Novel Resistance Determinants from Cucumber PI 197085 Against Pseudoperonospora cubensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Field Evaluation of PI 197085 Resistance
2.3. Phenotypic Evaluation of the F2 Mapping Population Under Controlled Inoculation
2.4. DNA Marker Analysis and Linkage Map Construction
2.5. QTL Analysis and Identification of Candidate Genes
3. Results
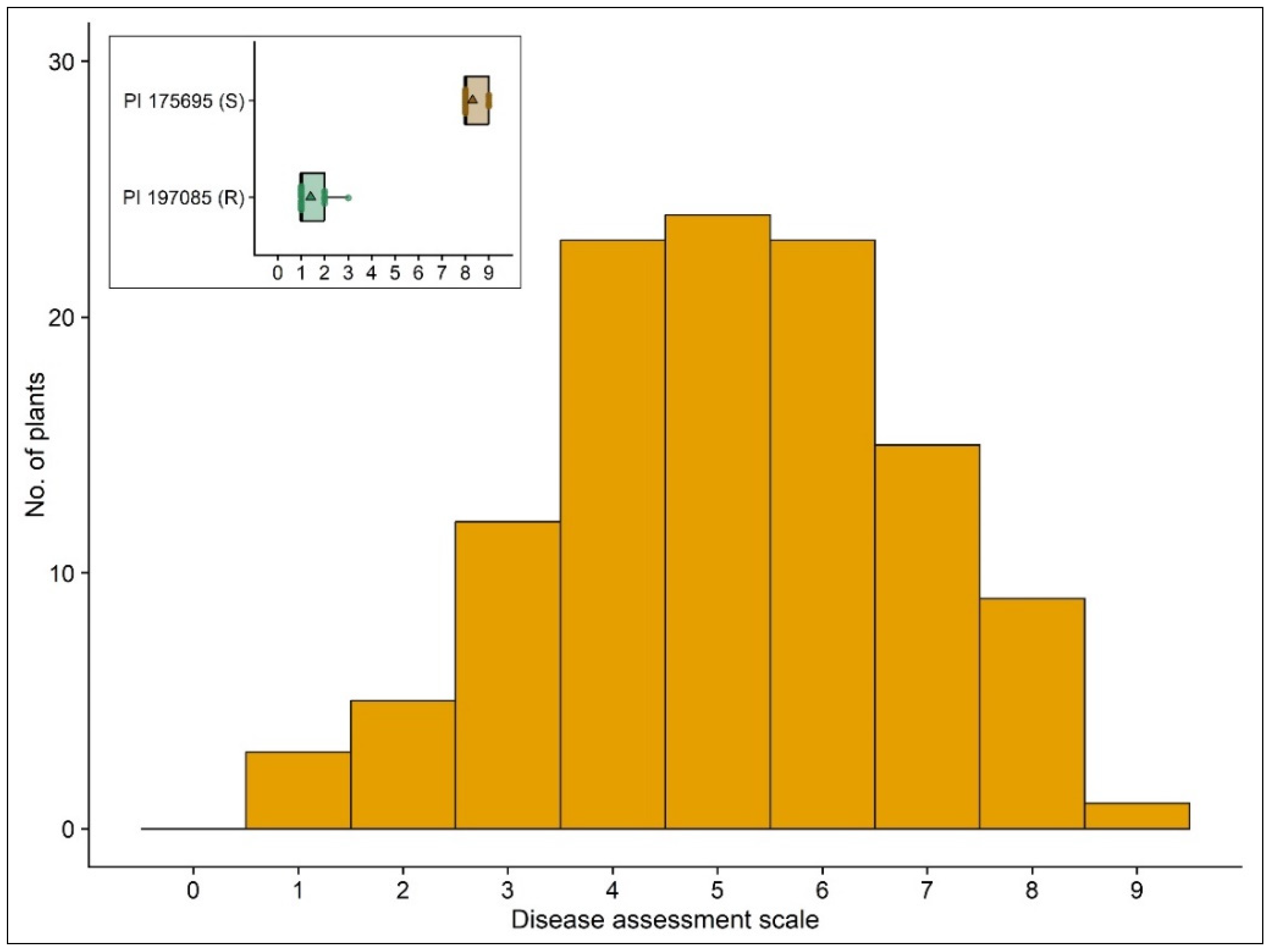
3.1. Field Performance of PI 197085 Resistance
3.2. Phenotypic Reaction of the Cucumber Plants to P. cubensis Inoculation
3.3. Construction of the Genetic Linkage Map
3.4. Identification of QTLs Underlying Resistance to P. cubensis
3.5. Prediction of Candidate Resistance Genes in Genomic Regions Harboring QTLs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AE | Additive Effect |
| BSA-seq | Bulked Segregant Analysis coupled with Sequencing |
| CAPS | Cleaved Amplified Polymorphic Sequence |
| cM | centiMorgan |
| CIM | Composite Interval Mapping |
| CuGenDBv2 | Cucurbit Genomics Database, version 2 |
| DE | Dominance Effect |
| DM | Downy Mildew |
| DSI | Disease Severity Index |
| GBS | Genotyping-by-Sequencing |
| GWAS | Genome-Wide Association Study (GWAS) |
| InDel | Insertion–Deletion polymorphism |
| ISSR | Inter-Simple Sequence Repeat |
| LG | Linkage Group |
| LOD | Logarithm of odds |
| Mb | Megabase(s) |
| NIL | Near-Isogenic Line(s) |
| PCR | Polymerase Chain Reaction |
| RAPD | Random Amplified Polymorphic DNA |
| RIL | Recombinant Inbred Line(s) |
| SNP | Single-Nucleotide Polymorphism |
| SCAR | Sequence-Characterized Amplified Region |
| SSR | Simple Sequence Repeat (also known as microsatellite marker) |
| TF | Transcription Factor(s) |
| QTL | Quantitative Trait Locus |
References
- Thomas, C.E. Downy mildew. In Compendium of Cucurbit Diseases; Zitter, T.A., Hopkins, D.L., Thomas, C.E., Eds.; American Phytopathological Society Press: St. Paul, MN, USA, 1996; pp. 25–27. [Google Scholar]
- Palti, J.; Cohen, Y. Downy mildew of Cucurbits (Pseudoperonospora Cubensis): The fungus and its hosts, distribution, epidemiology and control. Phytoparasitica 1980, 8, 109–147. [Google Scholar] [CrossRef]
- Savory, E.A.; Granke, L.L.; Quesada-Ocampo, L.M.; Varbanova, M.; Hausbeck, M.K.; Day, B. The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol. Plant Pathol. 2011, 12, 217–226. [Google Scholar] [CrossRef]
- Lebeda, A.; Cohen, Y. Cucurbit downy mildew (Pseudoperonospora cubensis)—Biology, ecology, epidemiology, host-pathogen interaction and control. Eur. J. Plant Pathol. 2011, 129, 157–192. [Google Scholar] [CrossRef]
- Cohen, Y.; Van den Langenberg, K.M.; Wehner, T.C.; Ojiambo, P.S.; Hausbeck, M.; Quesada-Ocampo, L.M.; Lebeda, A.; Sierotzki, H.; Gisi, U. Resurgence of Pseudoperonospora cubensis: The causal agent of cucurbit downy mildew. Phytopathology 2015, 105, 998–1012. [Google Scholar] [CrossRef]
- Wallace, E.C.; D’Arcangelo, K.N.; Quesada-Ocampo, L.M. Population analyses reveal two host-adapted clades of Pseudoperonospora cubensis, the causal agent of cucurbit downy mildew, on commercial and wild cucurbits. Phytopathology 2020, 110, 1578–1587. [Google Scholar] [CrossRef] [PubMed]
- Mirzwa-Mróz, E.; Zieniuk, B.; Yin, Z.; Pawełkowicz, M. Genetic insights and molecular breeding approaches for downy mildew resistance in cucumber (Cucumis sativus L.): Current progress and future prospects. Int. J. Mol. Sci. 2024, 25, 12726. [Google Scholar] [CrossRef]
