An Exploration of the Chemical Signals and Neural Pathways Driving the Attraction of Meloidogyne incognita and Caenorhabditis elegans to Favorable Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Culture Conditions
2.2. C. elegans Strains
2.3. Collection of Meloidogyne Incognita Eggs and Second-Stage Juveniles (J2s)
2.4. C. elegans Maintenance and Synchronization
2.5. Reagents
2.6. Preparation of Bacterial Suspension and Fermentation Broth of Bacteria
2.7. Phylogenetic Analysis
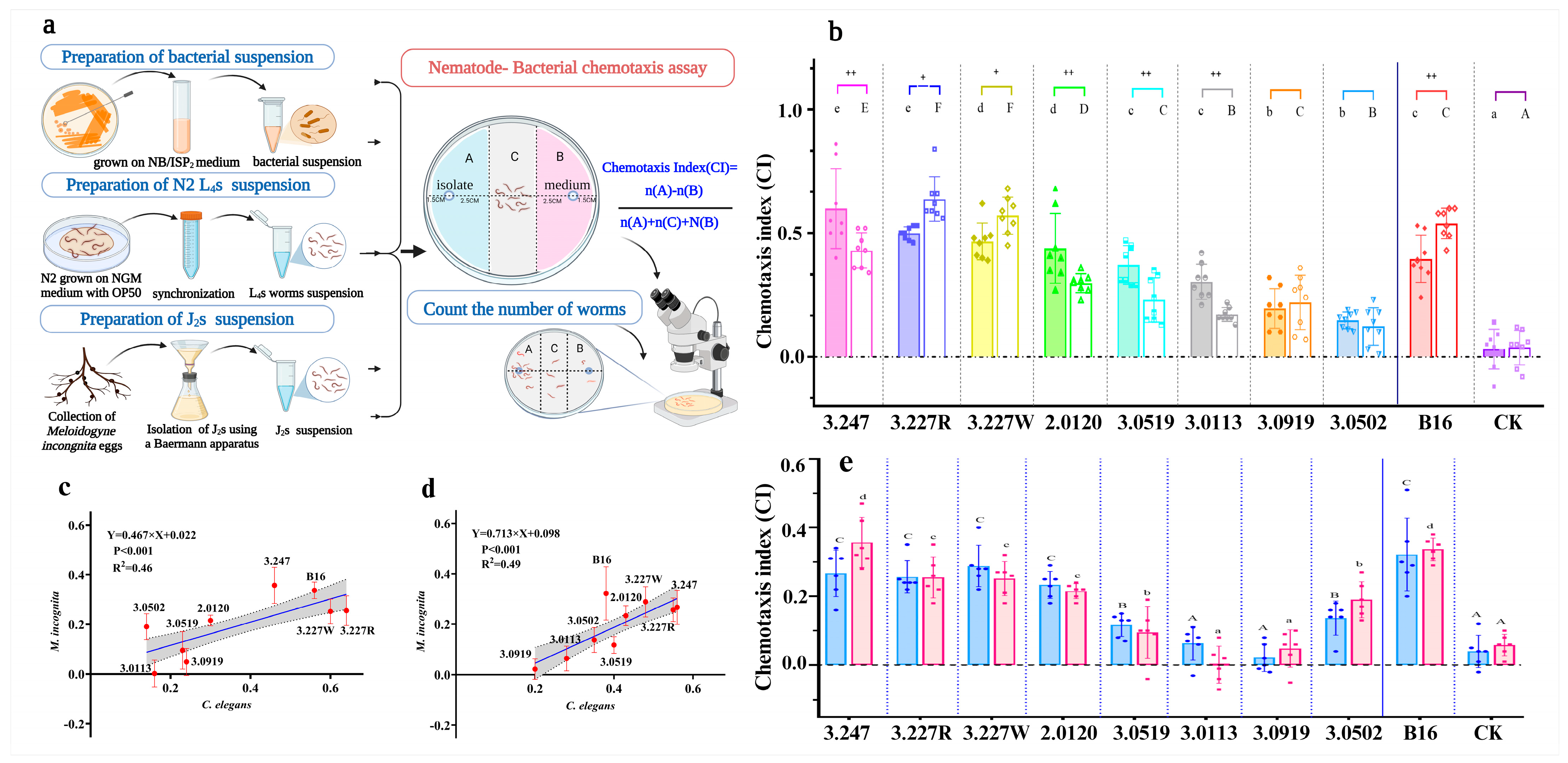
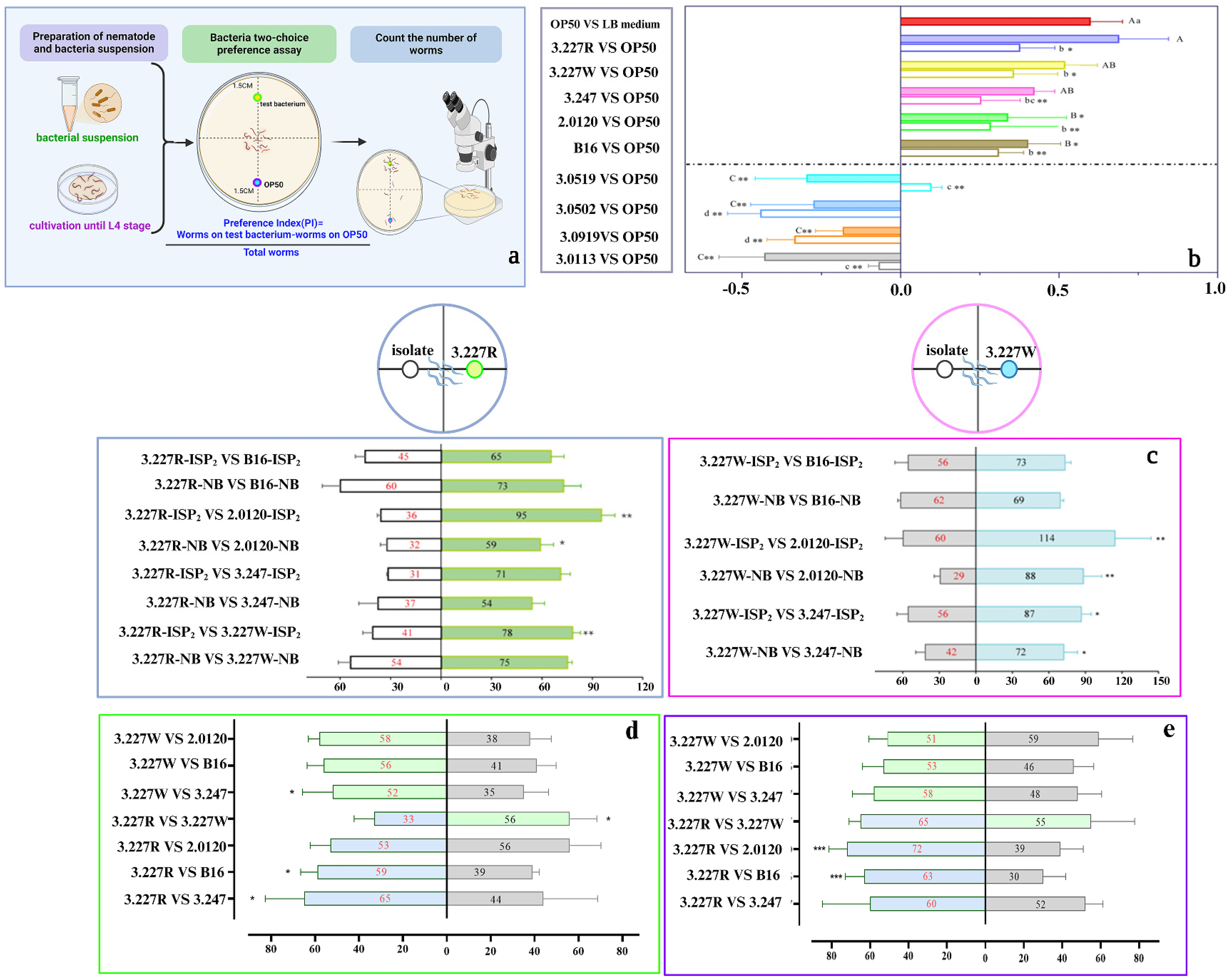
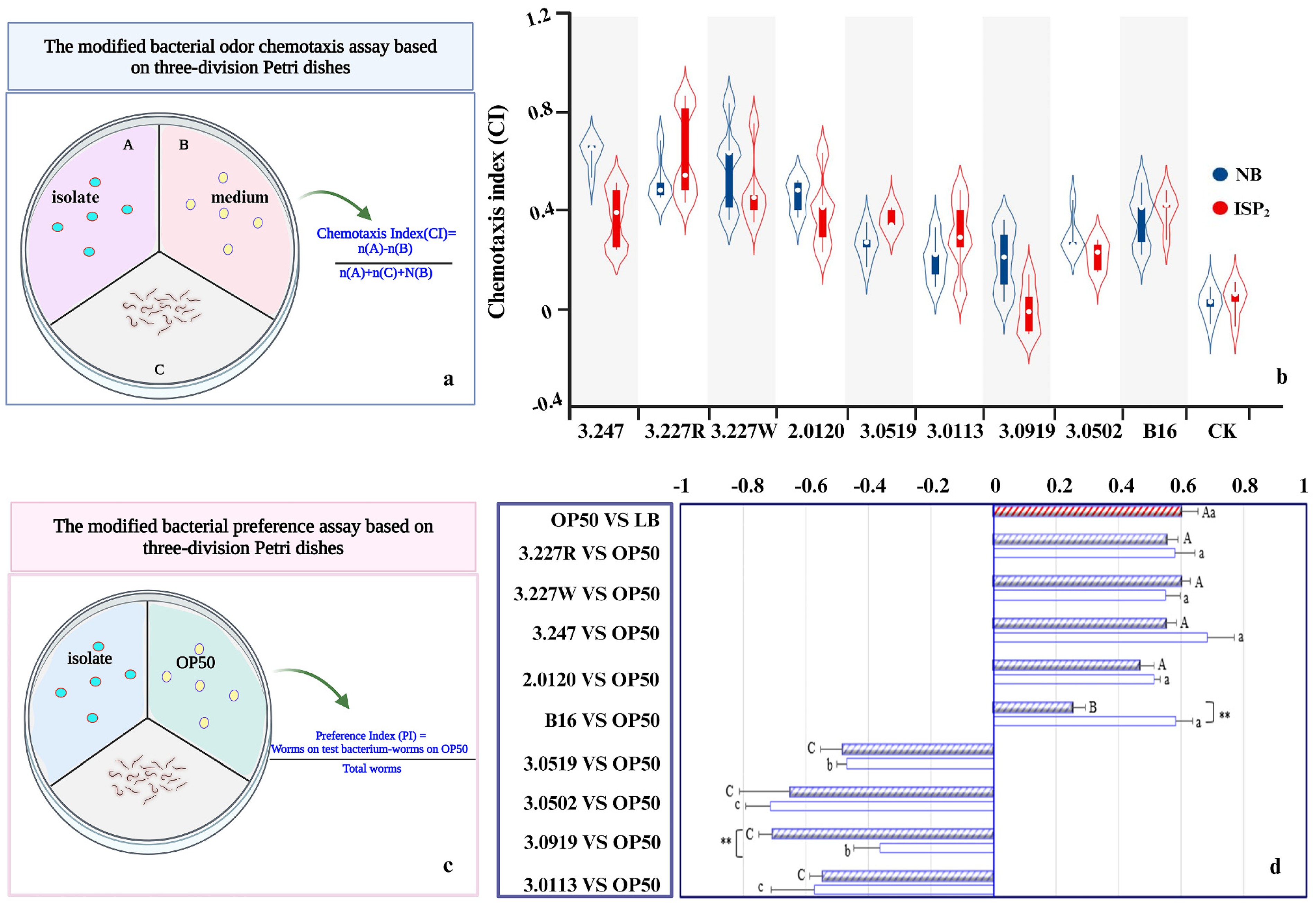
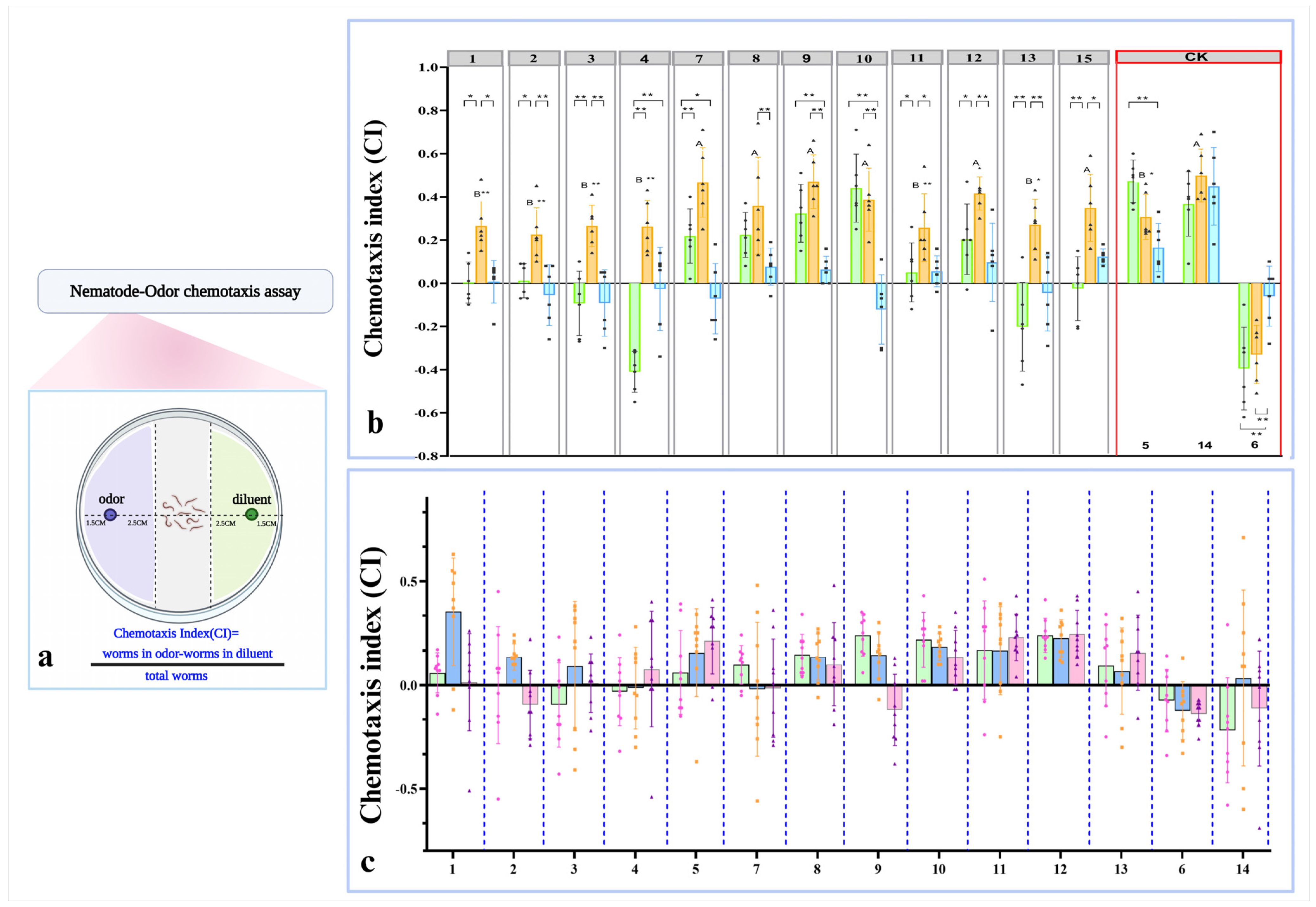
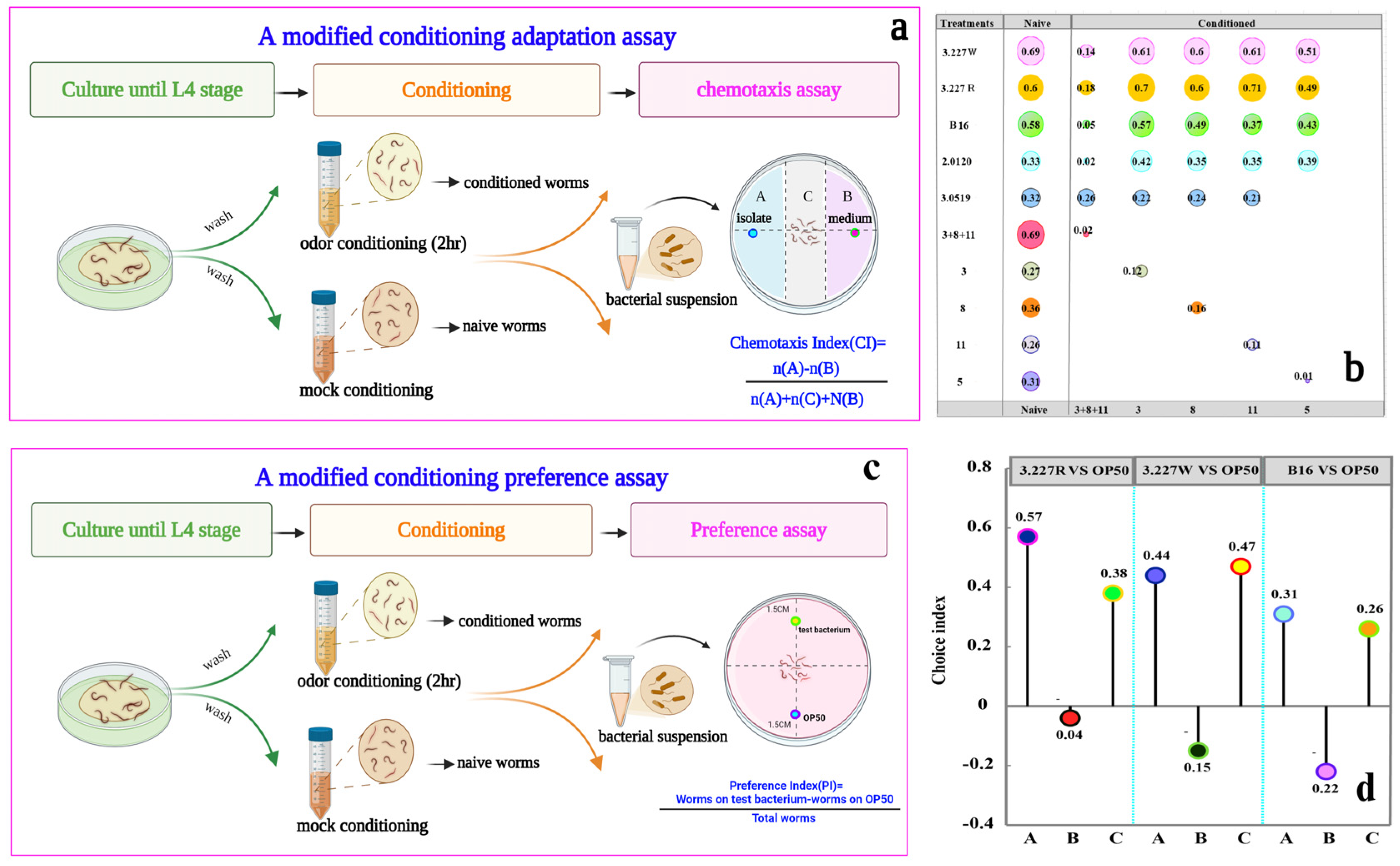
2.8. Nematode Behavioral Assays
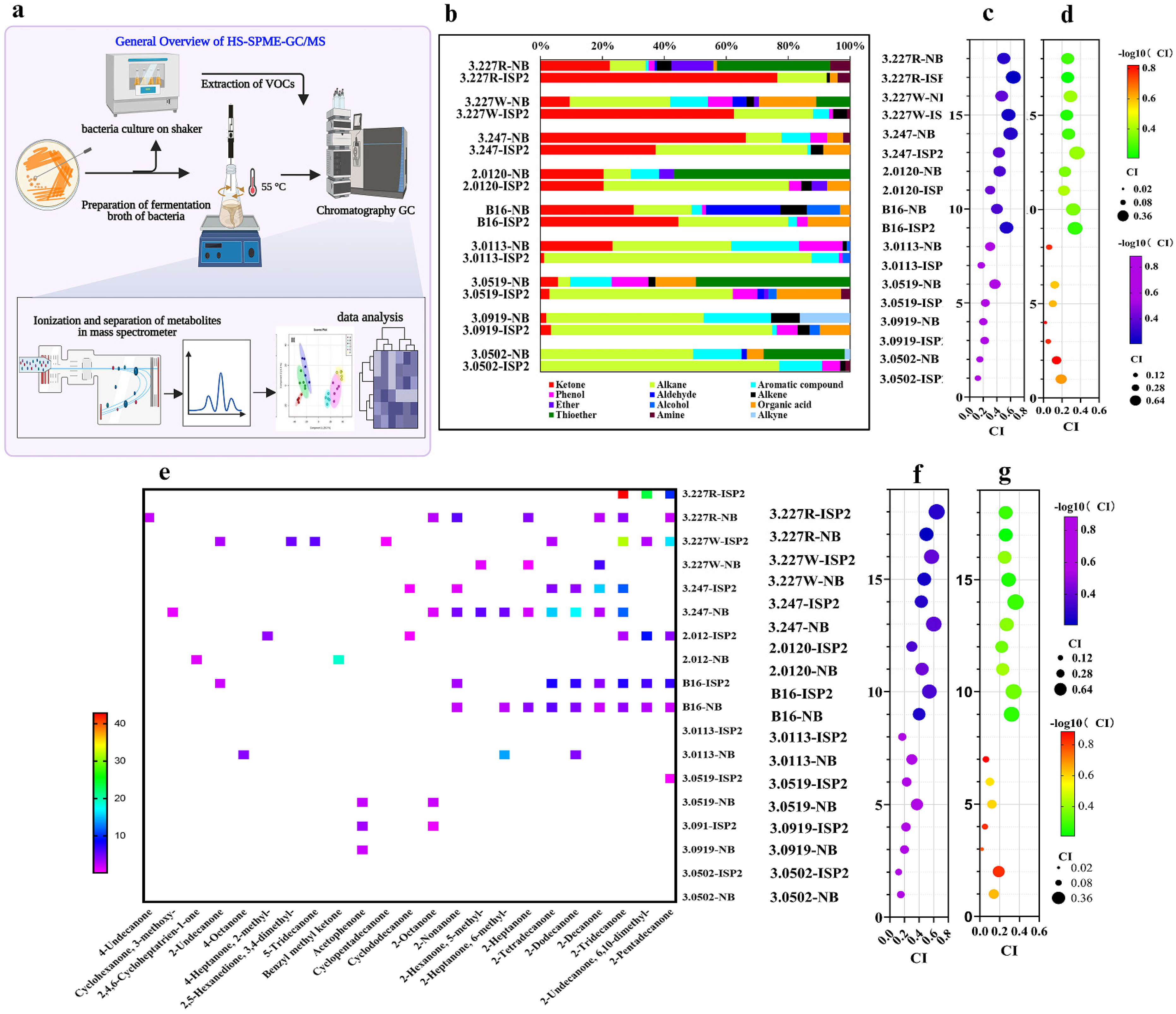
2.9. Analysis of Bacterial BVOCs by HS-SPME-GC-MS
2.10. Multivariate Data Analysis
2.11. Statistical Analysis
3. Results
3.1. Screening of Bi-Attractive Bacteria Toward Both C. elegans and M. incognita from Natural Environment
3.2. Volatile-Mediated Attraction of Nematodes by Preferred Bacteria
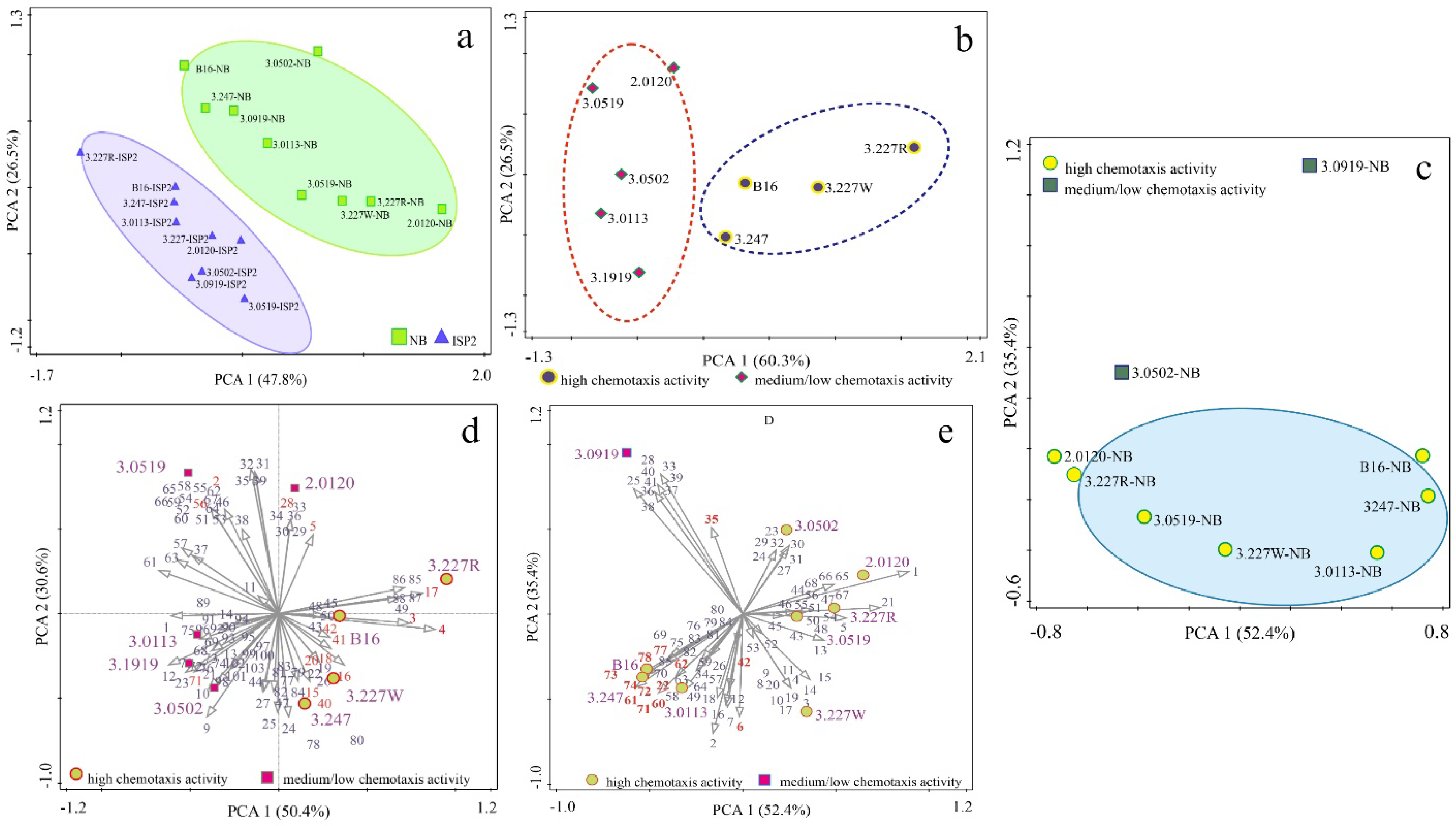
3.3. Identification of Volatile Profiles Released by Attractive Bacteria via HS-SPME-GC-MS
3.4. Potent Correlation Between the Strength of Attractive Activity of Bacteria and the Composition of BVOC Profiles
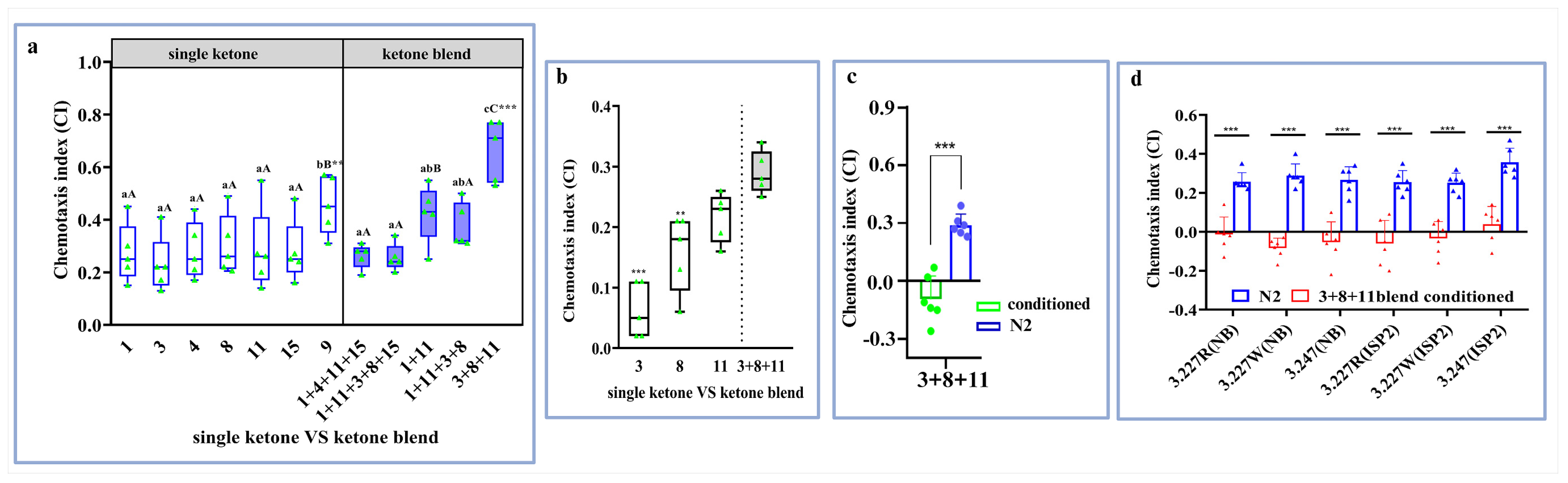
3.5. Heatmap Visualization for Contribution of Ketone Volatiles to the Attraction of Investigated Bacteria to Studied Nematodes
3.6. Ketone Volatile Blend Contributes to Attractiveness of C. elegans and M. incognita to the Preferred Bacteria
