Genome-Wide Identification of AP2/ERF Family Genes in Rubber Tree: Two HbAP2/ERF Genes Regulate the Expression of Multiple Natural Rubber Biosynthesis Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of AP2/ERFs in Rubber Tree (Hevea brasiliensis)
2.2. Phylogenetic Analysis
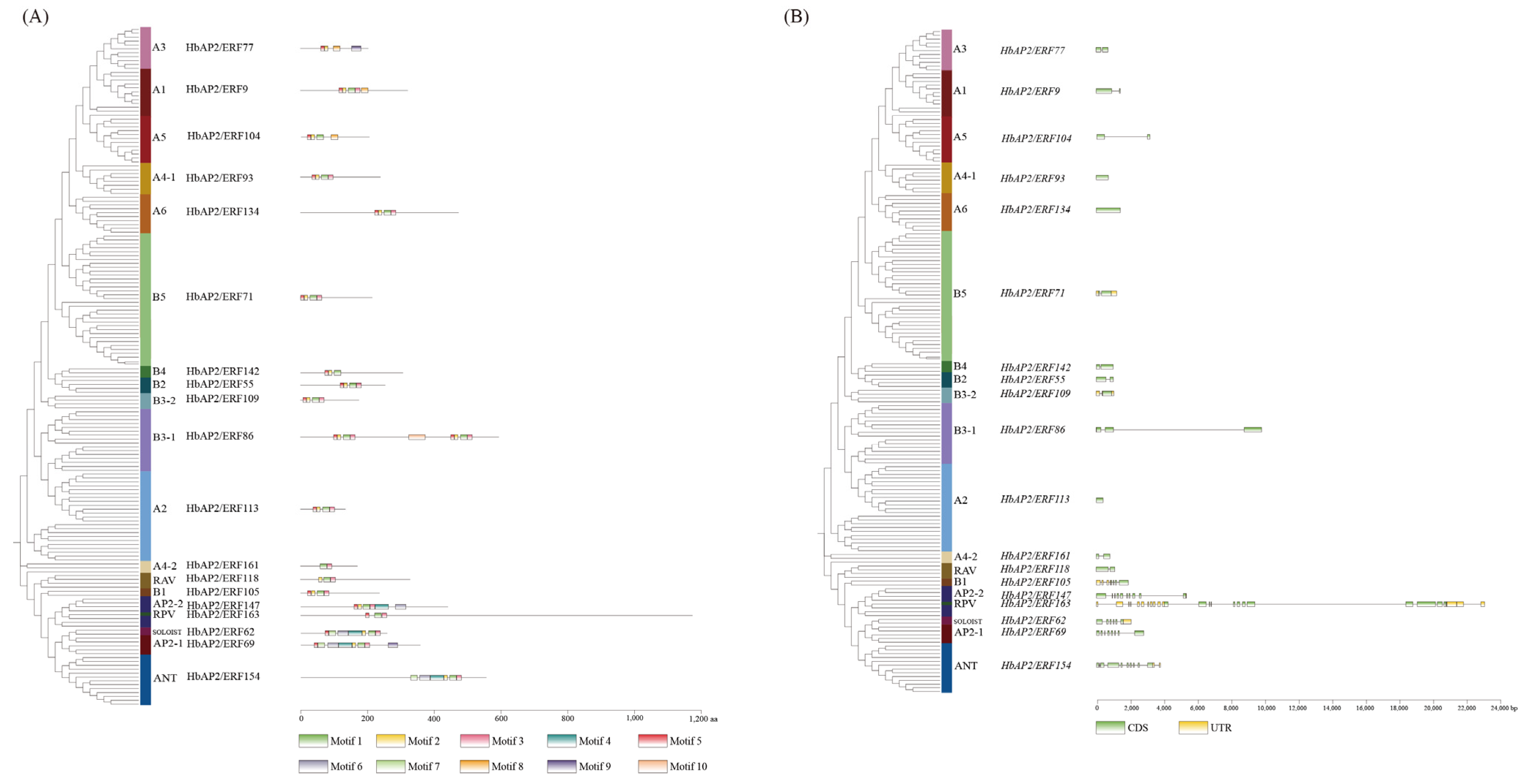
2.3. Chromosomal Localization, Conserved Motif, and Gene Structure Analysis
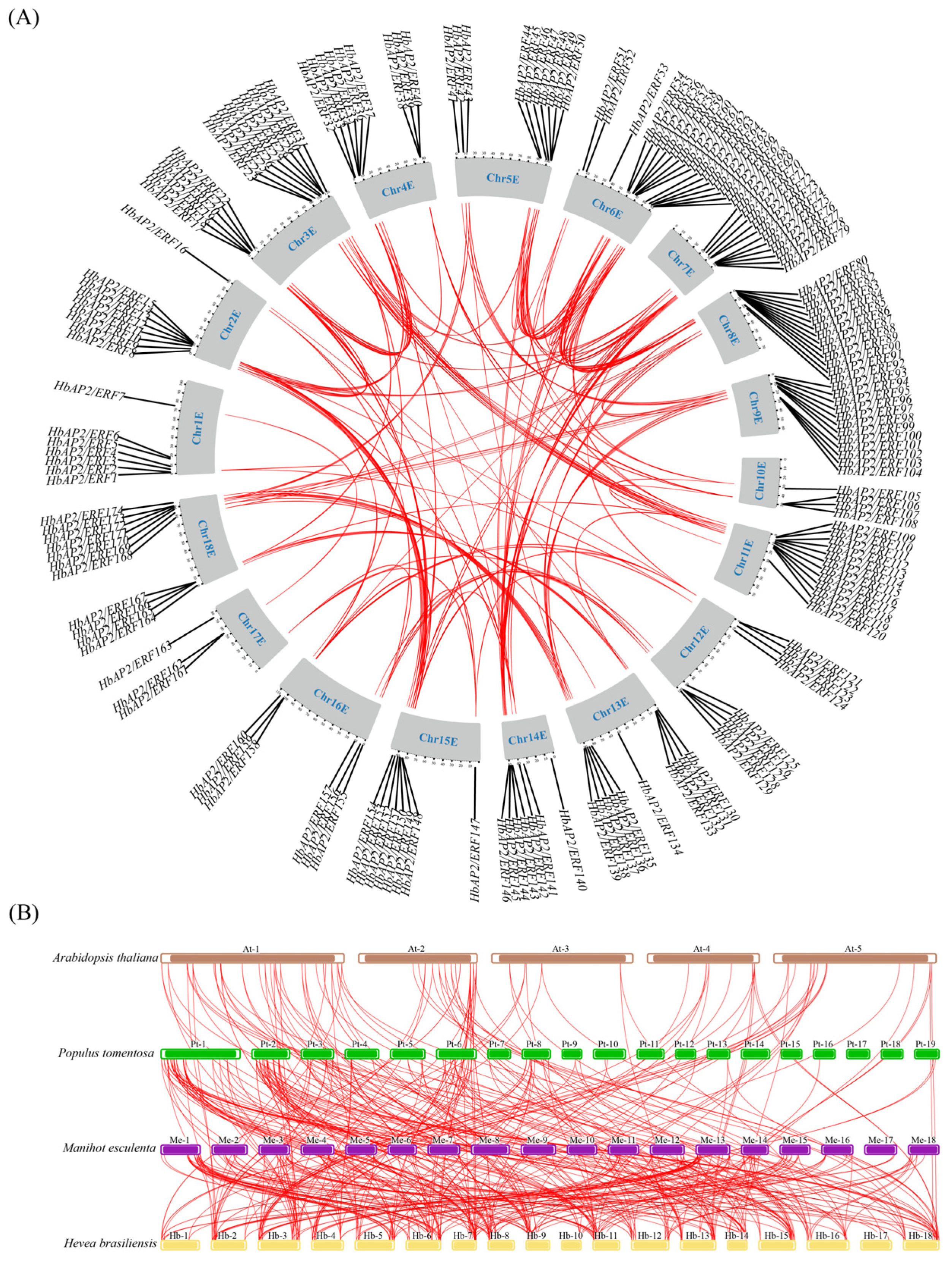
2.4. HbAP2/ERF Duplication and Synteny Analysis
2.5. Analysis of Cis-Regulatory Elements of HbAP2/ERFs
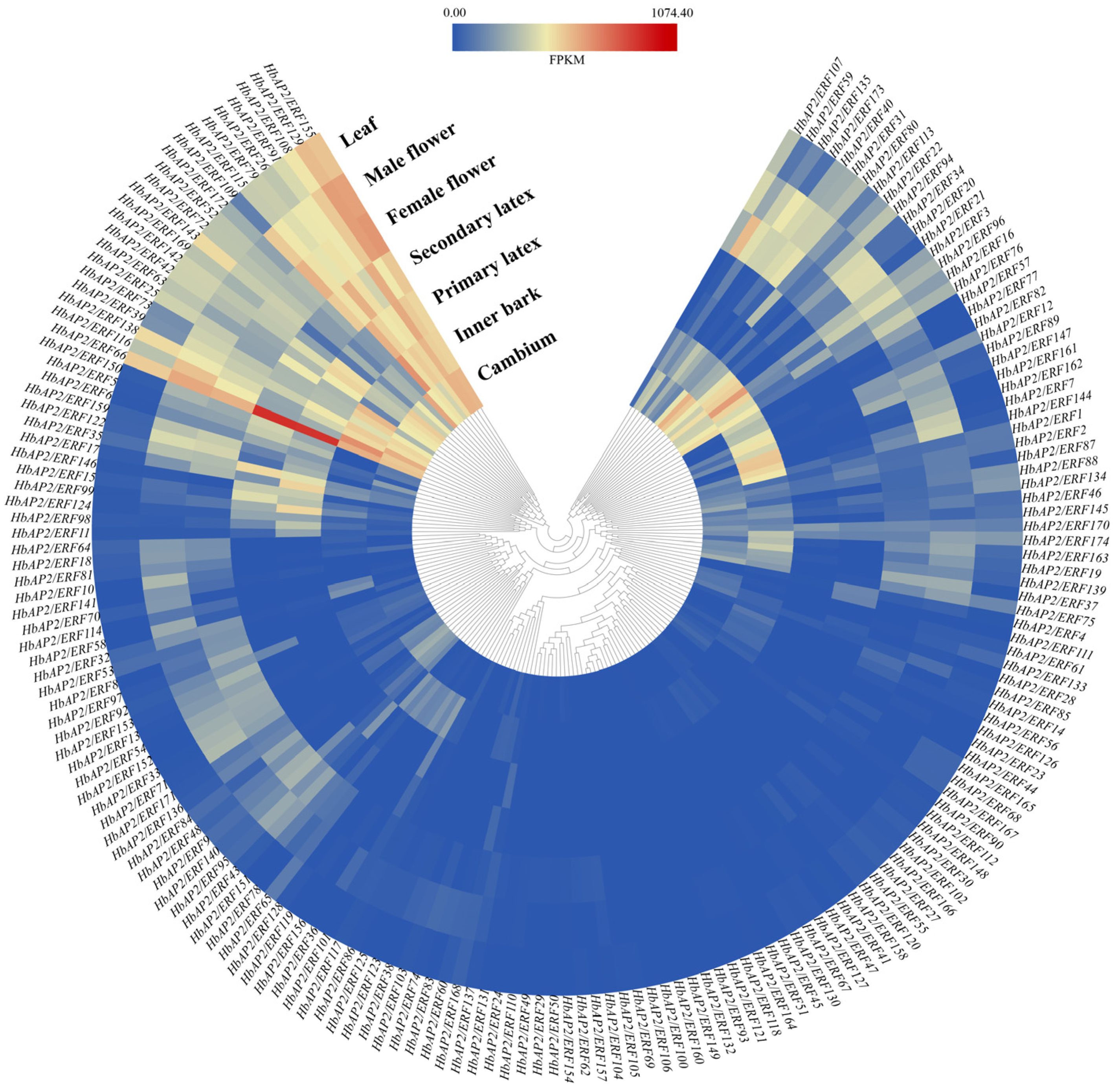
2.6. Tissue-Specific HbAP2/ERFs Expression Patterns
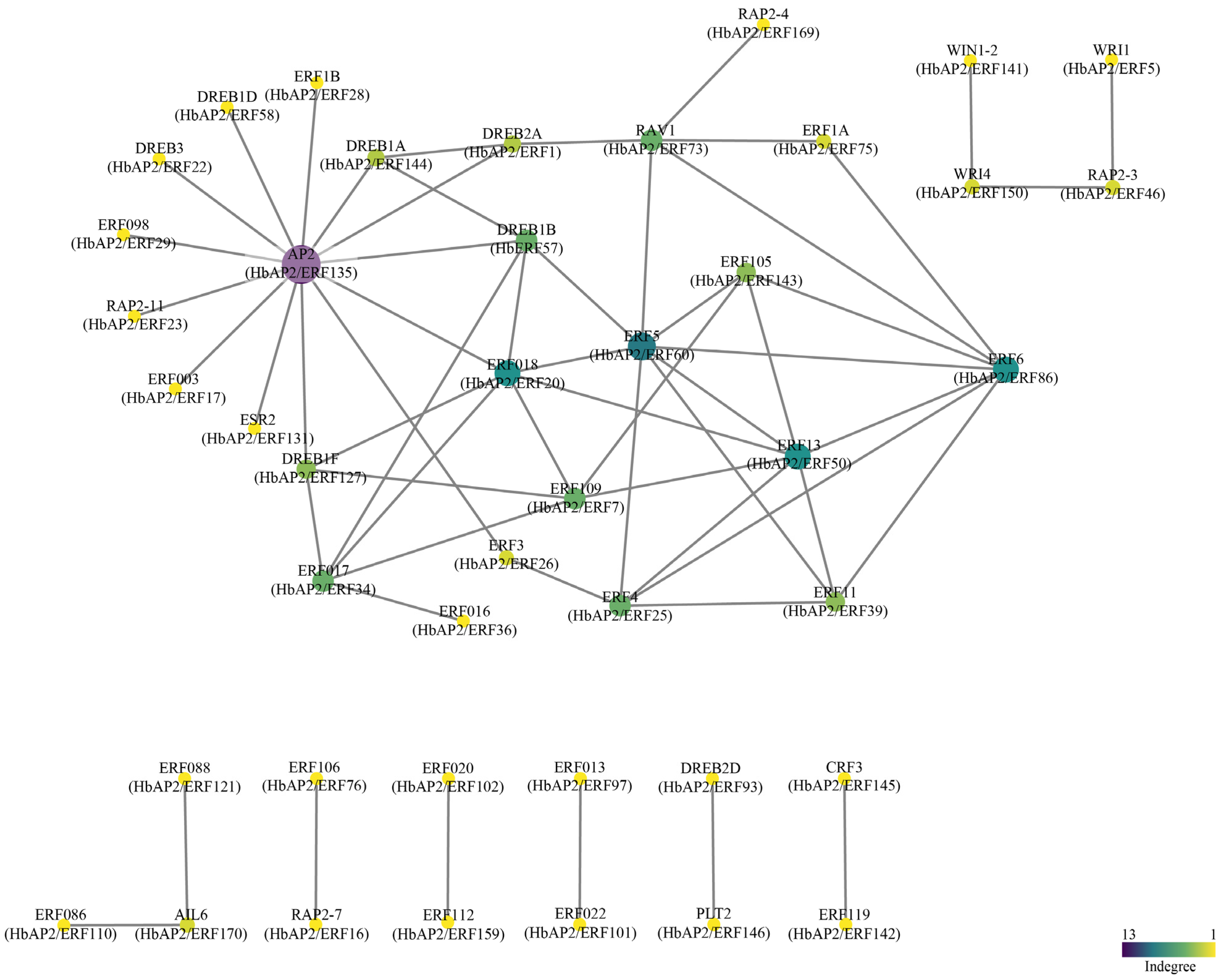
2.7. Protein–Protein Interactions of HbAP2/ERFs
2.8. Dual-Luciferase Reporter Assay
2.9. Statistical Analysis
3. Results
3.1. Identification and Characterization of HbAP2/ERFs in Rubber Tree
3.2. Phylogenetic Analysis of Rubber Tree HbAP2/ERFs
3.3. Duplication and Synteny Analysis of HbAP2/ERF Genes
3.4. Cis-Regulatory Element Analysis of HbAP2/ERFs
3.5. Tissue-Specific Expression Patterns of HbAP2/ERFs
3.6. Prediction of the HbAP2/ERF Protein–Protein Interaction Network
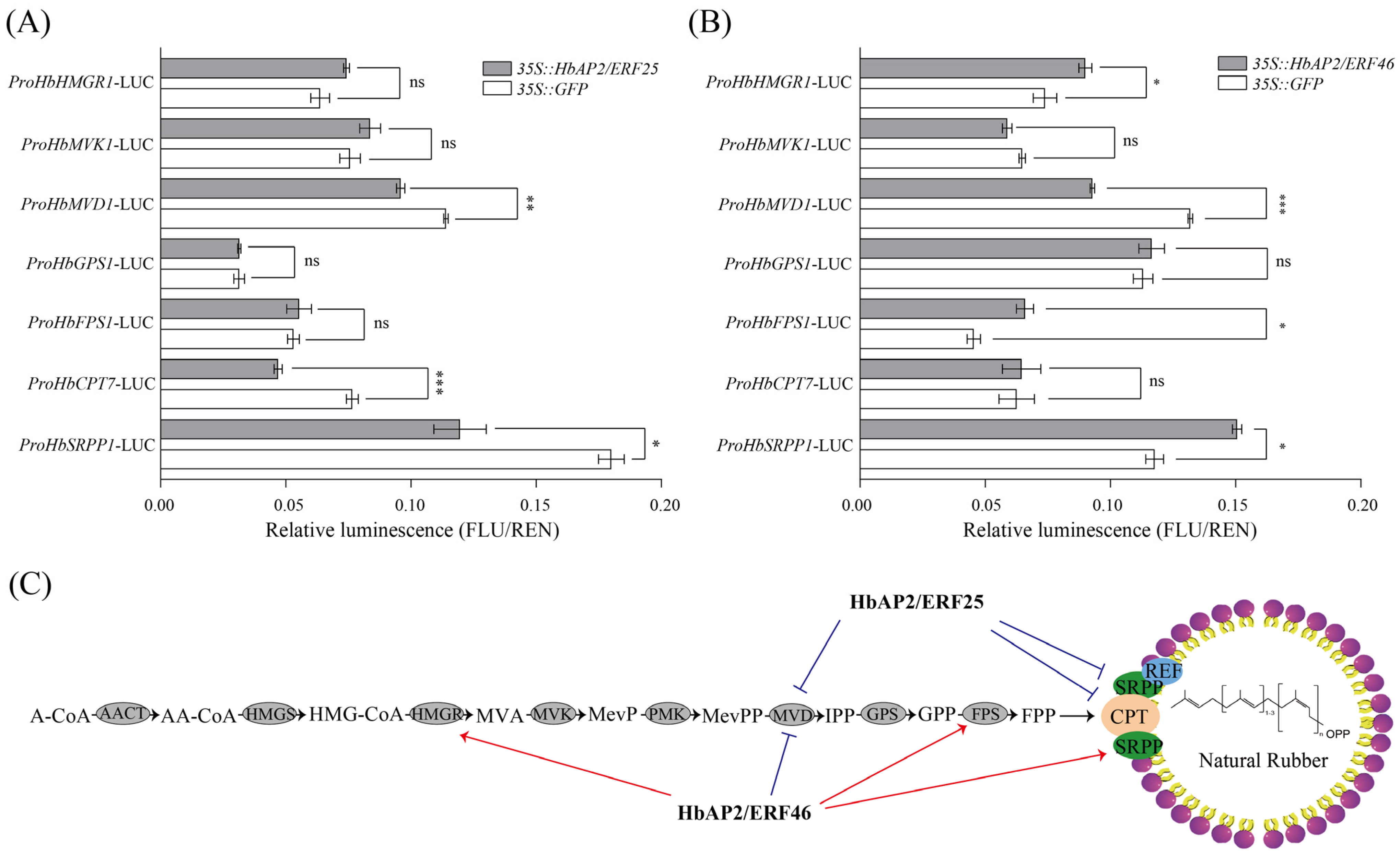
3.7. HbAP2/ERF25 and HbAP2/ERF46 Regulated Multiple NR Biosynthesis-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TF | Transcription factor |
| AP2/ERF | APETALA2/ethylene-responsive element-binding factor |
| DREB | Dehydration-responsive element binding |
| CBF | C-repeating binding factor |
| NR | Natural rubber |
| ABRE | ABA Responsive Element |
| CDS | Coding sequences |
| LTR | Low-temperature response |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| LUC | Luciferase |
| HMGR | 3-hydroxy-3-methylglutaryl coenzyme A reductase |
| MVK | Mevalonate kinase |
| MVD | Mevalonate diphosphate decarboxylase |
