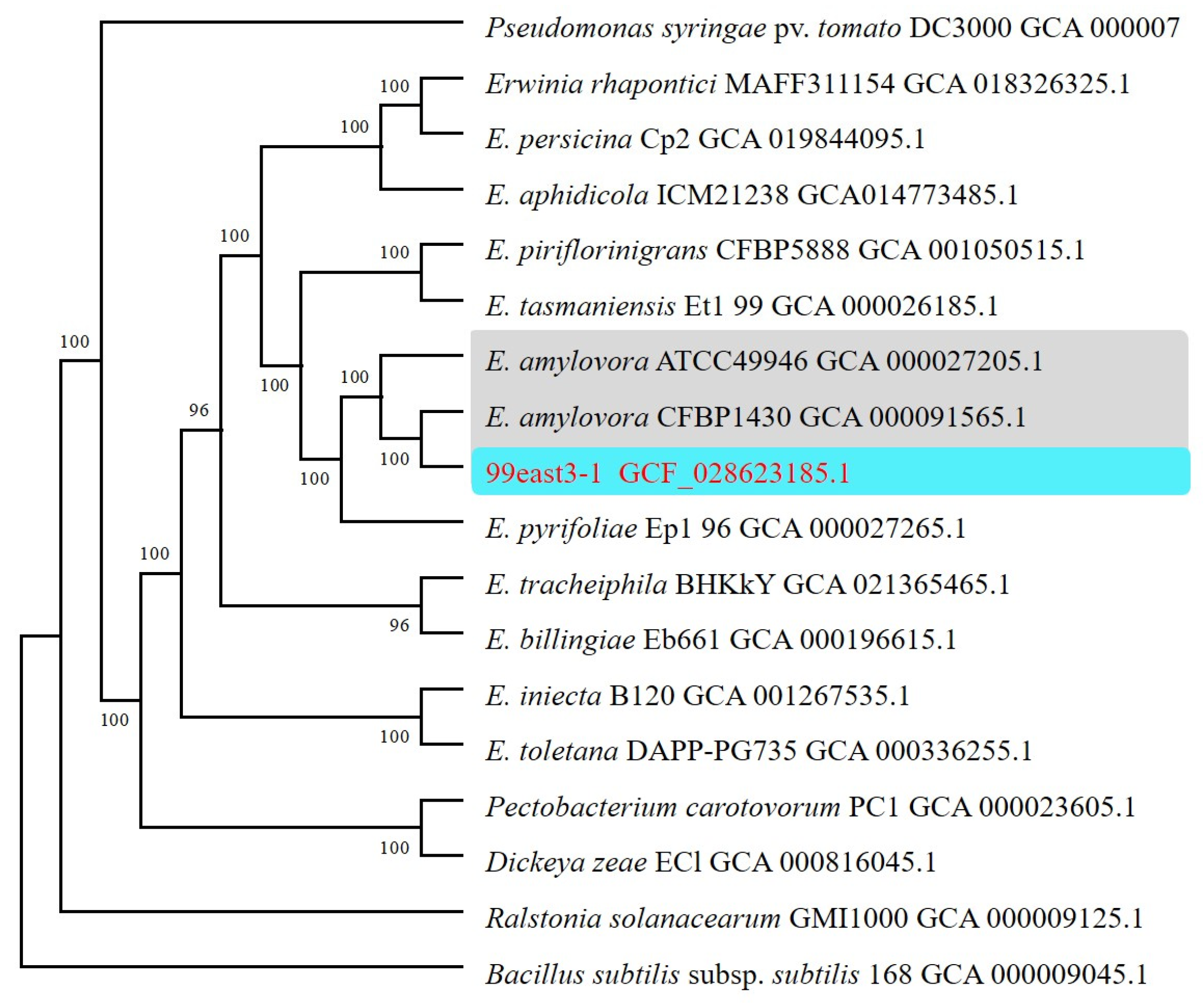
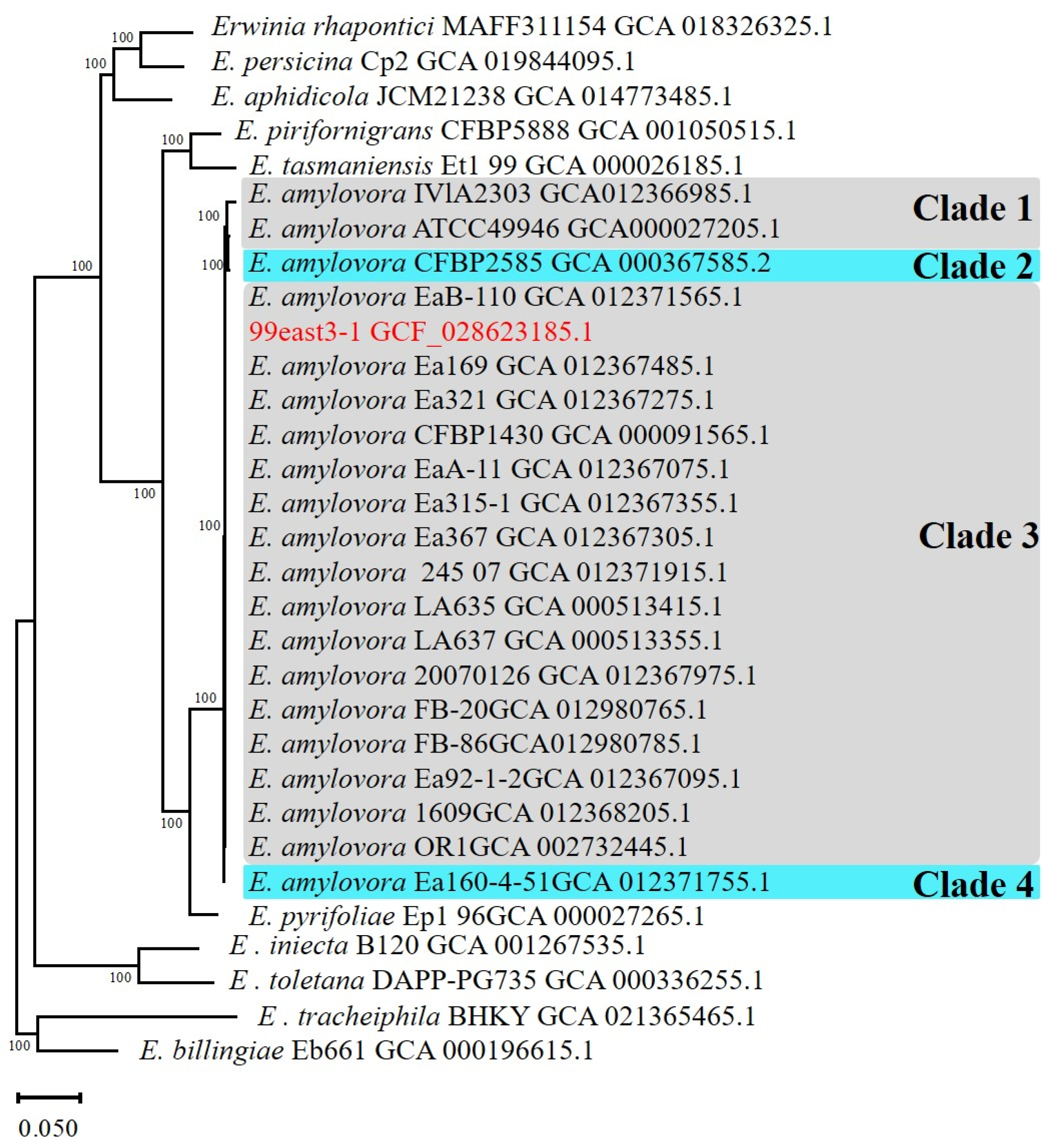
3.1. Phylogenetic Analysis
3.2. CRR1 Genotypes of E. amylovora Isolated from Xinjiang
3.3. Design of Specific Primers
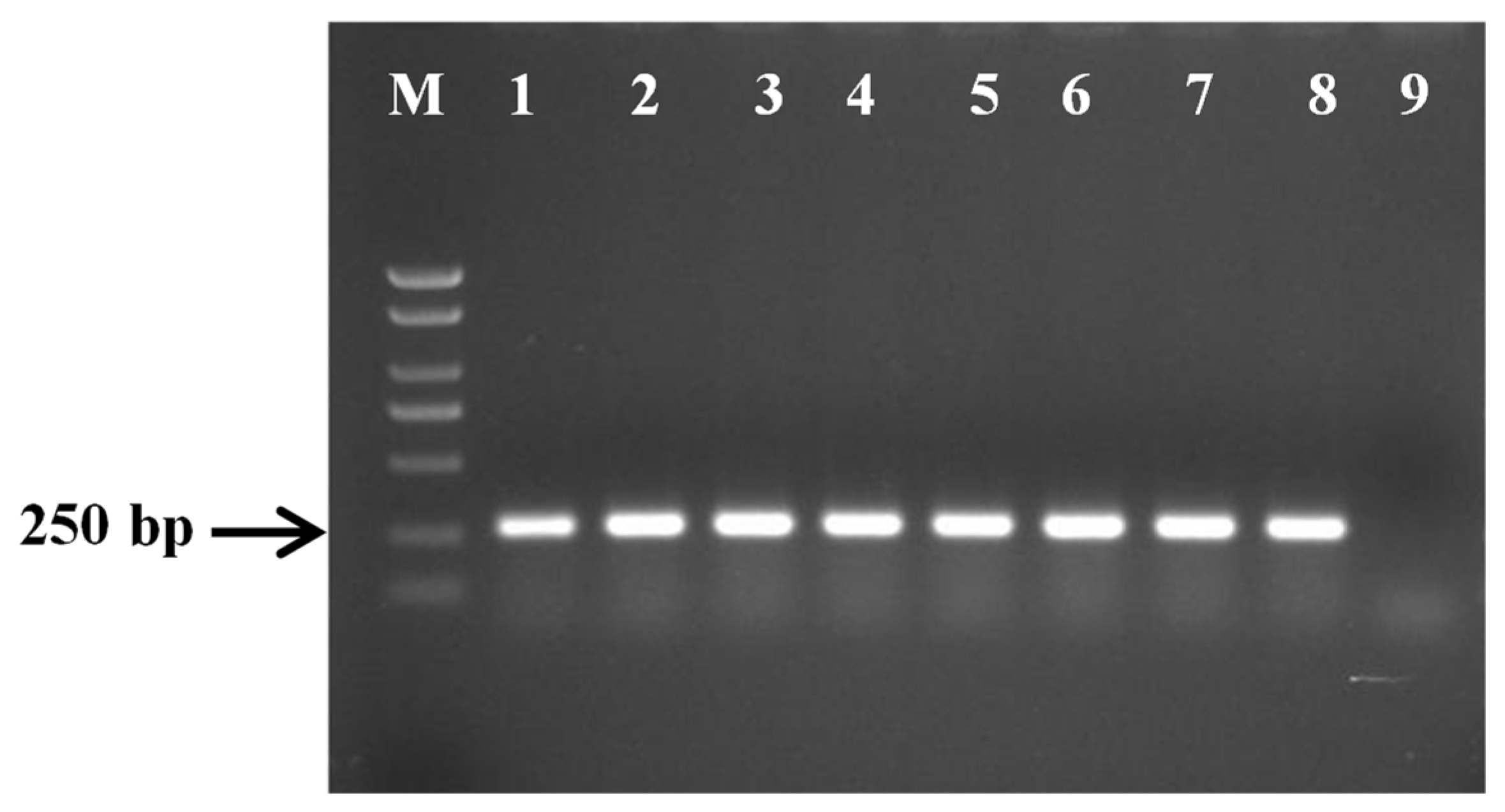
3.4. Specificity of PCR Primer
3.5. Optimization of the qPCR Detection System
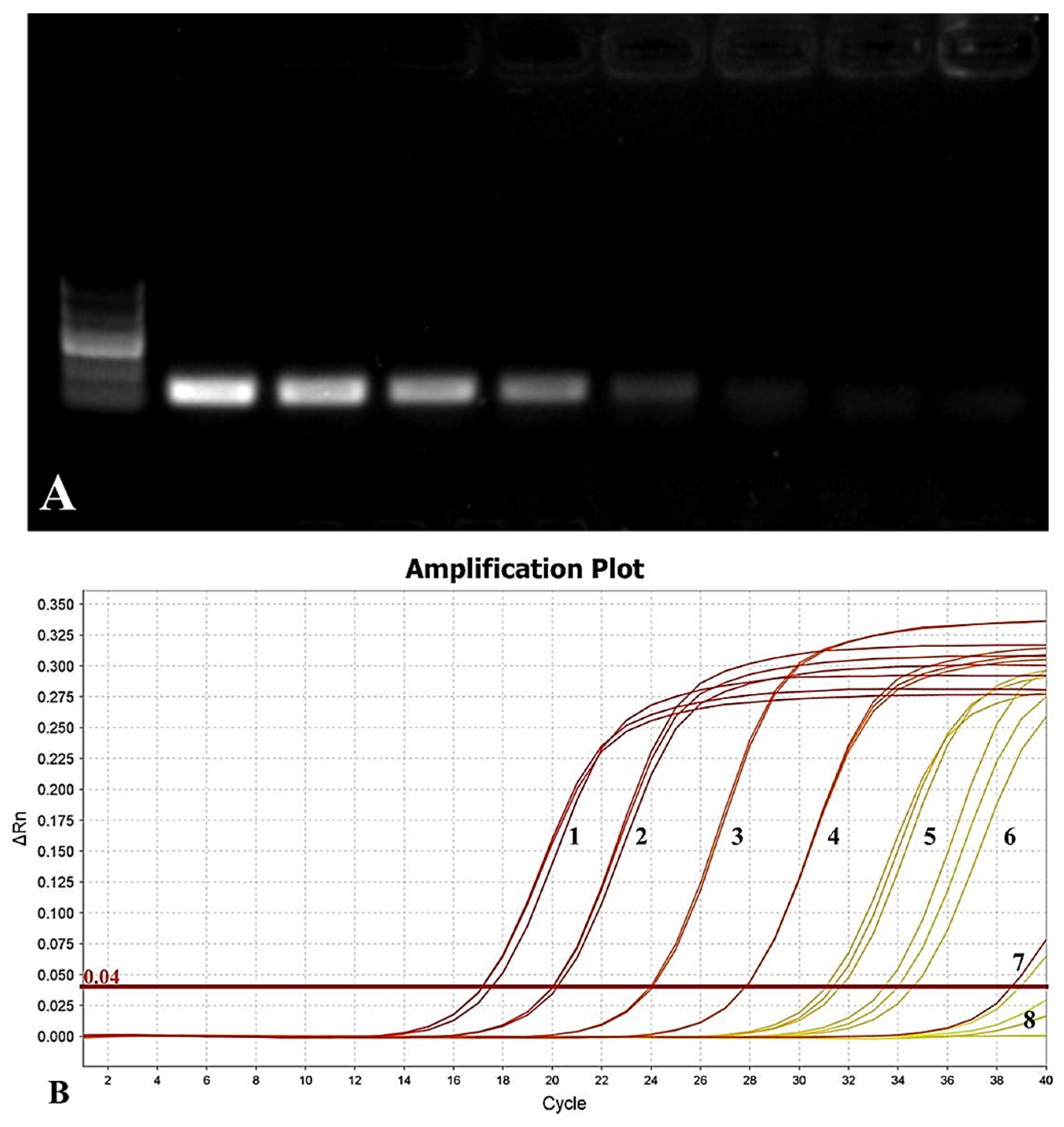
3.6. Sensitivity of Primers
3.6.1. Sensitivity Detection by Using Bacterial Suspension as a Template
3.6.2. Sensitivity Detection Using E. amylovora DNA as a Template
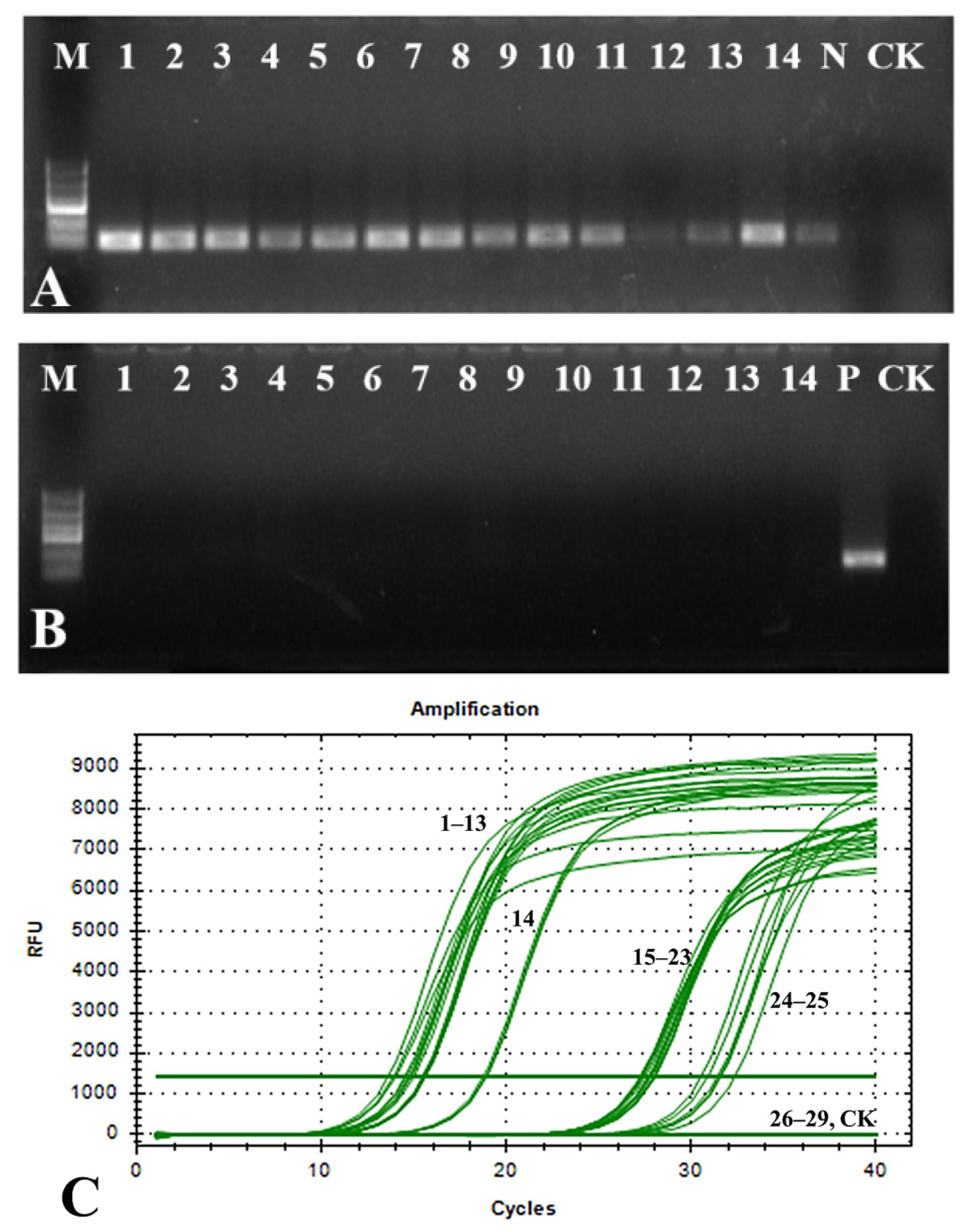
3.7. Detection of Infected Plant Tissues
3.7.1. Detection Using Total DNA Extracted from Infected Plant Tissues
3.7.2. Detection of Plant Tissue Grinding Suspension as Templates
3.7.3. Application of qPCR Detection System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, S.Q.; Zhang, L.; Wang, M.S.; Liu, W.W.; Cui, R.G.; Du, B.; Song, L.J. Effect of different pretreatment methods on soluble dietary fiber macromolecules extracted from Korla fragrant pear (Pyrus sinkiangensis Yü): Structure, physicochemical properties, and biological activity. Int. J. Biol. Macromol. 2025, 302, 140901. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, Y.; Long, H.; Yu, Z.; Huang, M.; Liu, X. Biodiversity and oenological property analysis of non-Saccharomyces yeasts isolated from Korla fragrant pears (Pyrus sinkiangensis Yu). Fermentation 2022, 8, 388. [Google Scholar] [CrossRef]
- Ham, H.; Lee, S.W.; Lee, Y.H. Genetic diversity and genotype distribution of Erwinia amylovora in Korea. Plant Pathol. J. 2025, 41, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, A.; Finelli, F.; Mazzoli, G.L. A severe unforeseen outbreak of fire blight in the Emilia-Romagna Region. Acta Hortic. 1999, 489, 171–176. [Google Scholar] [CrossRef]