3.7. Ketone Odor Blend Adaptation Contributes to Bacterial Adaptation in C. elegans
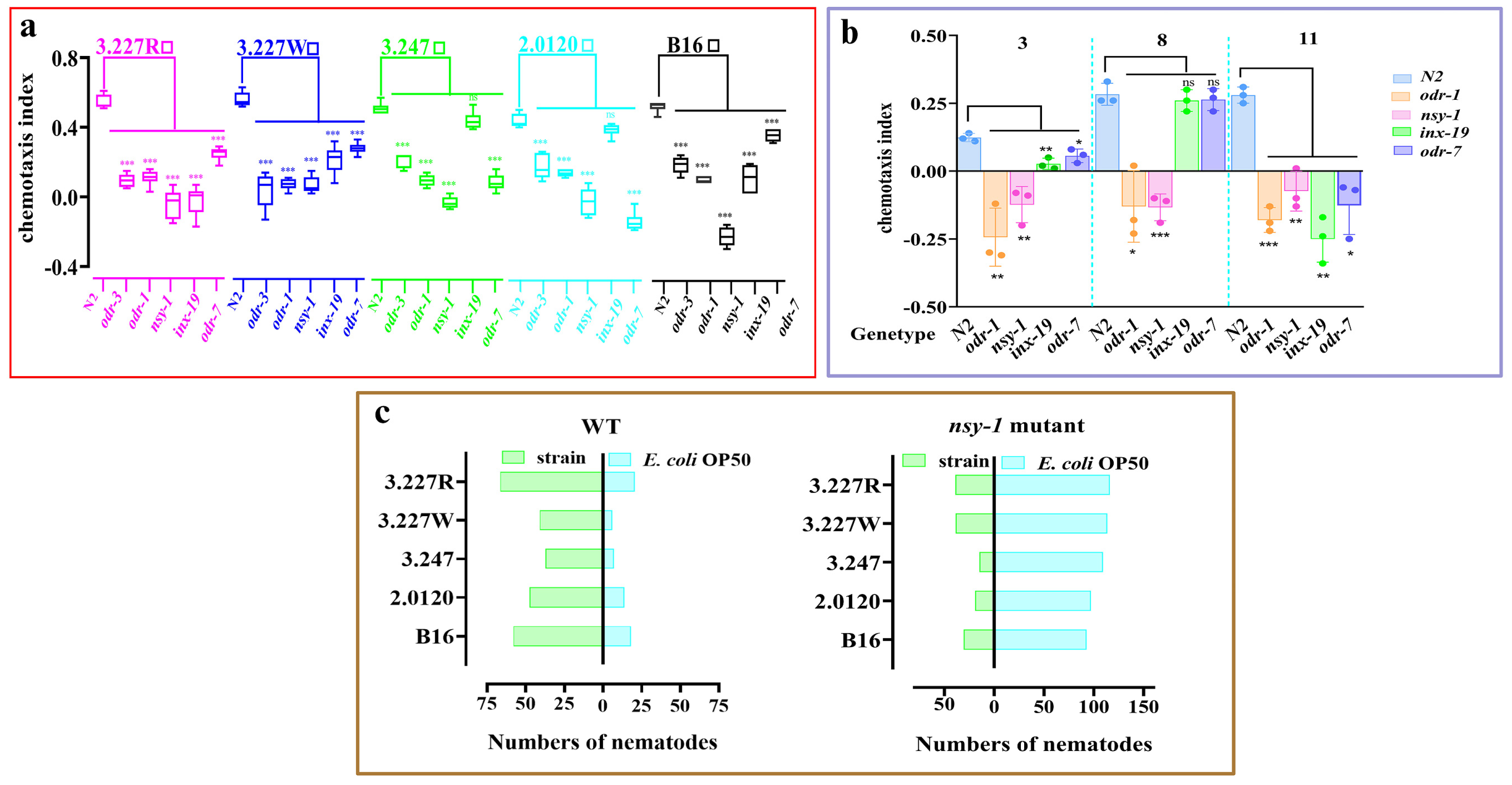
3.8. Identification of Sensory Neurons Involved in Detection of the Preferred Bacteria and Their Released Ketone Volatiles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BVOCs | bacterial volatile organic compounds |
| CGC | Caenorhabditis elegans Genetics Center |
| CI | chemotaxis index |
| DD | Dimethyl disulfide |
| HS-SPME-GC-MS | headspace solid-phase microextraction coupled with gas chromatography mass spectrometry |
| IAA | Isoamyl alcohol |
| ISP2 medium | International Streptomyces Project-2 Medium |
| J2s | second-stage juveniles |
| NA medium | nutrient agar medium |
| NB medium | nutrient broth medium |
| NMDS | nonmetric multidimensional scaling |
| OP50 | Escherichia coli OP50 |
| PCA | principal component analysis |
| PI | preference index |
| RKNs | root-knot nematodes |
References
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Pal-omares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 plant parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, G.; Magliano, M.; Deleury, E.; Abad, P.; Rosso, M.N. Transcriptome analysis of root knot nematode functions induced in the early stages of parasitism. New. Phytol. 2007, 176, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Kirwa, H.K.; Murungi, L.K.; Beck, J.J.; Torto, B. Elicitation of differential responses in the root-knot nematode Meloidogyne incognita to tomato root exudate cytokinin, flavonoids, and alkaloids. J. Agric. Food Chem. 2018, 66, 11291–11300. [Google Scholar] [CrossRef] [PubMed]