| GPS | Geranyl pyrophosphate synthase |
| FPS | Farnesyl pyrophosphate synthase |
| CPT | Cis-prenyltransferase |
| SRPP | Small rubber particle protein |
References
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF Super-Family Transcription Factors in Plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Shi, N.; Du, X.; Huang, T.; Zhang, Y.; Zhao, C.; Zhao, K.; Lin, Z.; Ma, D.; Li, Q.; et al. Bioinformatics Analysis and Expression Profiling Under Abiotic Stress of the DREB Gene Family in Glycyrrhiza uralensis. Int. J. Mol. Sci. 2025, 26, 9235. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Lan, Y.; Lin, M.H.; Zhou, H.K.; Ying, S.; Chen, M. Genome-Wide Identification and Transcriptional Analysis of AP2/ERF Gene Family in Pearl Millet (Pennisetum glaucum). Int. J. Mol. Sci. 2024, 25, 2470. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Zhang, H.R.; Wang, S.P.; Zhao, X.; Dong, S.J.; Chen, J.H.; Sun, Y.Q.; Sun, Q.W.; Liu, Q.G. Genome-Wide Identification and Comprehensive Analysis of the AP2/ERF Gene Family in Prunus sibirica under Low-Temperature Stress. BMC Plant Biol. 2024, 24, 883. [Google Scholar] [CrossRef]
- Li, W.; Geng, Z.W.; Zhang, C.P.; Wang, K.T.; Jiang, X.Q. Whole-Genome Characterization of Rosa chinensis AP2/ERF Transcription Factors and Analysis of Negative Regulator RcDREB2B in Arabidopsis. BMC Genom. 2021, 22, 90. [Google Scholar] [CrossRef]
- Zhuang, J.; Cai, B.; Peng, R.H.; Zhu, B.; Jin, X.F.; Xue, Y.; Gao, F.; Fu, X.Y.; Tian, Y.S.; Zhao, W.; et al. Genome-Wide Analysis of the AP2/ERF Gene Family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef]