- Albanese, D.; Cainelli, C.; Gualandri, V.; Larger, S.; Pindo, M.; Donati, C. Genome sequencing provides new insights on the distribution of Erwinia amylovora lineages in northern Italy. Environ. Microbiol. Rep. 2022, 14, 584–590. [Google Scholar] [CrossRef]
- Song, J.Y.; Lee, S.J.; Nam, M.; Lee, J.H.; Choi, E.J.; Cho, Y.R.; Lee, B.; Kim, J.F. Genomic epidemiology of Erwinia amylovora strains that caused the fire blight outbreak in Korea. Plant Dis. 2025. [Google Scholar] [CrossRef]
- CABI/EPPO (CAB International/European and Mediterranean Plant Protection Organization). EPPO Reporting Service; No. 2017/001; European and Mediterranean Plant Protection Organization: Paris, France, 2017. [Google Scholar]
- Rezzonico, F.; Bobushova, S.; Gaganidze, D.; Konurbaeva, M.; Mukhanov, S.; Jordan, S.; Sadunishvili, T.; Drenova, N.; Smits, T.H.M.; Doolotkeldieva, T. Epidemiological description of fire blight introduction patterns to Central Asia and the Caucasus region based on CRISPR spacer typing and genome analysis. Phytopathol. Res. 2024, 6, 66. [Google Scholar] [CrossRef]
- Gong, P.J.; Sun, W.B.; Yi, J.P.; Han, J.; Liu, M.L.; Han, C.Y.; Ming, L.; Wang, B.W.; Luo, M.; Zhao, Y.C.; et al. Comparative genomic analysis of Chinese Erwinia amylovora strains reveals genetic variations, plasmid diversity, and potential common ancestor. iMetaOmics 2025, 2, e44. [Google Scholar] [CrossRef]
- Drenova, N.V.; Isin, M.M.; Dzhaimurzina, A.A.; Zharmukhamedova, G.A.; Aitkulov, A.K. Bacterial fire blight in the Republic of Kazakhstan. Plant Health Res. Pract. 2013, 3, 44–48. [Google Scholar]