- Shivakumara, T.N.; Dutta, T.K.; Chaudhary, S.; von Reuss, S.H.; Williamson, V.M.; Rao, U. Homologs of Caenorhabditis elegans chemosensory genes have roles in behavior and chemotaxis in the root-knot nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 2019, 32, 876–887. [Google Scholar] [CrossRef]
- Bargmann, C.I. Chemosensation in C. elegans. In WormBook, the Online Review of C. elegans Biology; Community TCeR, Ed.; WormBook: Pasadena, CA, USA, 2007. [Google Scholar]
- Choi, J.I.; Yoon, K.-H.; Kalichamy, S.S.; Yoon, S.-S.; Lee, J.I. A natural odor attraction between lactic acid bacteria and the nematode Caenorhabditis elegans. ISME J. 2016, 10, 558–567. [Google Scholar] [CrossRef]
- Worthy, S.E.; Haynes, L.; Chambers, M.; Bethune, D.; Kan, E.; Chung, K.; Ota, R.; Taylor, C.J.; Glater, E.E. Identification of attractive odorants released by preferred bacterial food found in the natural habitats of C. elegans. PLoS ONE 2008, 13, e0201158. [Google Scholar] [CrossRef]
- Worthy, S.E.; Rojas, G.L.; Taylor, C.J.; Glater, E.E. Identification of odor blend used by Caenorhabditis elegans for pathogen recognition. Chem. Senses 2018, 43, 169–180. [Google Scholar] [CrossRef]
- Kim, D.H.; Flavell, S.W. Host-microbe interactions and the behavior of Caenorhabditis elegans. J. Neurogenet. 2020, 34, 500–509. [Google Scholar] [CrossRef]
- Diyapoglu, A.; Oner, M.; Meng, M. Application potential of bacterial volatile organic compounds in the control of root-knot nematodes. Molecules 2022, 27, 4355. [Google Scholar] [CrossRef]