- Thakur, R.P.; Mathur, K. Downy mildews of India. Crop Prot. 2002, 21, 333–345. [Google Scholar] [CrossRef]
- Abdelfatah, A.; Mazrou, Y.S.A.; Arafa, R.A.; Makhlouf, A.H.; El-Nagar, A. Control of cucumber downy mildew disease under greenhouse conditions using biocide and organic compounds via induction of the antioxidant defense machinery. Sci. Rep. 2025, 15, 11705. [Google Scholar] [CrossRef]
- Cohen, Y.; Rubin, A.E. Mating type and sexual reproduction of Pseudoperonospora cubensis, the downy mildew agent of cucurbits. Eur. J. Plant Pathol. 2012, 132, 577–592. [Google Scholar] [CrossRef]
- Cohen, Y.; Rubin, A.E.; Galperin, M. Host preference of mating type in Pseudoperonospora cubensis, the downy mildew causal agent of cucurbits. Plant Dis. 2013, 97, 292. [Google Scholar] [CrossRef]
- Rani, R.; Negi, P.; Sharma, S.; Jain, S. Occurrence of oosporic stage of Pseudoperonospora cubensis on cucumber, in Punjab, India: A first report. Crop Prot. 2022, 155, 105939. [Google Scholar] [CrossRef]
- Kikway, I.; Keinath, A.P.; Ojiambo, P.S. Field occurrence and overwintering of oospores of Pseudoperonospora cubensis in the Southeastern United States. Phytopathology 2022, 112, 1946–1955. [Google Scholar] [CrossRef]
- Runge, F.; Choi, Y.-J.; Thines, M. Phylogenetic investigations in the genus Pseudoperonospora reveal overlooked species and cryptic diversity in the P. cubensis species cluster. Eur. J. Plant Pathol. 2011, 129, 135–146. [Google Scholar] [CrossRef]
- Lebeda, A.; Křístková, E.; Sedláková, B. Pathotypes and races of Pseudoperonospora cubensis: Two concepts of virulence differentiation. Plant Pathol. 2024, 73, 2537–2547. [Google Scholar] [CrossRef]
- Thomas, A.; Carbone, I.; Choe, K.; Quesada-Ocampo, L.M.; Ojiambo, P.S. Resurgence of cucurbit downy mildew in the United States: Insights from comparative genomic analysis of Pseudoperonospora cubensis. Ecol. Evol. 2017, 7, 6231–6246. [Google Scholar] [CrossRef] [PubMed]
- Kozik, E.U.; Klosińska, U.; Call, A.D.; Wehner, T.C. Heritability and genetic variance estimates for resistance to downy mildew in cucumber accession Ames 2354. Crop Sci. 2013, 53, 177–182. [Google Scholar] [CrossRef]
- Sun, Z.; Yu, S.; Hu, Y.; Wen, Y. Biological control of the cucumber downy mildew pathogen Pseudoperonospora cubensis. Horticulturae 2022, 8, 410. [Google Scholar] [CrossRef]
- Call, A.D.; Criswell, A.D.; Wehner, T.C.; Klosinska, U.; Kozik, E.U. Screening cucumber for resistance to downy mildew caused by Pseudoperonospora cubensis (Berk. and Curt.) Rostov. Crop Sci. 2012, 52, 577–592. [Google Scholar] [CrossRef]
- Ojiambo, P.S.; Gent, D.H.; Quesada-Ocampo, L.M.; Hausbeck, M.K.; Holmes, G.J. Epidemiology and population biology of Pseudoperonospora cubensis: A model system for management of downy mildews. Annu. Rev. Phytopathol. 2015, 53, 223–246. [Google Scholar] [CrossRef]
- Bai, Z.; Yuan, X.; Cai, R.; Liu, L.; He, H.; Zhou, H.; Pan, J. QTL analysis of downy mildew resistance in cucumber. Prog. Nat. Sci. 2008, 18, 706–710. [Google Scholar]
- Pang, X.; Zhou, X.; Wan, H.; Chen, J. QTL mapping of downy mildew resistance in an introgression line derived from interspecific hybridization between cucumber and Cucumis hystrix. J. Phytopathol. 2013, 161, 536–543. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Sakata, Y.; Sugiyama, M.; Fukino, N. Identification of quantitative trait loci for downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 2014, 198, 265–276. [Google Scholar] [CrossRef]