- Duan, C.; Argout, X.; Gébelin, V.; Summo, M.; Dufayard, J.; Leclercq, J.; Kuswanhadi; Piyatrakul, P.; Pirrello, J.; Rio, M.; et al. Identification of the Hevea brasiliensis AP2/ERF Superfamily by RNA Sequencing. BMC Genom. 2013, 14, 30. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis Flower and Seed Development by the Homeotic Gene APETALA2 (AP2). Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef]
- Virág, E.; Tóth, B.B.; Kutasy, B.; Nagy, Á.; Pákozdi, K.; Pallos, J.P.; Kardos, G.; Hegedűs, G. Promoter Motif Profiling and Binding Site Distribution Analysis of Transcription Factors Predict Auto- and Cross-Regulatory Mechanisms in Arabidopsis Flowering Genes. Int. J. Mol. Sci. 2025, 26, 11152. [Google Scholar] [CrossRef]
- Virág, E.; Nagy, Á.; Tóth, B.B.; Kutasy, B.; Pallos, J.P.; Szigeti, Z.M.; Máthé, C.; Kardos, G.; Hegedűs, G. Master Regulatory Transcription Factors in β-Aminobutyric Acid-Induced Resistance (BABA-IR): A Perspective on Phytohormone Biosynthesis and Signaling in Arabidopsis Thaliana and Hordeum Vulgare. Int. J. Mol. Sci. 2024, 25, 9179. [Google Scholar] [CrossRef]
- Krizek, B.A. AINTEGUMENTA and AINTEGUMENTA-LIKE6 Regulate Auxin-Mediated Flower Development in Arabidopsis. BMC Res. Notes 2011, 4, 176. [Google Scholar] [CrossRef] [PubMed]
- Hirota, A.; Kato, T.; Fukaki, H.; Aida, M.; Tasaka, M. The Auxin-Regulated AP2/EREBP Gene PUCHI Is Required for Morphogenesis in the Early Lateral Root Primordium of Arabidopsis. Plant Cell 2007, 19, 2156–2168. [Google Scholar] [CrossRef] [PubMed]
- Kitomi, Y.; Ito, H.; Hobo, T.; Aya, K.; Kitano, H.; Inukai, Y. The Auxin Responsive AP2/ERF Transcription Factor CROWN ROOTLESS5 (CRL5) Is Involved in Crown Root Initiation in Rice through the Induction of OsRR1, a type-A Response Regulator of Cytokinin Signaling. Plant J. 2011, 67, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two Transcription Factors, DREB1 and DREB2, with an EREBP/AP2 DNA Binding Domain Separate Two Cellular Signal