- Chaisson, K.E.; Hallem, E.A. Chemosensory behaviors of parasites. Trends Parasitol. 2012, 28, 427–436. [Google Scholar] [CrossRef]
- Ferkey, D.M.; Sengupta, P.; L’Etoile, N.D. Chemosensory signal transduction in Caenorhabditis elegans. Genetics 2021, 217, iyab004. [Google Scholar] [CrossRef] [PubMed]
- Nunn, L.R.; Juang, T.D.; Beebe, D.J.; Wheeler, N.J.; Zamanian, M. A high-throughput sensory assay for parasitic and free-living nematodes. Integr. Biol. 2023, 15, zyad010. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.C.; Bae, Y.; Lee, S.J. The role of mRNA quality control in the aging of Caenorhabditis elegans. Mol. Cells 2023, 46, 664–671. [Google Scholar] [CrossRef]
- Kim, D.Y.; Moon, K.M.; Heo, W.; Du, E.J.; Park, C.G.; Cho, J.; Hahm, J.H.; Suh, B.C.; Kang, K.; Kim, K. A FMRFamide-like neuropeptide FLP-12 signaling regulates head locomotive behaviors in Caenorhabditis elegans. Mol. Cells 2024, 47, 100124. [Google Scholar] [CrossRef]
- Porta-de-la-Riva, M.; Fontrodona, L.; Villanueva, A.; Cerón, J. Basic Caenorhabditis elegans methods: Synchronization and observation. J. Vis. Exp. 2012, 10, e4019. [Google Scholar] [CrossRef]
- Zhai, Y.; Shao, Z.; Cai, M.; Zheng, L.; Li, G.; Huang, D.; Cheng, W.; Thomashow, L.S.; Weller, D.M.; Yu, Z.; et al. Multiple modes of nematode control by volatiles of Pseudomonas putida 1A00316 from Antarctic soil against Meloidogyne incognita. Front. Microbio 2018, 9, 253. [Google Scholar] [CrossRef]