- Doolotkeldieva, T.; Bobusheva, S. Fire blight disease caused by Erwinia amylovora on Rosaceae plants in Kyrgyzstan and biological agents to control this disease. Adv. Microbiol. 2016, 6, 831–851. [Google Scholar] [CrossRef]
- Park, D.H.; Yu, J.G.; Oh, E.J.; Han, K.S.; Mi, C.Y.; Lee, S.J. First report of fire blight disease on Asian pear caused by Erwinia amylovora in Korea. Plant Dis. 2016, 100, 1946. [Google Scholar] [CrossRef]
- Sun, W.; Gong, P.; Zhao, Y.; Ming, L.; Zeng, Q.; Liu, F. Current Situation of Fire Blight in China. Phytopathology 2023, 113, 2143–2151. [Google Scholar] [CrossRef] [PubMed]
- Maltseva, E.R.; Zharmukhamedova, G.A.; Jumanova, Z.K.; Naizabayeva, D.A.; Berdygu-lova, Z.A.; Dmitriyeva, K.A.; Tezekbayeva, B.; Khassein, A.; Skiba, Y.A.; Malakhova, N.P.; et al. Fire blight cases in Almaty region of Kazakhstan in the proximity of wild apple distribution area. J. Plant Pathol. 2023, 106, 971–978. [Google Scholar] [CrossRef]
- Eunjung, R.; Madison, E.D.; Leslie, M.E.; Julianne, H.G. Whole genome sequences of eight Erwinia amylovora phages isolated from South Korea. Microbiol. Resour. Announc. 2025, 14, e0106224. [Google Scholar] [CrossRef]
- Talhi, L.; Barbé, S.; Herrero, I.N.; Sebaihia, M.; Noales, E.M. Intraspecific diversity of Erwinia amylovora strains from northern Algeria. BMC Microbiol. 2024, 24, 389. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, Y.; Wu, S.; Gu, X.M.; Liu, X.R.; Tian, Y.L.; Hu, B.S.; Zhao, Y.F. Genome analysis reveals Erwinia amylovora in China is closely related to those of Kyrgyzstan and Kazakhstan. Phytopathol. Res. 2025, 7, 31. [Google Scholar] [CrossRef]