Transduction Pathways in Drought- and Low-Temperature-Responsive Gene Expression, Respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low Temperature Regulation of the Arabidopsis CBF Family of AP2 Transcriptional Activators as an Early Step in Cold-induced COR Gene Expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef]
- Yang, H.; Yu, C.; Yan, J.; Wang, X.H.; Chen, F.; Zhao, Y.; Wei, W. Overexpression of the Jatropha curcas JcERF1 Gene Coding an AP2/ERF-Type Transcription Factor Increases Tolerance to Salt in Transgenic Tobacco. Biochemistry 2014, 79, 1226–1236. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, Y.; Wu, J.; Cheng, Q.; Li, W.; Fan, S.; Jiang, L.; Xu, Z.; Kong, F.; Zhang, D.; et al. Overexpression of GmERF5, a New Member of the Soybean EAR Motif-Containing ERF Transcription Factor, Enhances Resistance to Phytophthora sojae in Soybean. J. Exp. Bot. 2015, 66, 2635–2647. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Chang, X.; Qi, D.Y.; Dong, L.D.; Wang, G.J.; Fan, S.J.; Jiang, L.Y.; Cheng, Q.; Chen, X.; Han, D.; et al. A Novel Soybean ERF Transcription Factor, GmERF113, Increases Resistance to Phytophthora sojae Infection in Soybean. Front. Plant Sci. 2017, 8, 299. [Google Scholar] [CrossRef]
- Li, D.; Liu, Y.; Chen, G.L.; Yan, Y.; Bai, Z.Q. The SmERF1b-like Regulates Tanshinone Biosynthesis in Salvia Miltiorrhiza Hairy Root. AoB PLANTS 2024, 16, plad086. [Google Scholar] [CrossRef]
- Li, D.C.; Liu, L.; Li, X.H.; Wei, G.; Cai, Y.P.; Sun, X.; Fan, H.H. DoAP2/ERF89 Activated the Terpene Synthase Gene DoPAES in Dendrobium Officinale and Participated in the Synthesis of β-Patchoulene. Peer J. 2024, 12, e16760. [Google Scholar] [CrossRef]