- Yoshida, K.; Hirotsu, T.; Tagawa, T.; Oda, S.; Wakabayashi, T.; Iino, Y.; Ishihara, T. Odour concentration-dependent ol-factory preference change in C. elegans. Nat. Commun. 2012, 3, 739. [Google Scholar] [CrossRef]
- Glater, E.E.; Rockman, M.V.; Bargmann, C.I. Multigenic, Natural Variation Underlies Caenorhabditis elegans Olfactory Preference for the Bacterial Pathogen Serratia marcescens. G3 Genes Genomes Genet. 2014, 4, 265–276. [Google Scholar] [CrossRef]
- Hsueh, Y.-P.; Gronquist, M.R.; Schwarz, E.M.; Nath, R.D.; Lee, C.-H.; Gharib, S.; Schroeder, F.C.; Sternberg, P.W. Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. eLife 2017, 6, e20023. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, H.; Bargmann, C.I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 2005, 438, 179–184. [Google Scholar] [CrossRef]
- Yamada, K.; Hirotsu, T.; Matsuki, M.; Butcher, R.A.; Tomioka, M.; Ishihara, T.; Clardy, J.; Kunitomo, H.; Iino, Y. Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science 2010, 329, 1647–1650. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Chen, Z.; Zeng, L.; Yang, X.; Huang, D.; Zhai, Y.; Cai, M.; Zheng, L.; Thomashow, L.S.; Weller, D.M.; et al. Control of Meloidogyne incognita in three- dimensional model systems and pot experiments by the attract-and-kill effect of furfural acetone. Plant Dis. 2021, 105, 2169–2176. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Huang, X.; Zhang, L.; Xu, J.; Yang, D.; Wei, K.; Niu, X.; An, Z.; Bennett, J.W.; Zou, C.; et al. A trojan horse mechanism of bacterial pathogenesis against nematodes. Proc. Natl. Acad. Sci. USA 2010, 107, 16631–16636. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, C.I. Neurobiology of the Caenorhabditis elegans genome. Science 1998, 11, 2028–2033. [Google Scholar] [CrossRef]