- Hyeonheui, H.; Suk, D.P. Novel approach toward the understanding of genetic diversity based on the two types of amino acid repeats in Erwinia amylovora. Sci. Rep. 2023, 13, 17876. [Google Scholar]
- Rico, A.; Führer, M.E.; Ortiz-Barredo, A.; Murillo, J. Polymerase chain reaction fingerprinting of Erwinia amylovora has a limited phylogenetic value but allows the design of highly specific molecular markers. Phytopathology 2008, 98, 260–269. [Google Scholar] [CrossRef]
- Momol, M.T.; Momol, E.A.; Lamboy, W.F.; Norelli, J.L.; Beer, S.V.; Aldwinckle, H.S. Charac-terization of Erwinia amylovora strains using random amplified polymorphic DNA fragments (RAPDs). J. Appl. Microbiol. 1997, 82, 389–398. [Google Scholar] [CrossRef]
- Jock, S.; Donat, V.; López, M.M.; Bazzi, C.; Geider, K. Following spread of ffre blight in Western, Central and Southern Europe by molecular differentiation of Erwinia amylovora strains with PFGE analysis. Environ. Microbiol. 2002, 4, 106–114. [Google Scholar] [CrossRef]
- Jock, S.; Wensing, A.; Pulawska, J.; Drenova, N.; Dreo, T.; Geider, K. Molecular analyses of Erwinia amylovora strains isolated in Russia, Poland, Slovenia and Austria describing further spread of fire blight in Europe. Microbiol. Res. 2013, 168, 447–454. [Google Scholar] [CrossRef]
- Bühlmann, A.; Dreo, T.; Rezzonico, F.; Pothier, J.F.; Smits, T.H.; Ravnikar, M.; Frey, J.E.; Duffy, B. Phylogeography and population structure of the biologically invasive phytopathogen Erwinia amylovora inferred using minisatellites. Environ. Microbiol. 2014, 16, 2112–2125. [Google Scholar] [CrossRef] [PubMed]