- Olfati, J.-A.; Samizadeh, H.; Peyvast, G.-A.; Khodaparast, S.A.; Rabiei, B. Dominant variance has an important role in downy mildew resistance in cucumber. Hortic. Environ. Biotechnol. 2011, 52, 422–426. [Google Scholar] [CrossRef]
- Van Vliet, G.J.A.; Meijsing, W.D. Relation in the inheritance of resistance to Pseudoperonospora cubensis Rost. and Sphaerotheca fuliginea Poll. in cucumber (Cucumis sativus L.). Euphytica 1977, 26, 793–796. [Google Scholar] [CrossRef]
- van Vliet, G.J.A.; Meysing, W.D. Inheritance of resistance to Pseudoperonospora cubensis Rost. in cucumber (Cucumis sativus L.). Euphytica 1974, 23, 251–255. [Google Scholar] [CrossRef]
- Zhang, S.P.; Liu, M.M.; Miao, H.; Zhang, S.Q.; Yang, Y.H.; Xie, B.Y.; Wehner, T.C.; Gu, X.F. Chromosomal mapping and QTL analysis of resistance to downy mildew in Cucumis sativus. Plant Dis. 2013, 97, 245–251. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Zhu, W.; Qin, X.; Xu, J.; Cheng, C.; Lou, Q.; Li, J.; Chen, J. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor. Appl. Genet. 2018, 131, 2229–2243. [Google Scholar] [CrossRef]
- Wang, Y.; Tan, J.; Wu, Z.; VandenLangenberg, K.; Wehner, T.C.; Wen, C.; Zheng, X.; Owens, K.; Thornton, A.; Bang, H.H.; et al. STAYGREEN, STAY HEALTHY: A loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol. 2019, 221, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; VandenLangenberg, K.; Wehner, T.C.; Kraan, P.A.G.; Suelmann, J.; Zheng, X.; Owens, K.; Weng, Y. QTL mapping for downy mildew resistance in cucumber inbred line WI7120 (PI 330628). Theor. Appl. Genet. 2016, 129, 1493–1505. [Google Scholar] [CrossRef]
- Wang, Y.; VandenLangenberg, K.; Wen, C.; Wehner, T.C.; Weng, Y. QTL mapping of downy and powdery mildew resistances in PI 197088 cucumber with genotyping-by-sequencing in RIL population. Theor. Appl. Genet. 2018, 131, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Szczechura, W.; Staniaszek, M.; Klosinska, U.; Kozik, E.U. Molecular analysis of new sources of resistance to Pseudoperonospora cubensis (Berk. et Curt.) Rostovzev in cucumber. Russ. J. Genet. 2015, 51, 974–979. [Google Scholar] [CrossRef]
- Berg, J.A.; Hermans, F.W.K.; Beenders, F.; Abedinpour, H.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. The amino acid permease (AAP) genes CsAAP2A and SlAAP5A/B are required for oomycete susceptibility in cucumber and tomato. Mol. Plant Pathol. 2021, 22, 658–672. [Google Scholar] [CrossRef]
- Berg, J.A.; Hermans, F.W.K.; Beenders, F.; Lou, L.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. Analysis of QTL DM4.1 for downy mildew resistance in cucumber reveals multiple subQTL: A novel RLK as candidate gene for the most important subQTL. Front. Plant Sci. 2020, 11, 569876. [Google Scholar] [CrossRef] [PubMed]
- Innark, P.; Panyanitikoon, H.; Khanobdee, C.; Samipak, S.; Jantasuriyarat, C. QTL identification for downy mildew resistance in cucumber using genetic linkage map based on SSR markers. J. Genet. 2020, 99, 81. [Google Scholar] [CrossRef]
- Li, L.; He, H.; Zou, Z.; Li, Y. QTL analysis for downy mildew resistance in cucumber inbred line PI 197088. Plant Dis. 2018, 102, 1240–1245. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, H.; Liu, P.; Miao, H.; Bai, Y.; Gu, X.; Zhang, S. Identification of novel loci and candidate genes for cucumber downy mildew resistance using GWAS. Plants 2020, 9, 1659. [Google Scholar] [CrossRef] [PubMed]