- Choi, W.; Ryu, S.E.; Cheon, Y.; Park, Y.J.; Kim, S.; Kim, E.; Koo, J.; Choi, H.; Moon, C.; Kim, K. A single chemosensory GPCR is required for a concentration-dependent behavioral switching in C. elegans. Curr. Biol. 2022, 32, 398–411. [Google Scholar] [CrossRef]
- Yang, W.; Wu, T.; Tu, S.; Qin, Y.; Shen, C.; Li, J.; Choi, M.K.; Duan, F.; Zhang, Y. Redundant neural circuits regulate olfactory integration. PLoS Genet. 2022, 18, e1010029. [Google Scholar] [CrossRef]
- Rengarajan, S.; Hallem, E.A. Olfactory circuits and behaviors of nematodes. Curr. Opin. Neurobiol. 2016, 41, 136–148. [Google Scholar] [CrossRef]
- Bargmann, C.I.; Hartwieg, E.; Horvitz, H.R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 1993, 74, 515–527. [Google Scholar] [CrossRef]
- Roayaie, K.; Crump, J.G.; Sagasti, A.; Bargmann, C.I. The Gα protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 1998, 20, 55–67. [Google Scholar] [CrossRef]
- Noelle, D.L.; Bargmann, C.I. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 2000, 25, 575–586. [Google Scholar] [CrossRef]
- Sengupta, P.; Colbert, H.A.; Bargmann, C.I. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell 1994, 79, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Troemel, E.R. Chemosensory signaling in C. elegans. Bioessays 1999, 21, 1011–1020. [Google Scholar] [CrossRef]