- Parcey, M.; Gayder, S.; Morley-Senkler, V.; Bakkeren, G.; Úrbez-Torres, J.R.; Ali, S. Comparative genomic analysis of Erwinia amylovora reveals novel insights in phylogenetic arrangement, plasmid diversity, and streptomycin resistance. Genomics 2020, 112, 3762–3772. [Google Scholar] [CrossRef]
- Kurz, M.; Carnal, S.; Dafny-Yelin, M.; Mairesse, O.; Rezzonico, F. Tracking the dissemination of Erwinia amylovora in the Eurasian continent using a PCR targeted on the duplication of a single CRISPR spacer. Phytopathol. Research. 2021, 3, 18. [Google Scholar] [CrossRef]
- Fei, N.; Yang, Y.; Song, B.; Zhu, X.; Guan, W.; Zhao, T. Complete Genome Sequence of Erwinia amylovora Strain 99east-3-1, Isolated from Pyrus sinkiangensis in China. Microbiol. Resour. Announc. 2023, 12, e0016123. [Google Scholar] [CrossRef] [PubMed]
- Ordax, M.; Piquer-Salcedo, J.E.; Santander, R.D.; Sabater-Muñoz, B.; Biosca, E.G.; López, M.M.; Marco-Noales, E. Medfly Ceratitis capitata as potential vector for fire blight pathogen Erwinia amylovora: Survival and transmission. PLoS ONE 2015, 10, e0127560. [Google Scholar] [CrossRef] [PubMed]
- Sedlak, J.; Paprstein, F.; Korba, J. Development of in vitro system for testing of pome fruit resistance to fire blight. In Proceedings of the XII International Workshop on Fire Blight, Warsaw, Poland, 16–20 August 2010; Volume 896, pp. 375–379. [Google Scholar]
- Chen, C.; Chen, J.; Hu, B.S.; Jiang, Y.H.; Liu, F.Q. Potential distribution of alien invasive species and risk assessment: A case study of Erwinia amylovora in China. Agric. Sci. China 2007, 6, 688–695. (In Chinese) [Google Scholar] [CrossRef]