- Win, K.T.; Vegas, J.; Zhang, C.; Song, K.; Lee, S. QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor. Appl. Genet. 2017, 130, 199–211. [Google Scholar] [CrossRef]
- Barnes, W.; Epps, W. An unreported type to resistance to cucumber downy mildew. Plant Dis. Report. 1954, 38, 620. [Google Scholar]
- Jenkins, J.M., Jr. Downy mildew resistance in cucumbers. J. Hered. 1942, 33, 35–38. [Google Scholar] [CrossRef]
- Kłosińska, U. Genetyczne i Anatomiczne Podstawy Odporności Ogórka na Mączniaka Rzekomego (Pseudoperonospora cubensis Berk. et Curt.). Ph.D. Thesis, National Institute of Horticultural Research, Skierniewice, Poland, 2013. [Google Scholar]
- Call, A.D.; Criswell, A.D.; Wehner, T.C.; Ando, K.; Grumet, R. Resistance of cucumber cultivars to a new strain of cucurbit downy mildew. HortScience 2012, 47, 171–178. [Google Scholar] [CrossRef]
- VandenLangenberg, K.M. Studies on Downy Mildew Resistance in Cucumber (Cucumis sativus L.). Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 2015. [Google Scholar]
- Caldwell, D.; Chan, E.; de Vries, J.; Joobeur, T.; King, J.; Reina, A.; Shetty, N. Methods and Compositions for Identifying Downy Mildew Resistant Cucumber Plants. U.S. Patent 8,809,622, 28 April 2011. [Google Scholar]
- Hammer, R.S.; Cohen, Y. Non-sikkim cucumber accessions resistant to downy mildew (Pseudoperonospora cubensis). Seeds 2025, 4, 8. [Google Scholar] [CrossRef]
- Chen, T.; Katz, D.; Ben Naim, Y.; Hammer, R.; Ben Daniel, B.H.; Rubin, A.E.; Cohen, Y. Isolate-dependent inheritance of resistance against Pseudoperonospora cubensis in cucumber. Agronomy 2020, 10, 1086. [Google Scholar] [CrossRef]
- Wehner, T.C.; Shetty, N.V. Downy mildew resistance of the cucumber germplasm collection in North Carolina field tests. Crop Sci. 1997, 37, 1331–1340. [Google Scholar] [CrossRef]
- Holmes, G.J.; Ojiambo, P.S.; Hausbeck, M.K.; Quesada-Ocampo, L.; Keinath, A.P. Resurgence of cucurbit downy mildew in the United States: A watershed event for research and extension. Plant Dis. 2015, 99, 428–441. [Google Scholar] [CrossRef]
- Ding, G.; Qin, Z.; Zhou, X.; Fan, J. RAPD and SCAR markers linked to downy mildew resistance genes in cucumber. Acta Bot. Boreali-Occident. Sin. 2007, 27, 1747–1751. [Google Scholar]
- Sharma, B.A.; Rana, R.S.; Lata, H.; Thakur, A.; Sharma, A. Mapping quantitative trait loci (QTLs) for resistance to downy mildew and powdery mildew in cucumber (Cucumis sativus L.). J. Plant Biochem. Biotechnol. 2025, 1–15. [Google Scholar] [CrossRef]
- Zhuo, D.; Zicheng, Z.; Yane, S.; Yahang, L.; Xiaobing, M.; Haonan, C. Molecular genetic basis of resistance to downy mildew in cucumber and melon. J. Plant Pathol. 2024, 106, 499–506. [Google Scholar] [CrossRef]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef]
- Jenkins, S.F., Jr.; Wehner, T.C. A system for the measurement of foliar diseases of cucumber. Cucurbit Genet. Coop. Rep. 1983, 6, 10–12. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Wickham, H. Data analysis. In ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 189–201. [Google Scholar]
- Clarke, E.; Sherrill-Mix, S.; Dawson, C. Ggbeeswarm: Categorical Scatter (Violin Point) Plots; R Package Version 0.7.2. 2023. Available online: https://CRAN.R-project.org/package=ggbeeswarm (accessed on 13 August 2025).
- Wilke, C. Cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’; R Package Version 1.1.3. 2025. Available online: https://github.com/wilkelab/cowplot (accessed on 13 August 2025).