- Nie, Z.Y.; Kang, G.J.; Yan, D.; Qin, H.D.; Yang, L.F.; Zeng, R.Z. Downregulation of HbFPS1 Affects Rubber Biosynthesis of Hevea brasiliensis Suffering from Tapping Panel Dryness. Plant J. 2023, 113, 504–520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.X.; Wu, S.H.; Chao, J.Q.; Yang, S.G.; Bao, J.; Tian, W.M. Genome-Wide Identification and Expression Analysis of MYC Transcription Factor Family Genes in Rubber Tree (Hevea brasiliensis Muell. Arg.). Forests 2022, 13, 531. [Google Scholar] [CrossRef]
- Yan, Y.T.; Liang, C.L.; Liu, X.; Tan, Y.C.; Lu, Y.L.; Zhang, Y.Y.; Luo, H.L.; He, C.Z.; Cao, J.; Tang, C.R.; et al. Genome-Wide Association Study Identifies Candidate Genes Responsible for Inorganic Phosphorus and Sucrose Content in Rubber Tree Latex. Trop. Plants 2023, 2, 24. [Google Scholar] [CrossRef]
- Wu, S.H.; Zhang, S.X.; Chao, J.Q.; Deng, X.M.; Chen, Y.Y.; Shi, M.; Tian, W.M. Transcriptome Analysis of the Signalling Networks in Coronatine-Induced Secondary Laticifer Differentiation from Vascular Cambia in Rubber Trees. Sci. Rep. 2016, 6, 36384. [Google Scholar] [CrossRef]
- Chao, J.; Wu, S.; Shi, M.; Xu, X.; Gao, Q.; Du, H.; Gao, B.; Guo, D.; Yang, S.; Zhang, S.; et al. Genomic insight into domestication of rubber tree. Nat. Commun. 2023, 14, 4651. [Google Scholar] [CrossRef]
- Xie, J.M.; Chen, Y.R.; Cai, G.J.; Cai, R.L.; Hu, Z.; Wang, H. Tree Visualization by One Table (tvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Sherf, B.A.; Navarro, S.L.; Hannah, R.R.; Wood, K.V. Dual-LuciferaseTM Reporter Assay: An Advanced Co-Reporter Technology Integrating Firefly and Renilla Luciferase Assays. Promega Notes 1996, 57, 2–8. [Google Scholar]
- Zhu, J.H.; Qu, L.; Zeng, L.W.; Wang, Y.; Li, H.L.; Peng, S.Q.; Guo, D. Genome-Wide Identification of HbVQ Proteins and Their Interaction with HbWRKY14 to Regulate the Expression of HbSRPP in Hevea brasiliensis. BMC Genom. 2025, 26, 53. [Google Scholar] [CrossRef] [PubMed]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 Domain of APETALA2 Defines a Large New Family of DNA Binding Proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef] [PubMed]