- Hu, B.; Lu, L.; Liu, F.; Xu, Z.; Chen, J.; Jiao, G. Detection of Erwinia amylovora by indirect immuno-fluorescent staining and co-agglutination methods. J. Nanjing Agric. Univ. 2003, 26, 41–45. (In Chinese) [Google Scholar]
- Bereswill, S.; Bugert, P.; Bruchmüller, I.; Geider, K. Identification of the fire blight pathogen, Erwinia amylovora, by PCR assays with chromosomal DNA. Appl. Environ. Microbiol. 1995, 61, 2636–2642. [Google Scholar] [CrossRef]
- Gottsberger, R. Development and evaluation of a real-time PCR assay targeting chromosomal DNA of Erwinia amylovora. Lett. Appl. Microbiol. 2010, 51, 285–292. [Google Scholar] [CrossRef]
- Yang, Y.; Qiao, P.; Wang, T.; Ji, W.; Fei, N.; Zhang, L.; Guan, W.; Zhao, T. Further Characterization of Host Preference of Acidovorax citrulli Based on Growth Competition between Group I and Group II Strains. Horticulturae 2022, 8, 1173. [Google Scholar] [CrossRef]


| No. of Strain | Bacteria Species | Host | Source |
|---|---|---|---|
| SYD617-3-2 | Erwinia amylovora | Pyrus sinkiangensis | Xinjiang Province, China |
| 618-2-2 | E. amylovora | P. sinkiangensis | Xinjiang Province, China |
| 99east-3-1 | E. amylovora | P.s sinkiangensis | Xinjiang Province, China |
| 617SYD-2-2 | E. amylovora | P. sinkiangensis | Xinjiang Province, China |
| S617-2-2 | E. amylovora | P. sinkiangensis | Xinjiang Province, China |
| Ea-F1 | E. amylovora | P.s sinkiangensis | Xinjiang Province, China |
| Ea-F2 | E. amylovora | P.s sinkiangensis | Xinjiang Province, China |
| Ea-J | E. amylovora | P. sinkiangensis | Xinjiang Province, China |