- Chiba, N.; Suwabe, K.; Nunome, T.; Hirai, M. Development of microsatellite markers in melon (Cucumis melo L.) and their application to major cucurbit crops. Breed. Sci. 2003, 53, 21–27. [Google Scholar] [CrossRef]
- Fukino, N.; Yoshioka, Y.; Kubo, N.; Hirai, M.; Sugiyama, M.; Sakata, Y.; Matsumoto, S. Development of 101 novel SSR markers and construction of an SSR-based genetic linkage map in cucumber (Cucumis sativus L.). Breed. Sci. 2008, 58, 475–483. [Google Scholar] [CrossRef]
- Danin-Poleg, Y.; Reis, N.; Tzuri, G.; Katzir, N. Development and characterization of microsatellite markers in Cucumis. Theor. Appl. Genet. 2001, 102, 61–72. [Google Scholar] [CrossRef]
- Horejsi, T.; Staub, J.E.; Thomas, C. Linkage of random amplified polymorphic DNA markers to downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 2000, 115, 105–113. [Google Scholar] [CrossRef]
- Tan, J.; Wang, Y.; Dymerski, R.; Wu, Z.; Weng, Y. Sigma factor binding protein 1 (CsSIB1) is a putative candidate of the major-effect QTL dm5.3 for downy mildew resistance in cucumber (Cucumis sativus). Theor. Appl. Genet. 2022, 135, 4197–4215. [Google Scholar] [CrossRef] [PubMed]
- Fazio, G.; Staub, J.E.; Chung, S.M. Development and characterization of PCR markers in cucumber. J. Am. Soc. Hortic. Sci. 2002, 127, 545–557. [Google Scholar] [CrossRef]
- Dar, A.A.; Mahajan, R.; Lay, P.; Sharma, S. Genetic diversity and population structure of Cucumis sativus L. by using SSR markers. 3 Biotech 2017, 7, 307. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.A.; Appiano, M.; Santillán Martínez, M.; Hermans, F.W.K.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 2015, 15, 243. [Google Scholar] [CrossRef]
- Staub, J.E.; Chung, S.-M.; Fazio, G. Conformity and genetic relatedness estimation in crop species having a narrow genetic base: The case of cucumber (Cucumis sativus L.). Plant Breed. 2005, 124, 44–53. [Google Scholar] [CrossRef]
- Wang, Y. Genetic Architecture of Downy Mildew (Pseudoperonospora cubensis) Resistance in Cucumber (Cucumis sativus L.). Ph.D. Thesis, University of Wisconsin, Madison, WI, USA, 2017. [Google Scholar]
- Ren, Y.; Zhang, Z.; Liu, J.; Staub, J.E.; Han, Y.; Cheng, Z.; Li, X.; Lu, J.; Miao, H.; Kang, H.; et al. An integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE 2009, 4, e5795. [Google Scholar] [CrossRef]
- Kõressaar, T.; Lepamets, M.; Kaplinski, L.; Raime, K.; Andreson, R.; Remm, M. Primer3_masker: Integrating masking of template sequence with primer design software. Bioinformatics 2018, 34, 1937–1938. [Google Scholar] [CrossRef]
- Van Ooijen, J. JoinMap®, 4.0. Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V.: Wageningen, The Netherlands, 2006.
