Abstract
Understanding and evaluating directed acyclic graphs (DAGs) is crucial for causal discovery, particularly in high-dimensional and small-sample datasets such as microbial abundance data. This study introduces DAGMetrics, an R package designed to comprehensively evaluate and compare DAGs. The package provides descriptive and comparative metrics, streamlining the assessment of outputs from various structure learning algorithms. It was applied to datasets generated for potato tubers and soils from different terroirs (continental and island) and stages (at harvest and post-harvest). Using a comprehensive set of descriptive and comparative metrics, DAGMetrics facilitated model selection by identifying balanced and robust DAGs. The PC algorithm with Spearman correlation produced DAGs with moderate complexity and high stability across scaling and transformation setups. Additionally, the package enabled detailed exploration of the Markov blanket space, revealing small Markov blankets (up to seven nodes) and numerous isolated nodes. Identified matching edges between Markov blankets across different terroirs and stages aligned with known microbial interactions, highlighting the package’s utility in facilitating the discovery of biologically meaningful relationships. This study illustrates the utility of DAGMetrics in providing objective and reproducible tools for DAG evaluation along with its potential application in agronomic and other domains involving complex structured data.
1. Introduction
Over the past several years, causal discovery has gained popularity in various fields, including economics [1,2], climate science [3], and biology [4,5,6,7]. More recently, its application has been extended to agronomy. For instance, causal discovery has been successfully employed in proteogenomics to identify cause-and-effect relationships between gene and protein modules [8]. These findings highlight the potential of causal discovery to enhance molecular data exploration and advance the study of key biological processes in plants. Another example is the study of interaction networks in olive leaves, where causal discovery combining transcriptomic and proteomic data has revealed critical differences between primed and non-primed tissues [9]. Causal discovery offers a framework that can model causal relationships among variables. Unlike traditional statistical methods, which identify associations without specifying the direction of influence, causal models can explicitly determine cause-and-effect relationships.
Directed acyclic graphs (DAGs) have become powerful tools for representing causal relationships among variables. However, effective evaluation and comparison of DAGs remains a significant challenge across disciplines. As highlighted in [10], it is important to perform DAG evaluation and comparison in a robust manner, as computationally derived causal relationships require further validation across different conditions, datasets, and contexts to ensure their persistence and stability. Similarly, ref. [11] emphasized the lack of suitable metrics for evaluating DAGs, particularly when the true structure of relationships is unknown. In microbial data analysis, which is often characterized by high dimensionality and limited sample sizes, ensuring the reliability of model evaluation is a critical concern. Structure learning algorithms can yield significantly different results due to variations in algorithmic approaches and underlying assumptions. The absence of a ground-truth DAG in most real-world applications further complicates the evaluation of these algorithms. While existing tools offer extensive functionality for structure learning, they often lack user-friendly and comprehensive approaches for comparing DAGs or analyzing their local structures in detail. To address these challenges, a tool is needed for systematically comparing DAGs by assessing their structural complexity and analyzing differences in local and global structures.
In this paper, we introduce DAGMetrics, an R package designed to facilitate the comprehensive evaluation and comparison of DAGs. The package provides a suite of descriptive and comparative metrics to assess both global and local structures. Descriptive metrics such as the number of edges and colliders offer insights into DAG complexity, while comparative metrics evaluate structural similarity. In addition to numerical outputs, DAGMetrics includes visualization tools that enable users to intuitively explore structural differences and similarities.
We demonstrate the functionality of DAGMetrics using real-world agronomic datasets. For example, the package is applied to microbial abundance data collected from potato tubers at the harvesting and post-harvesting stages in both island and continental regions. By comparing DAGs learned from these datasets, we uncover differences in microbial interactions and potential causal relationships. The package’s ability to analyze local structures makes it particularly well suited for agronomic applications and other fields dealing with high-dimensional datasets.
The rest of this paper is organized as follows. In Section 2, we discuss the materials and methods used in this work, including the main concepts of causal discovery, the functionality of the DAGMetrics package, and the preprocessing and statistical analyses applied to the datasets. In Section 3, we present the results, highlighting insights gained and achievements made using the package. In Section 4, we discuss DAGMetrics’ features and the obtained results. Finally, we conclude the paper in Section 5 by summarizing the contributions of this work, its implications for agronomy, and future research directions.
2. Materials and Methods
2.1. DAGs, Bayesian Networks, CPDAGs, and Markov Blankets
A DAG is a visual representation of relationships between variables, with arrows (edges) indicating the direction of influence or potential causality. DAGs are widely used in many scientific fields to aid in understanding and communicating assumptions about how variables may affect each other. A DAG is a graph in which all edges are directed and there are no cycles. The absence of cycles ensures that when starting from any node and following the direction of the edges, it is impossible to return to the same node. In the context of causal discovery and inference, DAGs encode causal relationships between variables [12]. The absence of an edge between two nodes indicates conditional independence between the corresponding variables given a specific set of other variables.
DAGs are a powerful framework for representing Bayesian networks, which model the joint probability distribution of a set of random variables [13]. A Bayesian network is defined by a DAG , where V is the set of nodes corresponding to random variables and E is the set of directed edges that encode conditional dependencies between these variables. If we break down DAGs by triplets of connected nodes , and , three main structures can be identified: chains (), forks (), and colliders (). The first two exhibit conditional independence between and given , while in a collider, conditioning on induces dependence between and . For more details on the main three-node structures, the reader is referred to Section S1 of the Supplementary Material.
A completed partially-directed acyclic graph (CPDAG) represents a Markov equivalence class of DAGs [4] and encodes the same conditional independencies, with directed edges indicating consistent causal relationships across all equivalent DAGs and undirected edges signifying directional uncertainty. For more details, refer to Section S2 of the Supplementary Material.
The Markov blanket of a node in a DAG is the minimal set of nodes that renders conditionally independent of all other nodes in the graph [2]. In other words, the behavior of a target node can be fully explained by the nodes in its Markov blanket. The Markov blanket of a node consists of its parents, its children, and the other parents of its children. In high-dimensional datasets such as microbial abundance in potatoes, analyzing the global network may be impractical due to its complexity. Instead, the focus often shifts to specific nodes of interest and their immediate connections as captured by their Markov blankets.