| Ea-8 | E. amylovora | P. sinkiangensis | Plant Protection Institute of Xinjiang Academy of Agricultural Sciences |
| Ea-11 | E. amylovora | P. sinkiangensis | Plant Protection Institute of Xinjiang Academy of Agricultural Sciences |
| Ea-14 | E. amylovora | P. sinkiangensis | Plant Protection Institute of Xinjiang Academy of Agricultural Sciences |
| Ea-38 | E. amylovora | P.s sinkiangensis | Plant Protection Institute of Xinjiang Academy of Agricultural Sciences |
| Ea-39 | E. amylovora | P. sinkiangensis | Plant Protection Institute of Xinjiang Academy of Agricultural Sciences |
| Ea-61 | E. amylovora | P.s sinkiangensis | Plant Protection Institute of Xinjiang Academy of Agricultural Sciences |
| CICC 10449 | E. piriflorinigrans | Pyrus | China Center of Industrial Culture Collection, CICC |
| Tm10 | Ralstonia solanacearum | Nicotiana tabacum | Lab collection |
| RsLB | R. solanacearum | Solanum lycopersicum | Lab collection |
| psl8 | Pseudomonas syringae pv. lachrymans | Cucumis sativus | Lab collection |
| psJL | P. syringae pv. lachrymans | C. melo | Jilin Province, China |
| YN1 | Xanthomonas oryzae pv. oryzae | Oryza sativa | Yunnan Province, China |
| GD414 | X. oryzae pv. oryzae | O. sativa | Guangdong Province, China |
| Pxl1 | X. campestris pv. vesicatoria | Solanum lycopersicum | Lab collection |
| H1 | X. campestris pv. vesicatoria | Capsicum annuum | Lab collection |
| ZY1 | Bacillus amyloliquefaciens | C. annuum | Beijing, China |
| DH5α | Escherichia coli | — | Lab collection |
| RH05 | E. coli | — | Lab collection |
| Aac5 | Acidovorax citrulli | Citrullus lanatus | Taiwan Province, China |
| pslb65 | A. citrulli | C. melo | Xinjiang Province, China |