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Li, Q.; Li, H.; Huang, W.; Xu, Y.; Zhou, Q.; Wang, S.; Ruan, J.; Huang, S.; Zhang, Z. A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). GigaScience 2019, 8, giz072. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Basten, C.; Zeng, Z. Windows QTL Cartographer, 2.5; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 2012.
- Wang, Y.; Bo, K.; Gu, X.; Pan, J.; Li, Y.; Chen, J.; Wen, C.; Ren, Z.; Ren, H.; Chen, X.; et al. Molecularly tagged genes and quantitative trait loci in cucumber with recommendations for QTL nomenclature. Hortic. Res. 2020, 7, 3. [Google Scholar] [CrossRef]
- Yu, J.; Wu, S.; Sun, H.; Wang, X.; Tang, X.; Guo, S.; Zhang, Z.; Huang, S.; Xu, Y.; Weng, Y.; et al. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2022, 51, D1457–D1464. [Google Scholar] [CrossRef]
- Polat, İ.; Baysal, Ö.; Mercati, F.; Kitner, M.; Cohen, Y.; Lebeda, A.; Carimi, F. Characterization of Pseudoperonospora cubensis isolates from Europe and Asia using ISSR and SRAP molecular markers. Eur. J. Plant Pathol. 2014, 139, 641–653. [Google Scholar] [CrossRef]
- Nowicki, M.; Hadziabdic, D.; Trigiano, R.N.; Boggess, S.L.; Kanetis, L.; Wadl, P.A.; Ojiambo, P.S.; Cubeta, M.A.; Spring, O.; Thines, M.; et al. “Jumping Jack”: Genomic microsatellites underscore the distinctiveness of closely related Pseudoperonospora cubensis and Pseudoperonospora humuli and provide new insights into their evolutionary past. Front. Microbiol. 2021, 12, 686759. [Google Scholar] [CrossRef]
- Staub, J.E.; Meglic, V. Molecular genetic markers and their legal relevance for cultivar discrimination: A case study in cucumber. HortTechnology 1993, 3, 291–300. [Google Scholar] [CrossRef]
- Miao, H.; Zhang, S.; Wang, X.; Zhang, Z.; Li, M.; Mu, S.; Cheng, Z.; Zhang, R.; Huang, S.; Xie, B.; et al. A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 2011, 182, 167–176. [Google Scholar] [CrossRef]
- Greeff, C.; Roux, M.; Mundy, J.; Petersen, M. Receptor-like kinase complexes in plant innate immunity. Front. Plant Sci. 2012, 3, 202. [Google Scholar] [CrossRef]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.V.; Jugulam, M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Narváez-Barragán, D.A.; Tovar-Herrera, O.E.; Guevara-García, A.; Serrano, M.; Martinez-Anaya, C. Mechanisms of plant cell wall surveillance in response to pathogens, cell wall-derived ligands and the effect of expansins to infection resistance or susceptibility. Front. Plant Sci. 2022, 13, 969343. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Biswas, A.; Dey, S.; Bhattacharjee, T.; Chakrabarty, S. Cytochrome P450 gene families: Role in plant secondary metabolites production and plant defense. J. Xenobiotics 2023, 13, 402–423. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Wang, Z.; Xu, F.; Yang, Y.; Fang, J.; Wu, J.; Pan, J.; Wang, Q.; Xu, L. Mapping QTL and identifying candidate genes for resistance to brown stripe in highly allo-autopolyploid modern sugarcane. Horticulturae 2025, 11, 922. [Google Scholar] [CrossRef]