- Wes, P.D.; Bargmann, C.I. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 2001, 410, 698–701. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, S.H.; Chronis, N.; Tsunozaki, M.; Gray, J.M.; Ramot, D.; Goodman, M.B.; Bargmann, C.I. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 2007, 450, 63–70. [Google Scholar] [CrossRef]
- Lin, A. Functional imaging and quantification of multineuronal olfactory responses in C. elegans. Sci. Adv. 2023, 9, eade1249. [Google Scholar] [CrossRef]
- Dong, L.; Li, X.; Huang, L.; Gao, Y.; Zhong, L.; Zheng, Y.; Zuo, Y. Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts Mi-flp-18 expression to block infection. J. Exp. Bot. 2014, 65, 131–141. [Google Scholar] [CrossRef]
- Giang, T.; He, J.; Belaidi, S.; Scholz, H. Key odorants regulate food attraction in Drosophila melanogaster. Front. Behav. Neurosci. 2017, 11, 160. [Google Scholar] [CrossRef]
- Wooding, M.; Naudé, Y.; Rohwer, E.; Bouwer, M. Controlling mosquitoes with semiochemicals: A review. Parasites Vectors 2020, 13, 80. [Google Scholar] [CrossRef]
- Taniguchi, G.; Uozumi, T.; Kiriyama, K.; Kamizaki, T.; Hirotsu, T. Screening of odor-receptor pairs in Caenorhabditis elegans reveals different receptors for high and low odor concentrations. Sci. Signal. 2014, 7, ra39. [Google Scholar] [CrossRef]
- Werner, K.M.; Perez, L.J.; Ghosh, R.; Semmelhack, M.F.; Bassler, B.L. Caenorhabditis elegans recognizes a bacterial quorum-sensing signal molecule through the AWCON neuron. J. Biol. Chem. 2014, 289, 26566–26573. [Google Scholar] [CrossRef]
- Terra, W.C.; Lopes de Paula, L.; de Brum, D.; Campos, V.P.; de Oliveira, D.F.; De Souza, J.T. Chemotaxis in Root-Knot Nematodes. In Root-Galling Disease of Vegetable Plants; Springer Nature: Singapore, 2023; pp. 85–115. [Google Scholar] [CrossRef]