2.2. Structure Learning and Evaluation
The process of inferring a DAG from data is referred to as structure learning. Structure learning algorithms, including constraint-based methods (e.g., the PC algorithm [12]) and score-based methods (e.g., Tabu Search [14]), aim to uncover underlying causal relationships by leveraging conditional independence tests or optimizing scoring functions. These methods provide the foundation for automated DAG construction, and have gained popularity in fields such as agronomy, where high-dimensional datasets require advanced methodological tools.
Evaluating DAGs is an important step in assessing their structure in terms of complexity and similarity. We distinguish between two categories of metrics: descriptive and comparative.
2.2.1. Descriptive Metrics
Descriptive metrics provide information about the structure and complexity of a DAG. Key metrics include the following:
- Number of Edges: Measures connectivity and complexity of a DAG.
- Number of Colliders: Counts the number of colliders. In the case of CPDAGs, only edges involved in colliders are typically directed, as they represent causal relationships consistent across all equivalent DAGs. This metric serves as an indicator of the graph’s complexity and the presence of potential causal relationships inferred by the structure learning algorithm.
- Number of Root Nodes: Nodes with no incoming edges.
- Number of Leaf Nodes: Nodes with no outgoing edges.
- Number of Isolated Nodes: Nodes with no connections.
- Degree Metrics: These metrics relate to a specific node as compared to the full structure. They include in-degree, out-degree, and total degree for nodes, highlighting the importance and connectivity of individual nodes.
2.2.2. Comparative Metrics
Comparative metrics evaluate the similarity between two DAGs. They facilitate the assessment of multiple DAGs to identify similarity in algorithm’s outputs, detect temporal changes in causal structures, or evaluate algorithm performance on synthetic data.
Common metrics include:
- Structural Hamming Distance (SHD) [15]: Quantifies the number of edge modifications (additions, deletions, reversals) needed to transform one DAG into another. The SHD is widely used in benchmarking studies [16,17] and for evaluation of new algorithms [18,19,20].
- Precision, Recall, and F1 Score: Useful for comparisons of DAGs, providing insights into proportion of correctly identified edges, completeness of identified edges, and overall similarity assessment. These metrics are used by scholars to compare their novel algorithms [21,22,23].
- True Positive, False Positive, and False Negative Counts: These identify matches and mismatches in edges between DAGs in counts [24].
These metrics are often scattered across multiple packages, with descriptive metrics missing in many of them. As a result, researchers frequently need to switch between packages or even programming languages, and must manually create custom scripts to achieve a comprehensive evaluation. These fragmented workflows introduce inefficiencies and increase the likelihood of errors.
These limitations underscore the need for a unified tool that integrates descriptive and comparative metrics into a single framework. By streamlining the evaluation process and providing intuitive visualization options, such a tool can eliminate inefficiencies, reduce errors, and enable researchers to focus on extracting meaningful insights rather than dealing with technical challenges.
2.3. DAGMetrics: An R Package for Evaluating and Comparing DAGs
DAGMetrics is an R package designed for the evaluation and comparison of DAGs. It provides a comprehensive suite of descriptive and comparative metrics, covering all the aforementioned metrics for both the global network and the Markov blankets of individual nodes. Supplementary Table S1 presents the availability of these metrics in the DAGMetrics package compared to other widely used R and Python packages. Additionally, the package includes intuitive visualization features to enhance interpretability and facilitate analysis.
The package offers two primary plotting options:
- 1.
- Single DAG Visualization: Users can visualize either the full network or the Markov blankets of specified nodes.
- 2.
- Side-by-Side Comparison: This feature allows for direct comparison of two DAGs or the Markov blankets of specified nodes. In these visualizations, selected nodes and matching edges between the two DAGs are highlighted in green, making it easier to identify shared structures and differences.
These visualization tools are particularly valuable for focusing on specific areas of interest. Researchers can specify a list of nodes, enabling a targeted examination of local structures. This capability is especially useful in high-dimensional datasets, where analyzing the global network can be overwhelming. By breaking the network into manageable parts, the DAGMetrics package enhances interpretability and provides deeper insights into complex datasets.
DAGMetrics (version 1.0.0) is available on GitHub at https://github.com/averinpa/DAGMetrics (accessed on 15 January 2025). A detailed workflow diagram (Supplementary Figure S1) and user guide, including sample code, sample output (Supplementary Table S2, Supplementary Figures S2–S5), and a comprehensive overview of the package’s functionality, are provided in Section S4 of the Supplementary Material. Example use cases and the main functions employed in this study are available as Jupyter Notebooks in the DAGMetrics GitHub repository.
2.4. Data Description and Preprocessing
The DAGMetrics package was applied to two metagenomic datasets consisting of soil and potato tuber microbial diversity from two diverse terroirs: continental (Lakoma, Thessaloniki) and island (Naxos, Aegean Sea). In particular, the tuber dataset represented microbiome data from potato tubers of continental and island terroirs at harvest and post-harvest, essentially as previously described [10]. The soil dataset consisted of soil samples originating from the same contrasting terroirs, namely, Naxos and Lakoma [25]. Raw data were obtained from the National Centre for Biotechnology Information (NCBI) sequence read archive (SRA) under BioProject accession numbers PRJNA854325 (for the tuber dataset) and PRJNA970975 (for the soil dataset).
After filtering and preprocessing (see Supplementary Material, Section S5: Dataset Characteristics and Filtering, including Supplementary Table S3, which details the number of samples and bacterial taxonomic units across datasets, and Supplementary Table S4, which shows the percentage of variables that were normally distributed before and after transformation), six datasets were prepared for both island and continental terroirs. These included combinations of the original, scaled, and transformed versions ready for further structure learning.
2.5. Model Selection
For the tuber datasets (island and continental terroirs at harvest and post-harvest stages) and the soil datasets (island and continental terroirs), two constraint-based algorithms and two score-based algorithms were applied, each with various hyperparameter settings.