- Zhao, Y.; Su, C. Mapping quantitative trait loci for yield-related traits and predicting candidate genes for grain weight in maize. Sci. Rep. 2019, 9, 16112. [Google Scholar] [CrossRef]
- Yun, P.; Zhang, C.; Ma, T.; Xia, J.; Zhou, K.; Wang, Y.; Li, Z. Identification of qGL4.1 and qGL4.2, two closely linked QTL controlling grain length in rice. Mol. Breed. 2024, 44, 11. [Google Scholar] [CrossRef] [PubMed]
 , whereas additional QTLs from other linkage mapping studies are also indicated as blocks accompanied by their respective citations in the legend [21,22,23,27,28,30,31,32,35,36,38,43,44,49]. Vertical lines mark the association signals reported by Liu et al. [37] using GWAS. Physical positions are given in Mb.
, whereas additional QTLs from other linkage mapping studies are also indicated as blocks accompanied by their respective citations in the legend [21,22,23,27,28,30,31,32,35,36,38,43,44,49]. Vertical lines mark the association signals reported by Liu et al. [37] using GWAS. Physical positions are given in Mb.
 , whereas additional QTLs from other linkage mapping studies are also indicated as blocks accompanied by their respective citations in the legend [21,22,23,27,28,30,31,32,35,36,38,43,44,49]. Vertical lines mark the association signals reported by Liu et al. [37] using GWAS. Physical positions are given in Mb.
, whereas additional QTLs from other linkage mapping studies are also indicated as blocks accompanied by their respective citations in the legend [21,22,23,27,28,30,31,32,35,36,38,43,44,49]. Vertical lines mark the association signals reported by Liu et al. [37] using GWAS. Physical positions are given in Mb.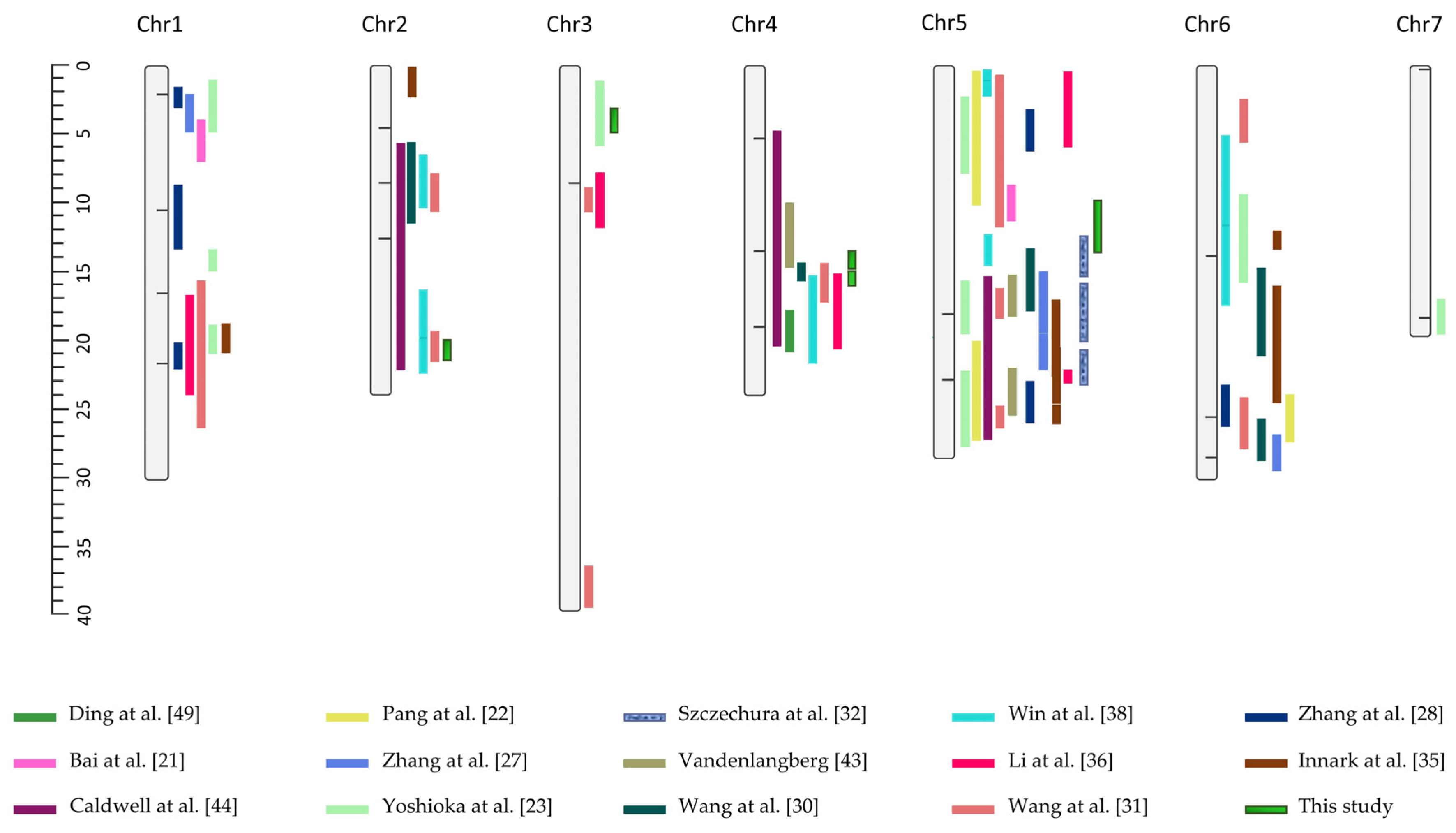



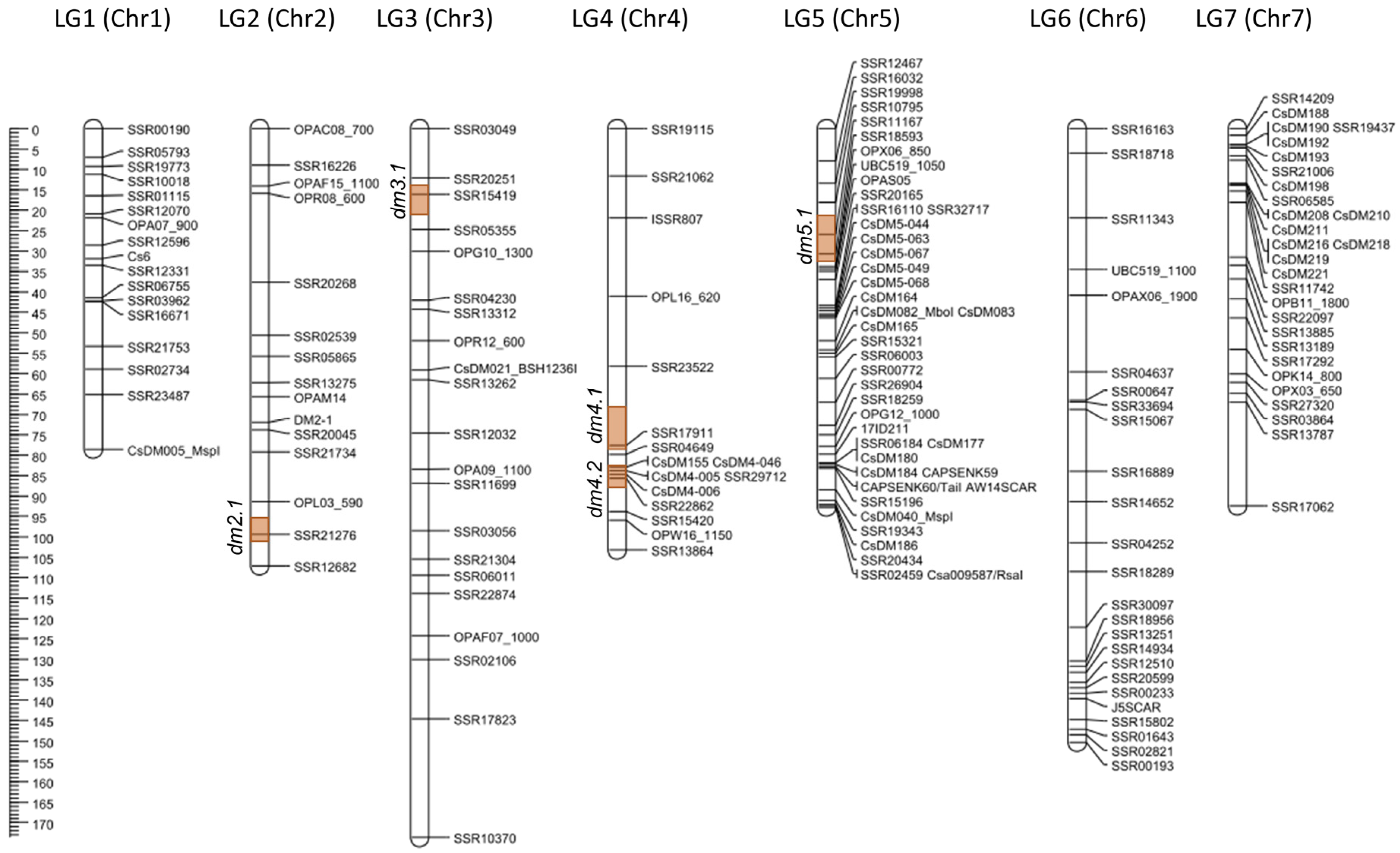
| Linkage Group | Corresponding Chromosome | Marker Loci | Map Length (cM) | Mean Inter-Marker Distance (cM) * |
|---|---|---|---|---|
| LG1 | 1 | 17 | 78.62 | 4.91 |
| LG2 | 2 | 15 | 107.12 | 7.65 |
| LG3 | 3 | 21 | 173.64 | 8.68 |
| LG4 | 4 | 16 | 103.24 | 6.88 |
| LG5 | 5 | 42 | 92.82 | 2.26 |
| LG6 | 6 | 25 | 150.28 | 6.26 |
| LG7 | 7 | 28 | 92.43 | 3.42 |
| Total | - | 164 | 798.14 | - |
| Average | - | 23.4 | 114.02 | 5.43 |
| QTL | Chr | Peak (cM) | LOD Value | Significant Loci | 1-LOD Support Interval | AE | DE | R2 (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Left Marker | Left Position | Right Marker | Right Position | ||||||||
| dm2.1 | 2 | 99.01 | 7.78 | SSR21276 | - | 95.6 | - | 101.2 | −0.844 | 0.127 | 14.7 |