- Drews, G.N.; Bowman, J.L.; Meyerowitz, E.M. Negative Regulation of the Arabidopsis Homeotic Gene AGAMOUS by the APETALA2 Product. Cell 1991, 65, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Bertran Garcia De Olalla, E.; Cerise, M.; Rodríguez-Maroto, G.; Casanova-Ferrer, P.; Vayssières, A.; Severing, E.; López Sampere, Y.; Wang, K.; Schäfer, S.; Formosa-Jordan, P.; et al. Coordination of Shoot Apical Meristem Shape and Identity by APETALA2 during Floral Transition in Arabidopsis. Nat. Commun. 2024, 15, 6930. [Google Scholar] [CrossRef]
- Fujimoto, S.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis Ethylene-Responsive Element Binding Factors Act as Transcriptional Activators or Repressors of GCC Box–Mediated Gene Expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [CrossRef]
- Zhuang, J.; Chen, J.M.; Yao, Q.H.; Xiong, F.; Sun, C.C.; Zhou, X.R.; Zhang, J.; Xiong, A. Discovery and Expression Profile Analysis of AP2/ERF Family Genes from Triticum aestivum. Mol. Biol. Rep. 2011, 38, 745–753. [Google Scholar] [CrossRef]
- Guo, B.J.; Wei, Y.F.; Xu, R.B.; Lin, S.; Luan, H.Y.; Lv, C.; Zhang, X.Z.; Song, X.Y.; Xu, R.G. Genome-Wide Analysis of APETALA2/Ethylene-Responsive Factor (AP2/ERF) Gene Family in Barley (Hordeum vulgare L.). PLoS ONE 2016, 11, e0161322. [Google Scholar] [CrossRef]
- Zhuang, J.; Deng, D.X.; Yao, Q.H.; Zhang, J.; Xiong, F.; Chen, J.M.; Xiong, A. Discovery, Phylogeny and Expression Patterns of AP2-like Genes in Maize. Plant Growth Regul. 2010, 62, 51–58. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Chen, X.; Xu, Z.; Guan, S.; Li, L.C.; Li, A.; Guo, J.; Mao, L.; Ma, Y. Phylogeny, Gene Structures, and Expression Patterns of the ERF Gene Family in Soybean (Glycine max L.). J. Exp. Bot. 2008, 59, 4095–4107. [Google Scholar] [CrossRef]