For the constraint-based algorithms, we used the PC algorithm [12] from the pcalg package [26] and the Inter-IAMB [27] algorithm from the bnlearn package [28]. Both algorithms were tested with three different correlation setups for calculating conditional independence tests: Kendall correlation, Spearman correlation, and Pearson correlation. The significance level () for the tests was set to 5%.
For the score-based algorithms, we applied Tabu Search [14] and Hill-Climbing. Prior to applying these algorithms, the data were discretized, following a common approach adopted by other researchers [4,6]. Two scoring metrics, BIC and AIC, were evaluated during the analysis.
This process resulted in a total of 240 DAGs for the tuber dataset and 120 DAGs for the soil dataset.
The model configurations were encoded as follows: the first letter denotes the algorithm family, with C for constraint-based and S for score-based methods; the number following the letter identifies the specific algorithm, with C1 corresponding to PC, C2 to Inter-IAMB, S1 to Tabu Search, and S2 to Hill-Climbing; the second digit represents the hyperparameter used (1 for Kendall, 2 for Spearman, and 3 for Pearson for the constraint-based algorithms; 1 for BIC and 2 for AIC for the score-based algorithms); the third digit specifies the scaling approach, with 1 for no scaling, 2 for Z-score normalization, and 3 for min–max scaling; and the fourth digit indicates the transformation applied, with 1 for no transformation and 2 for rank-based transformation. For consistency and easier navigation, the order of algorithm codes is kept the same across all figures.
The compare_dags function from the DAGMetrics package was used to calculate a comprehensive set of descriptive and comparative metrics in order to evaluate the resulting DAGs in terms of complexity and similarity.
To assess the complexity of the resulting DAGs, we utilized:
- The normalized number of edges.
- The normalized number of colliders.
- The normalized number of isolated nodes.
To ensure comparability of DAG across different stages and terroirs, we used normalized descriptive metrics, with normalization performed by dividing each metric by the number of nodes in a structure. These metrics provide a framework for comparing the complexity of DAGs across different settings.
For similarity evaluation, we employed the F1 score, which offers insights into the structural differences and overlaps between DAGs.
The most appropriate algorithm was chosen by selecting DAGs with moderate complexity and stability across different scaling and transformation setups.
2.6. Statistical Analysis
The selected DAGs were utilized to explore the Markov blanket structures within each terroir and stage. This analysis aimed to provide a detailed description of the Markov blanket space and to identify Markov blankets with matching causal relationships across terroirs (island and continent) and stages (harvest and post-harvest).
The compare_dags function from the DAGMetrics package was employed to calculate descriptive metrics for each Markov blanket. These metrics included:
- The number of edges.
- The number of colliders.
- The number of nodes.
- The number of isolated nodes.
Additionally, matching causal relationships were identified using the true positive count metric, which quantifies the shared causal connections between Markov blankets.
For an in-depth analysis of specific microbes, the vis_dag_comparison function was used. This function facilitated visual comparisons of the Markov blankets of microbes of interest by generating side-by-side network visualizations.
To further enhance the interpretability of the results, network visualizations were created using Cytoscape software (version 3.10.3) [29], which allows for the customization of network diagrams.
3. Results
This section presents results demonstrating how the DAGMetrics package facilitates the selection of the most appropriate model. This methodology may be of interest to researchers applying causal discovery algorithms to agronomic datasets. Further, we present the results of a detailed analysis of Markov blankets. Finally, we outline the estimated Markov blankets with matching edges and provide a detailed description of the identified and selected Markov blankets.
3.1. Model Selection
The complexity of the DAGs produced by score-based and constraint-based algorithms varied significantly. The detailed results are presented in Figure 1 for the tuber dataset. The corresponding information for the soil dataset is displayed in Supplementary Figure S6.
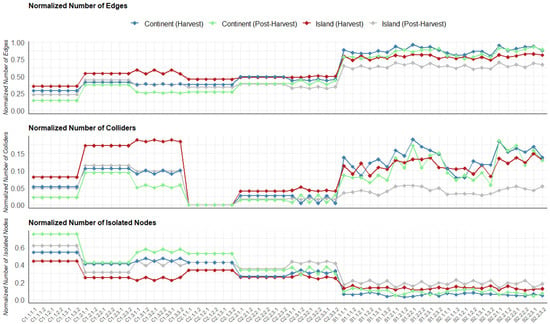
Figure 1.
Normalized number of edges, normalized number of colliders, and normalized number of isolated nodes for island (harvest and post-harvest) and continental (harvest and post-harvest) terroirs across models in the tuber dataset. The models are on the x-axis.
On average, score-based algorithms generated denser graphs with more edges, while constraint-based algorithms produced sparser DAGs for both the tuber and soil datasets. Additionally, the DAGs generated by constraint-based algorithms had a higher proportion of isolated nodes.
Interestingly, the number of colliders showed a mixed pattern; some constraint-based algorithms produced more colliders than score-based algorithms, while others produced fewer. The complexity of the DAGs learned by the two types of algorithms also varied depending on the hyperparameter settings. For instance, score-based algorithms using the BIC criterion produced less complex structures compared to those using the AIC criterion. Among constraint-based algorithms, those utilizing the Kendall correlation yielded the least complex graphs. For example, the structure of continental abundance at the post-harvest stage learned using the PC algorithm with Kendall correlation had 75% isolated nodes (Figure 1). Similarly, for the soil dataset, structures learned with the PC algorithm and Kendall correlation resulted in 79% and 85% isolated nodes for island and continental terroirs, respectively (Supplementary Figure S6). Constraint-based algorithms using the Spearman and Pearson correlations exhibited medium complexity compared to score-based algorithms and constraint-based algorithms using Kendall correlation. In terms of algorithm stability under different scaling and transformation setups, outputs from the PC and Inter-IAMB algorithms using the Spearman and Kendall correlations demonstrated consistent patterns. This trend was observed across both the tuber and soil datasets.