- Coke, M.C.; Bell, C.A.; Urwin, P.E. The Use of Caenorhabditis elegans as a Model for Plant-Parasitic Nematodes: What Have We Learned? Annu. Rev. Phytopathol. 2024, 62, 157–172. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, X.; Wang, W.; Wei, C.; Cen, H.; Deng, L.; Tan, D.; Mo, M.; Ma, L. An Exploration of the Chemical Signals and Neural Pathways Driving the Attraction of Meloidogyne incognita and Caenorhabditis elegans to Favorable Bacteria. Agronomy 2025, 15, 590. https://doi.org/10.3390/agronomy15030590
Qin X, Wang W, Wei C, Cen H, Deng L, Tan D, Mo M, Ma L. An Exploration of the Chemical Signals and Neural Pathways Driving the Attraction of Meloidogyne incognita and Caenorhabditis elegans to Favorable Bacteria. Agronomy. 2025; 15(3):590. https://doi.org/10.3390/agronomy15030590
Chicago/Turabian StyleQin, Xunda, Wuqin Wang, Chonglong Wei, Hao Cen, Liping Deng, Dandan Tan, Minghe Mo, and Li Ma. 2025. "An Exploration of the Chemical Signals and Neural Pathways Driving the Attraction of Meloidogyne incognita and Caenorhabditis elegans to Favorable Bacteria" Agronomy 15, no. 3: 590. https://doi.org/10.3390/agronomy15030590
APA StyleQin, X., Wang, W., Wei, C., Cen, H., Deng, L., Tan, D., Mo, M., & Ma, L. (2025). An Exploration of the Chemical Signals and Neural Pathways Driving the Attraction of Meloidogyne incognita and Caenorhabditis elegans to Favorable Bacteria. Agronomy, 15(3), 590. https://doi.org/10.3390/agronomy15030590