- Zhuang, J.; Yao, Q.H.; Xiong, A.; Zhang, J. Isolation, Phylogeny and Expression Patterns of AP2-Like Genes in Apple (Malus × domestica Borkh). Plant Mol. Biol. Rep. 2011, 29, 209–216. [Google Scholar] [CrossRef]
- Song, X.M.; Li, Y.; Hou, X.L. Genome-Wide Analysis of the AP2/ERF Transcription Factor Superfamily in Chinese Cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.L.; Huang, S.Y.; Chen, X.Z.; Zai, W.S.; Xiong, Z.L. Re-identification of AP2/ERF Transcription Factors in Search for Disease Resistance-related Genes in Tomato Plant. Fujian J. Agric. Sci. 2025, 40, 113–122. [Google Scholar] [CrossRef]
- Hammou, R.A.; Caid, M.B.E.; Harrouni, C.; Daoud, S. Germination Enhancement, Antioxidant Enzyme Activity, and Metabolite Changes in Late Argania Spinosa Kernels under Salinity. J. Arid Environ. 2023, 219, 105095. [Google Scholar] [CrossRef]
- Wang, Y.L.; Jiang, C.H.; Zhang, X.T.; Yan, H.M.; Yin, Z.G.; Sun, X.M.; Gao, F.H.; Zhao, Y.; Liu, W.; Han, S.C.; et al. Upland rice genomic signatures of adaptation to drought resistance and navigation to molecular design breeding. Plant Biotechnol. J. 2023, 22, 662–677. [Google Scholar] [CrossRef]
- Huang, J.Y.; Zhao, X.B.; Bürger, M.; Wang, Y.R.; Chory, J. Two Interacting Ethylene Response Factors Regulate Heat Stress Response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef]
- Li, H.L.; Wei, L.R.; Guo, D.; Wang, Y.; Zhu, J.H.; Chen, X.T.; Peng, S.Q. HbMADS4, a MADS-Box Transcription Factor from Hevea brasiliensis, Negatively Regulates HbSRPP. Front. Plant Sci. 2016, 7, 1709. [Google Scholar] [CrossRef]