The similarity between the DAGs produced by different structure learning algorithms was measured using the F1 score for each dataset. Detailed results for the tuber dataset in the island terroir at the harvest stage are presented in Figure 2, while Supplementary Figures S7 and S8 visualize the corresponding results for other terroirs and stages of the tuber and soil datasets. The results revealed a high level of dissimilarity between the outputs of the score-based and constraint-based algorithms (Figure 2 and Supplementary Figures S7 and S8). Additionally, variations were observed among the outputs of constraint-based algorithms (Figure 2 and Supplementary Figures S7 and S8). For instance, the DAGs learned with the PC algorithm using Pearson correlation differed from those using Spearman or Kendall correlation as well as from the structures produced by Inter-IAMB configurations (Figure 2 and Supplementary Figures S7 and S8).
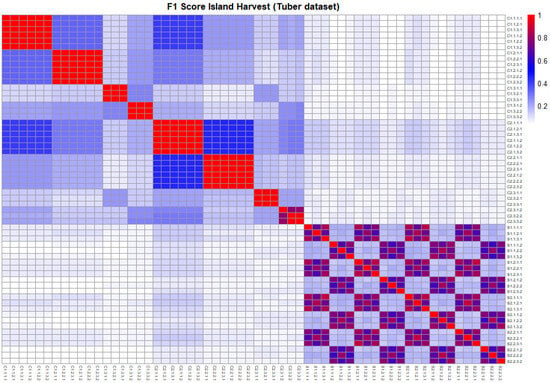
Figure 2.
Plot illustrating the similarity between the outputs of different structure learning algorithms, using the F1 score as a metric. The plot corresponds to the tuber dataset in the island terroir at the harvest stage.
The PC and Inter-IAMB algorithms using the Pearson correlation demonstrated consistency across normalization setups, whereas the same algorithms with the Spearman and Kendall correlations exhibited similarity across scaling and transformation setups. Score-based methods showed moderate similarity across different algorithms; however, notable differences were observed across transformation setups.
3.2. Analysis of Markov Blankets
The structures of the Markov blankets in the two datasets exhibited both similarities and differences. As shown in Figure 3 for the tuber dataset and Supplementary Figure S9 for the soil dataset, most Markov blankets contained no colliders in their structures. Colliders were rare in the soil dataset, with only a few instances having a single collider and none having more than one collider. In contrast, the tuber dataset showed a different pattern, with instances of up to three colliders across terroirs and stages. Notably, the island terroir even contained instances with four and six colliders (Figure 3). In the soil dataset, the Markov blankets most frequently had no edges, followed by those with one edge, with only a few instances having two or three edges. There was a significant gap in frequency between the number of Markov blankets with no edges and those with one edge (Supplementary Figure S9). In the tuber dataset, particularly in the continental terroir, Markov blankets with no edges were predominant, but the gap between those with no edges and one edge was smaller compared to the soil dataset. In the island tuber dataset, Markov blankets with one edge were more frequent than those with no edges. Markov blankets with two edges were slightly less frequent than those with one edge, although their frequencies were similar in the continental terroir and the post-harvest stage (Figure 3).
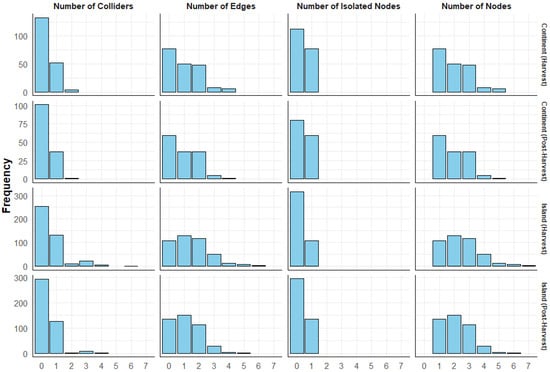
Figure 3.
Plot illustrating the distribution of Markov blankets in the tuber dataset. The horizontal panels represent different terroirs and stages, while the vertical panels correspond to different metrics. The frequency of the Markov blankets is displayed on the y-axis, while the x-axis represents the values of each metric. For example, the top-left panel shows the distribution of the number of colliders across Markov blankets in the continental harvest DAG. The results indicate that over 100 Markov blankets had no colliders in their structure, more than 50 Markov blankets contained one collider, and only a few Markov blankets had two colliders.
Regarding isolated nodes, the soil dataset across terroirs had more isolated nodes than the Markov blankets without isolated nodes (Supplementary Figure S9). In contrast, the tuber dataset contained fewer isolated nodes compared to the Markov blankets without isolated nodes (Figure 3). One-node Markov blankets were the most frequent structures in the soil dataset as well as in the continental terroir of the tuber dataset. However, the gap between the frequency of one-node and two-node structures was greater in the soil dataset. In the tuber dataset, structures with up to seven nodes were observed, whereas in the soil dataset the structures were limited to a maximum of four nodes. Three-node structures were rare in the soil dataset, but were as frequent as two-node structures in the tuber dataset (Figure 3 and Supplementary Figure S9).
Interestingly, there is a relationship between the number of edges, the number of isolated nodes, and the number of nodes in the structure. Markov blankets with no edges always have one isolated node, and the number of nodes in their local structure is always zero.
3.3. Discovered Local Structures
The DAGMetrics package facilitated the identification of local structures with matching edges. Figure 4 illustrates these local structures, highlighting matching edges for both island and continental terroirs across the harvest and post-harvest stages.
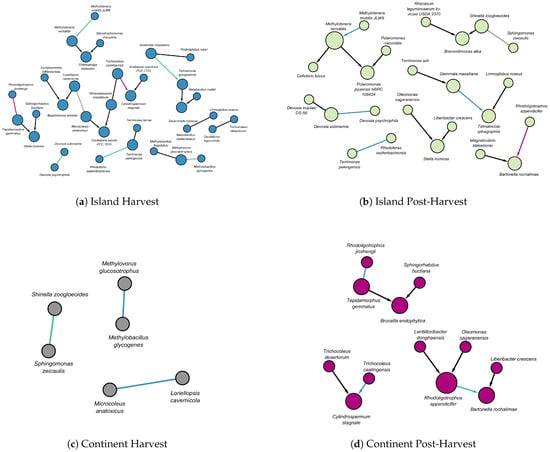
Figure 4.