| BNCC108207 | Salmonella typhimurium | — | Lab collection |
| Region | Sample Type (Number of Samples Without Typical Symptoms/Number of Samples with Typical Symptoms) | PCR (Number of Positive Samples/Total Number of Samples) | qPCR (Number of Positive Samples/Total Number of Samples) |
|---|---|---|---|
| Korla, Xinjiang | Leaves of pear (2/6) | 7/8 | 8/8 |
| Branches of pear (2/6) | 7/8 | 8/8 | |
| Blossoms of pear (2/6) | 7/8 | 8/8 | |
| Ili, Xinjiang | Leaves of pear (0/3) | 3/3 | 3/3 |
| Leaves of apricot (0/1) | 1/1 | 1/1 | |
| Leaves of apple (0/3) | 3/3 | 3/3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fei, N.; Song, B.; Yan, J.; Wei, H.; Zhao, T.; Guan, W.; Ji, W.; Yang, Y. Phylogenetic Diversity and Quantitative PCR Detection of Erwinia amylovora in Xinjiang, China. Agronomy 2025, 15, 1065. https://doi.org/10.3390/agronomy15051065
Fei N, Song B, Yan J, Wei H, Zhao T, Guan W, Ji W, Yang Y. Phylogenetic Diversity and Quantitative PCR Detection of Erwinia amylovora in Xinjiang, China. Agronomy. 2025; 15(5):1065. https://doi.org/10.3390/agronomy15051065
Chicago/Turabian StyleFei, Nuoya, Bo Song, Jianpei Yan, Haoyu Wei, Tingchang Zhao, Wei Guan, Weiqin Ji, and Yuwen Yang. 2025. "Phylogenetic Diversity and Quantitative PCR Detection of Erwinia amylovora in Xinjiang, China" Agronomy 15, no. 5: 1065. https://doi.org/10.3390/agronomy15051065
APA StyleFei, N., Song, B., Yan, J., Wei, H., Zhao, T., Guan, W., Ji, W., & Yang, Y. (2025). Phylogenetic Diversity and Quantitative PCR Detection of Erwinia amylovora in Xinjiang, China. Agronomy, 15(5), 1065. https://doi.org/10.3390/agronomy15051065