- Qu, L.; Li, H.L.; Guo, D.; Wang, Y.; Zhu, J.H.; Yin, L.Y.; Peng, S.Q. HbWRKY27, a Group IIe WRKY Transcription Factor, Positively Regulates HbFPS1 Expression in Hevea brasiliensis. Sci. Rep. 2020, 10, 20639. [Google Scholar] [CrossRef]
- Guo, D.; Li, H.L.; Zhu, J.H.; Wang, Y.; Peng, S.Q. HbTGA1, a TGA Transcription Factor From Hevea brasiliensis, Regulates the Expression of Multiple Natural Rubber Biosynthesis Genes. Front. Plant Sci. 2022, 13, 909098. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.; Sun, Y.; Cao, W.; Wu, S.; Deng, X.; Yang, S.; Shi, M.; Yuan, H.; Chao, J. Genome-Wide Identification of AP2/ERF Family Genes in Rubber Tree: Two HbAP2/ERF Genes Regulate the Expression of Multiple Natural Rubber Biosynthesis Genes. Agronomy 2025, 15, 2881. https://doi.org/10.3390/agronomy15122881
Du X, Sun Y, Cao W, Wu S, Deng X, Yang S, Shi M, Yuan H, Chao J. Genome-Wide Identification of AP2/ERF Family Genes in Rubber Tree: Two HbAP2/ERF Genes Regulate the Expression of Multiple Natural Rubber Biosynthesis Genes. Agronomy. 2025; 15(12):2881. https://doi.org/10.3390/agronomy15122881
Chicago/Turabian StyleDu, Xiaoyu, Yi Sun, Wenqing Cao, Shaohua Wu, Xiaomin Deng, Shuguang Yang, Minjing Shi, Hongmei Yuan, and Jinquan Chao. 2025. "Genome-Wide Identification of AP2/ERF Family Genes in Rubber Tree: Two HbAP2/ERF Genes Regulate the Expression of Multiple Natural Rubber Biosynthesis Genes" Agronomy 15, no. 12: 2881. https://doi.org/10.3390/agronomy15122881
APA StyleDu, X., Sun, Y., Cao, W., Wu, S., Deng, X., Yang, S., Shi, M., Yuan, H., & Chao, J. (2025). Genome-Wide Identification of AP2/ERF Family Genes in Rubber Tree: Two HbAP2/ERF Genes Regulate the Expression of Multiple Natural Rubber Biosynthesis Genes. Agronomy, 15(12), 2881. https://doi.org/10.3390/agronomy15122881