The above figure illustrates matching edges between DAGs across different terroirs and stages: (a) island harvest nodes are shown in blue, (b) island post-harvest nodes are shown in green, (c) continental harvest nodes are shown in gray, and (d) continental post-harvest nodes are shown in purple. In each panel, the edges are color-coded to indicate whether they match across different terroirs or stages. Black edges signify no match with any edge in another terroir or stage, blue edges indicate a match with the island harvest DAG, green edges correspond to matches with the island post-harvest DAG, gray edges represent matches with the continental harvest DAG, and purple edges indicate matches with the continental post-harvest DAG. For example, in (b), the purple edge between Rhodoligotrophos appendicifer and Bartonella rochalimae indicates that a corresponding edge can be found in a DAG with purple nodes (d). Directed edges represent causal influence, whereas undirected edges denote uncertainty in the direction of influence.
In the island terroir, matching edges were observed between the harvest (Figure 4a) and post-harvest (Figure 4b) stages; specifically:
- A directed edge from Methylotenera mobilis to Methylotenera versatilis.
- A directed edge from Gemmata massiliana to Telmatocola shagniphila.
In addition, two matching undirected relationships were identified between the harvest (Figure 4a) and post-harvest (Figure 4b) stages:
- Devosia psychrophila had an undirected relationship with Devosia submarina.
- Rhodoferax saidenbachensis had an undirected relationship with Terrimonas pekingensis.
A undirected relationship spanning both island and continental terroirs was also found between Loriellopsis cavernicola and Microcoleus anatoxicus during the harvest stage (Figure 4a,c).
The harvest stage in the island terroir (Figure 4a) had persistent relationships with the island post-harvest structures (Figure 4b) as well as with continental structures during both the harvest and post-harvest stages (Figure 4c,d). A notable direct relationship from Trichocoleus caatingensis to Cylindrospermum stagnale (Figure 4a,d) was found as well.
In the island terroir at post-harvest stage, apart from relationships with island structures at the harvest stage, additional persistent relationships were observed with continental structures at both the harvest and post-harvest stages:
- An undirected relationship between Shinella zoogloeoides and Sphingomonas zeicaulis was observed in both the island post-harvest stage and the continent harvest stage (Figure 4b,c).
- A directed relationship from Rhodoligotrophos appendicifer to Bartonella rochalimae was observed in both the island at post-harvest and continent at post-harvest stages (Figure 4b,d).
We identified not only local structures with matching edges but also local structures that differ between stages and terroirs. In Figure 5, the local structures of Rhizobium leguminosarum bv. viciae USDA 2370 are presented across stages and locations.
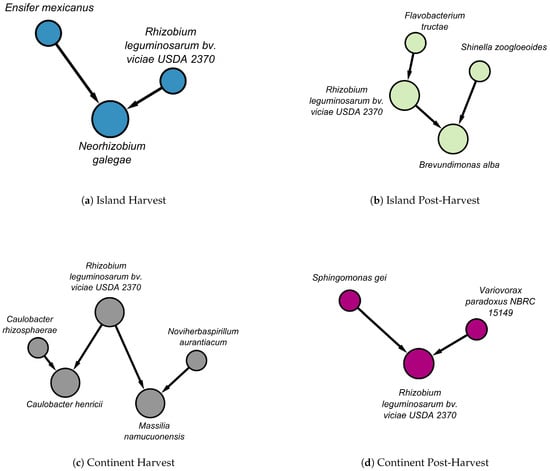
Figure 5.
The above figure represents the Markov blankets of Rhizobium leguminosarum bv. viciae USDA 2370 across different terroirs and stages in the tuber dataset: (a) nodes in the island terroir at harvest are shown in blue, (b) nodes in the island terroir at post-harvest are shown in green, (c) nodes in the continental terroir at harvest are shown in grey, and (d) nodes in the continental terroir at harvest are shown in purple. The edges in the panels have different colors. Black indicates that there is no match with any edge in a different terroir or stage. Directed edges represent causal influence, whereas undirected edges represent uncertainty about the direction of influence.
In the island terroir at the harvest stage, Rhizobium leguminosarum bv. viciae USDA 2370 and Ensifer mexicanus causally influence Neorhizobium galegae (Figure 5a). In contrast, at the post-harvest stage, Rhizobium leguminosarum bv. viciae USDA 2370 receives a directed edge from Flavobacterium tructae, which together with Shinella zoogloeoides has a causal influence on Brevundimonas alba (Figure 5b).
In the continental terroir, Rhizobium leguminosarum bv. viciae USDA 2370 and Caulobacter rhizosphaerae causally influence Caulobacter henricii. Additionally, together with Noviherbaspirillum aurantiacum, they both causally influence Massilia namucuonensis (Figure 5c). At the post-harvest stage, Rhizobium leguminosarum bv. viciae USDA 2370 receives directed influence from Sphingomonas gei and Variovorax paradoxus (Figure 5d).
We could not identify any matching edges between the island and continental terroirs in the soil dataset. However, there were some relationships that differed between the island and continental terroirs. For example, in the island structure, Nitrospira japonica has an undirected edge with Nitrobacter winogradskyi, whereas in continental terroir Nitrospira japonica and Limisphaera ngamtamarikiensis together exert a causal influence on Syntrophotalea carbinolica DSM (Supplementary Figure S10).
The number of reads for species in tubers from the island and continental terroirs at harvest and post-harvest are provided in Supplementary Table S5 along with the number of reads for species in soil that were discussed or presented in the figures.
4. Discussion
In this paper, we introduce the DAGMetrics R package for evaluating and comparing DAGs. DAGMetrics provides an intuitive and easy-to-use framework for researchers seeking detailed insights into the structure of causal networks. The package’s visualization tools facilitate the exploration of DAGs, enabling to highlight key structural differences efficiently. Additionally, its ability to compare local structures allows differences and similarities to be detected across different locations, time points, and stages of a system. Another significant advantage of DAGMetrics is its ability to visualize not only the Markov blanket of a selected node but also to drill down further to display Markov blankets of multiple nodes. This feature enables multiple local structures to be explored and compared simultaneously, promoting focus on relationships of interest without the need to display the full structure, which can be especially overwhelming in high-dimensional datasets where the full structure may consist of hundreds of nodes. By providing this functionality, DAGMetrics makes it easier to analyze causal structures.
The DAGMetrics package was applied to two distinct metagenomic datasets generated for the potato tuber and soil microbiome. DAGMetrics was able to streamline the comprehensive evaluation of outputs from various structure learning algorithms, aiding in the identification of the most appropriate DAGs for downstream analysis. Additionally, it facilitated the description of the Markov blanket space of the DAGs across different terroirs and stages. Finally, it enabled the identification of matching nodes and dissimilar Markov blankets in structures derived from different terroirs and stages.
From a model selection perspective, the comprehensive set of descriptive and comparative metrics provided by DAGMetrics allows users to reflect on the desired characteristics of the selected DAG. The absence of a ground-truth DAG and the resulting inability to assess the accuracy of the learned structures remain challenges. However, a detailed evaluation of the complexity, stability, and similarity of outputs of different structure learning algorithms with different scaling and transformation setups offers indirect indications of the robustness of the learned structures. Our analysis of two different datasets revealed consistent patterns in the behavior of different structure learning algorithms. For example, the PC algorithm with Spearman correlation produced balanced DAGs with moderate complexity and structural stability across various scaling and transformation techniques.
The description of the Markov blanket space provided insights into the studied agronomic microbial abundance datasets. Despite challenges such as small sample sizes and high dimensionality, the studied datasets shared structural characteristics. These included many isolated nodes, moderately small Markov blankets (up to seven nodes), and the existence of isolated substructures within the global structure.
The use of DAGs facilitated by the DAGMetrics package provided profound insights into the microbial dynamics within potato tubers at two physiological stages (at harvest and post-harvest) from two diverse terroirs (island and continental). In particular, our results on the directed relationships between species such as Methylotenera mobilis and Methylotenera versatilis may suggest potential metabolic, genetic, or ecological dependencies that are crucial for microbial stability and function. Both species belong to the family of Methylophilaceae. The former is involved in the denitrification pathway, a capability that is integral to its role in nitrogen cycling, while the latter is involved in methane metabolism, which is critical in the global carbon cycle and methane biotransformation [30,31]. Furthermore, these species can be found in similar environments, probably sharing some ecological niches, which suggests similarities in their metabolic capabilities and environmental adaptations that allow them to thrive in similar methylamine-rich environments [32]. Therefore, these interactions or associations make Methylotenera mobilis and Methylotenera versatilis critical, especially in environments where nutrient uptake and carbon cycling need to be stimulated [33]. Moreover, the persistent relationships we founf across different stages and regions, such as those between Devosia psychrophila and Devosia submarina, highlight the robustness of microbial associations that potentially underpin critical processes such as decomposition and nutrient cycling, supporting the contribution of intricate microbial associations to the multifunctionality of agricultural ecosystems [34]. Although both species belong to the same Devosia genus, they can be found in distinct environmental habitats, with Devosia psychrophile being adapted to cold environments and Devosia submarina to marine ecosystems (deep sea sediments). In our study, these species were more abundant in the island terroir, suggesting sophisticated genetic and metabolic adaptations that enable them to thrive in these habitats. The undirected relationship between Rhodoferax saidenbachensis and Terrimonas pekingensis likely plays a role in the decomposition of organic matter, as they are both found in environments that are rich in organic material [35,36], thereby contributing to the nutrient cycling necessary for ecosystem functioning. The cross-terroir relationship between Loriellopsis cavernicola and Microcoleus anatoxicus at the harvest stage underscores the remarkable ecological resilience and adaptability of these microbial species, highlighting their potential to sustain microbial community structures under varying environmental conditions. Both cyanobacterium species are integral to their respective ecosystems, with mat-forming Microcoleus anatoxicus being able to survive in extreme harsh conditions [37] and Loriellopsis cavernicola being initially identified in Greek and Spanish caves [38]. Additionally, the local structures between Rhizobium leguminosarum and several other bacteria, such as Neorhizobium galegae, Flavobacterium tructae, and Nitrospira spp., suggest a robust consortium of bacteria related to nitrogen fixation and organic matter decomposition [39]. By fostering these microbial interactions and ensuring a healthy soil microbiome, farmers can enhance soil quality, improve potato health and yield, and reduce dependency on chemical inputs. Integrating knowledge of these microbial roles into agricultural practices can lead to more sustainable and productive potato farming systems. By further exploring the matching edges obtained from the DAGMetrics package for both island and continental terroirs, the complex and dynamic nature of microbial communities of potato tubers can be further unraveled, providing insights into the potential of regulating ecosystem functions by manipulating healthy microbial interactions.
5. Conclusions
These findings highlight the ability of DAGMetrics to reveal meaningful structural relationships within complex microbial datasets, demonstrating its potential as a valuable tool for causal discovery in agronomic research and beyond. By providing a broad range of descriptive and comparative metrics along with intuitive visualization capabilities, DAGMetrics enables systematic evaluation of both global and local structures. The DAGMetrics package facilitates the identification of structural similarities and differences across different conditions. Moreover, its ability to compare DAGs across multiple datasets, track structural changes, and highlight matching connections makes it a powerful resource for researchers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15040987/s1, Supplementary Table S1: Comparison of metrics presented in DAGMetrics and existing Python and R packages; Supplementary Figure S1: Core functions of DAGMetrics package; Supplementary Table S2: compare dags function output; Supplementary Figure S2: Markov Blankets of N4 and N291; Supplementary Figure S3: Markov Blankets of N4, N291, and N256; Supplementary Figure S4: Markov blanket of N105 in both bn1 and bn2, with matching arcs highlighted; Supplementary Figure S5: Markov blankets of N105 and N33 in both bn1 and bn2, with matching arcs highlighted; Supplementary Table S3: Number of samples and bacteria taxonomic units across datasets; Supplementary Table S4: Percentage of variables that were normally distributed before and after rank-based transformation; Supplementary Figure S6: Normalised number of edges, normalised number of colliders and normalised number of isolated nodes for continental and island terroirs accross models in the soil dataset. On the x-axis the model configurations; Supplementary Figure S7: The similarity between the outputs of different structure learning algorithms, using the F1 score as a metric (soil dataset); Supplementary Figure S8: This plot illustrates the similarity between the outputs of different structure learning algorithms, using the F1 score as a metric (tuber dataset); Supplementary Figure S9: Distribution of Markov blankets in the soil dataset; Supplementary Figure S10: The figure represents Markov blankets of Nitrospira japonica across different terroirs in the soil dataset. Supplementary Table S5. Contains the number of reads for species.
Author Contributions
Conceptualization, P.A. and T.M.; methodology, P.A. and T.M.; software, P.A.; validation, I.M. and T.M.; formal analysis, P.A.; investigation, T.M., M.G., and I.M.; resources, I.M. and A.X.; data curation, P.A. and I.M.; writing—original draft preparation, P.A. and I.M.; writing—review and editing, T.M., M.G., I.M. and A.X.; visualization, P.A.; supervision, T.M.; project administration, T.M.; funding acquisition, T.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Raw data used in this study were obtained from the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA). The tuber dataset is available under BioProject accession number PRJNA854325 and the soil dataset is available under BioProject accession number PRJNA970975. These datasets can be accessed through the NCBI SRA database.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sadeghi, A.; Gopal, A.; Fesanghary, M. Causal Discovery in Financial Markets: A Framework for Nonstationary Time-Series Data. arXiv 2023, arXiv:2312.17375. [Google Scholar] [CrossRef]
- Addo, P.M.; Manibialoa, C.; McIsaac, F. Exploring nonlinearity on the CO2 emissions, economic production and energy use nexus: A causal discovery approach. Energy Rep. 2021, 7, 6196–6204. [Google Scholar] [CrossRef]
- Ebert-Uphoff, I.; Deng, Y. Causal discovery for climate research using graphical models. J. Clim. 2012, 25, 5648–5665. [Google Scholar] [CrossRef]
- Agrahari, R.; Foroushani, A.; Docking, T.R.; Chang, L.; Duns, G.; Hudoba, M.; Karsan, A.; Zare, H. Applications of Bayesian network models in predicting types of hematological malignancies. Sci. Rep. 2018, 8, 6951. [Google Scholar] [CrossRef]
- Friedman, N.; Linial, M.; Nachman, I.; Pe’er, D. Using Bayesian networks to analyze expression data. In Proceedings of the Fourth Annual International Conference on Computational Molecular Biology, Tokyo, Japan, 8–11 April 2000; pp. 127–135. [Google Scholar]
- Foroushani, A.; Agrahari, R.; Docking, R.; Chang, L.; Duns, G.; Hudoba, M.; Karsan, A.; Zare, H. Large-scale gene network analysis reveals the significance of extracellular matrix pathway and homeobox genes in acute myeloid leukemia: An introduction to the Pigengene package and its applications. BMC Med. Genom. 2017, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gaiteri, C.; Bodea, L.G.; Wang, Z.; McElwee, J.; Podtelezhnikov, A.A.; Zhang, C.; Xie, T.; Tran, L.; Dobrin, R.; et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 2013, 153, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Ganopoulou, M.; Michailidis, M.; Angelis, L.; Ganopoulos, I.; Molassiotis, A.; Xanthopoulou, A.; Moysiadis, T. Could Causal Discovery in Proteogenomics Assist in Understanding Gene–Protein Relations? A Perennial Fruit Tree Case Study Using Sweet Cherry as a Model. Cells 2021, 11, 92. [Google Scholar] [CrossRef]
- Skodra, C.; Michailidis, M.; Moysiadis, T.; Stamatakis, G.; Ganopoulou, M.; Adamakis, I.D.S.; Angelis, L.; Ganopoulos, I.; Tanou, G.; Samiotaki, M.; et al. Disclosing the molecular basis of salinity priming in olive trees using proteogenomic model discovery. Plant Physiol. 2023, 191, 1913–1933. [Google Scholar] [CrossRef]
- Boutsika, A.; Michailidis, M.; Ganopoulou, M.; Dalakouras, A.; Skodra, C.; Xanthopoulou, A.; Stamatakis, G.; Samiotaki, M.; Tanou, G.; Moysiadis, T.; et al. A wide foodomics approach coupled with metagenomics elucidates the environmental signature of potatoes. Iscience 2023, 26, 105917. [Google Scholar] [CrossRef]
- Brouillard, P.; Squires, C.; Wahl, J.; Kording, K.P.; Sachs, K.; Drouin, A.; Sridhar, D. The Landscape of Causal Discovery Data: Grounding Causal Discovery in Real-World Applications. arXiv 2024, arXiv:2412.01953. [Google Scholar]
- Spirtes, P.; Glymour, C.; Scheines, R. Causation, Prediction, and Search; MIT Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Pearl, J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference; Morgan Kaufmann: Burlington, MA, USA, 1988. [Google Scholar]
- Bouckaert, R.R. Bayesian Belief Networks: From Construction to Inference. Ph.D. Thesis, Universiteit Utrecht, Faculteit Wiskunde en Informatica, Utrecht, The Netherlands, 1995. [Google Scholar]
- Tsamardinos, I.; Brown, L.E.; Aliferis, C.F. The max-min hill-climbing Bayesian network structure learning algorithm. Mach. Learn. 2006, 65, 31–78. [Google Scholar] [CrossRef]
- Scutari, M.; Graafland, C.E.; Gutiérrez, J.M. Who learns better Bayesian network structures: Accuracy and speed of structure learning algorithms. Int. J. Approx. Reason. 2019, 115, 235–253. [Google Scholar] [CrossRef]
- Constantinou, A.C.; Liu, Y.; Chobtham, K.; Guo, Z.; Kitson, N.K. Large-scale empirical validation of Bayesian Network structure learning algorithms with noisy data. Int. J. Approx. Reason. 2021, 131, 151–188. [Google Scholar] [CrossRef]
- Zheng, X.; Aragam, B.; Ravikumar, P.K.; Xing, E.P. Dags with no tears: Continuous optimization for structure learning. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 3–8 December 2018; Volume 31. [Google Scholar]
- Fang, Z.; Zhu, S.; Zhang, J.; Liu, Y.; Chen, Z.; He, Y. On low-rank directed acyclic graphs and causal structure learning. IEEE Trans. Neural Netw. Learn. Syst. 2023, 35, 4924–4937. [Google Scholar] [CrossRef]
- Ng, I.; Ghassami, A.; Zhang, K. On the role of sparsity and dag constraints for learning linear dags. In Proceedings of the Advances in Neural Information Processing Systems, Virtual, 6–12 December 2020; Volume 33, pp. 17943–17954. [Google Scholar]
- Huang, B.; Zhang, K.; Zhang, J.; Ramsey, J.; Sanchez-Romero, R.; Glymour, C.; Schölkopf, B. Causal discovery from heterogeneous/nonstationary data. J. Mach. Learn. Res. 2020, 21, 1–53. [Google Scholar]
- Maeda, T.N.; Shimizu, S. RCD: Repetitive causal discovery of linear non-Gaussian acyclic models with latent confounders. In Proceedings of the International Conference on Artificial Intelligence and Statistics, PMLR, Online, 26–28 August 2020; pp. 735–745. [Google Scholar]
- Maeda, T.N.; Shimizu, S. Causal additive models with unobserved variables. In Proceedings of the Uncertainty in Artificial Intelligence, PMLR, Online, 27–30 July 2021; pp. 97–106. [Google Scholar]
- Shimizu, S.; Hoyer, P.O.; Hyvärinen, A.; Kerminen, A.; Jordan, M. A linear non-Gaussian acyclic model for causal discovery. J. Mach. Learn. Res. 2006, 7, 2003–2030. [Google Scholar]
- Boutsika, A.; Xanthopoulou, A.; Tanou, G.; Zacharatou, M.E.; Vernikos, M.; Nianiou-Obeidat, I.; Ganopoulos, I.; Mellidou, I. A microbiome survey of contrasting potato terroirs using 16S rRNA long-read sequencing. Plant Soil 2024, 505, 431–448. [Google Scholar] [CrossRef]
- Kalisch, M.; Mächler, M.; Colombo, D.; Maathuis, M.H.; Bühlmann, P. Causal inference using graphical models with the R package pcalg. J. Stat. Softw. 2012, 47, 1–26. [Google Scholar] [CrossRef]
- Tsamardinos, I.; Aliferis, C.F.; Statnikov, A.R.; Statnikov, E. Algorithms for large scale Markov blanket discovery. FLAIRS 2003, 2, 376–381. Available online: https://cdn.aaai.org/FLAIRS/2003/Flairs03-073.pdf (accessed on 25 January 2025).
- Scutari, M. Learning Bayesian networks with the bnlearn R package. arXiv 2009, arXiv:0908.3817. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Beck, D.A.; Vorobev, A.; Smalley, N.; Kunkel, D.D.; Lidstrom, M.E.; Chistoserdova, L. Novel methylotrophic isolates from lake sediment, description of Methylotenera versatilis sp. nov. and emended description of the genus Methylotenera. Int. J. Syst. Evol. Microbiol. 2012, 62, 106–111. [Google Scholar] [CrossRef]
- Mustakhimov, I.; Kalyuzhnaya, M.G.; Lidstrom, M.E.; Chistoserdova, L. Insights into denitrification in Methylotenera mobilis from denitrification pathway and methanol metabolism mutants. J. Bacteriol. 2013, 195, 2207–2211. [Google Scholar] [CrossRef] [PubMed]
- Salcher, M.M.; Schaefle, D.; Kaspar, M.; Neuenschwander, S.M.; Ghai, R. Evolution in action: Habitat transition from sediment to the pelagial leads to genome streamlining in Methylophilaceae. ISME J. 2019, 13, 2764–2777. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chen, A.; Sheng, R.; Hou, H.; Zhu, B.; Wei, W.; Zhang, W. Long-term chemical and organic fertilization induces distinct variations of microbial associations but unanimous elevation of soil multifunctionality. Sci. Total Environ. 2024, 931, 172862. [Google Scholar] [CrossRef]
- Jin, D.; Wang, P.; Bai, Z.; Jin, B.; Yu, Z.; Wang, X.; Zhuang, G.; Zhang, H. Terrimonas pekingensis sp. nov., isolated from bulking sludge, and emended descriptions of the genus Terrimonas, Terrimonas ferruginea, Terrimonas lutea and Terrimonas aquatica. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 5, 1658–1664. [Google Scholar] [CrossRef] [PubMed]
- Kaden, R.; Spröer, C.; Beyer, D.; Krolla-Sidenstein, P. Rhodoferax saidenbachensis sp. nov., a psychrotolerant, very slowly growing bacterium within the family Comamonadaceae, proposal of appropriate taxonomic position of Albidiferax ferrireducens strain T118T in the genus Rhodoferax and emended description of the genus Rhodoferax. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 4, 1186–1193. [Google Scholar]
- Junier, P.; Cailleau, G.; Fatton, M.; Udriet, P.; Hashmi, I.; Bregnard, D.; Corona-Ramirez, A.; di Francesco, E.; Kuhn, T.; Mangia, N.; et al. A cohesive Microcoleus strain cluster causes benthic cyanotoxic blooms in rivers worldwide. Water Res. X 2024, 24, 100252. [Google Scholar] [CrossRef]
- Lamprinou, V.; Hernández-Mariné, M.; Canals, T.; Kormas, K.; Economou-Amilli, A.; Pantazidou, A. Morphology and molecular evaluation of Iphinoe spelaeobios gen. nov., sp. nov. and Loriellopsis cavernicola gen. nov., sp. nov., two stigonematalean cyanobacteria from Greek and Spanish caves. Int. J. Syst. Evol. Microbiol. 2011, 61, 2907–2915. [Google Scholar] [CrossRef]
- Qadir, M.; Hussain, A.; Iqbal, A.; Shah, F.; Wu, W.; Cai, H. Microbial utilization to nurture robust agroecosystems for food security. Agronomy 2024, 14, 1891. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).