| dm3.1 | 3 | 17.01 | 9.78 | SSR15419 | SSR20251 | 13.9 | SSR05355 | 21.2 | −1.081 | −0.224 | 14.0 |
| dm4.1 | 4 | 77.01 | 6.60 | SSR17911 | - | 68.2 | SSR04649 | 79.0 | −0.823 | −0.089 | 9.8 |
| dm4.2 | 4 | 84.01 | 7.14 | SSR29712, CsDM4-006 | CsDM155 | 80.4 | SSR15420 | 88.5 | −0.877 | −0.091 | 10.4 |
| dm5.1 | 5 | 29.01 | 5.01 | SSR18593 | SSR11167 | 21.4 | OPX06_850 | 32.6 | −0.716 | −0.229 | 5.5 |
| QTL | Chr | Interval (Mb) | Total Genes | Annotated Genes | Representative Functional Categories |
|---|---|---|---|---|---|
| dm2.1 | 2 | 22.06–22.85 | 127 | 98 | Transcription factors (WRKY, NAC), receptor-like kinases (incl. LYK2), NLR-like proteins, membrane transporters (ABC, YSL), vesicle trafficking (SNARE), redox/stress enzymes, protein regulation (RING E3, BTB/POZ) |
| dm3.1 | 3 | 3.22–5.04 | 199 | 161 | Transcription factors (WRKY, ERF, MYB, bHLH), vacuolar ion transport), proteasome regulator, subtilisin-like proteases, cell-wall modification (pectin lyases, COBRA-like), ubiquitin-related proteins. |
| dm4.1 | 4 | 17.71–17.83 | 12 | 10 | Ethylene-responsive transcription factor (ERF014), receptor-like kinases (two RLKs), redox/thiol enzymes, and ubiquitin pathway components |
| dm4.2 | 4 | 18.20–18.55 | 27 | 21 | Receptor-like kinases (PRK4, LRK10L2, LR10-like), transcriptional regulators (VOZ1-like, MADS-box 23-like), glycosyltransferases, cytochrome P450s, sterol/brassinosteroid-related enzymes (incl. cytochrome P450), redox/electron-transfer components (NADH–cytochrome b5 reductase) |
| dm5.1 | 5 | 5.74–11.97 | 359 | 271 | Resistance proteins (N-like), receptor-like kinases (PSKR1, ERL1-like, cysteine-rich RLK), diverse transcription factors, vesicle trafficking, cell wall modification, redox enzymes, epigenetic regulators, RNA silencing, transporters |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szczechura, W.; Kłosińska, U.; Nowakowska, M.; Nowak, K.; Nowicki, M.; Kozik, E.U.; Tyrka, M. Novel Resistance Determinants from Cucumber PI 197085 Against Pseudoperonospora cubensis. Agronomy 2025, 15, 2633. https://doi.org/10.3390/agronomy15112633
Szczechura W, Kłosińska U, Nowakowska M, Nowak K, Nowicki M, Kozik EU, Tyrka M. Novel Resistance Determinants from Cucumber PI 197085 Against Pseudoperonospora cubensis. Agronomy. 2025; 15(11):2633. https://doi.org/10.3390/agronomy15112633
Chicago/Turabian StyleSzczechura, Wojciech, Urszula Kłosińska, Marzena Nowakowska, Katarzyna Nowak, Marcin Nowicki, Elżbieta U. Kozik, and Mirosław Tyrka. 2025. "Novel Resistance Determinants from Cucumber PI 197085 Against Pseudoperonospora cubensis" Agronomy 15, no. 11: 2633. https://doi.org/10.3390/agronomy15112633
APA StyleSzczechura, W., Kłosińska, U., Nowakowska, M., Nowak, K., Nowicki, M., Kozik, E. U., & Tyrka, M. (2025). Novel Resistance Determinants from Cucumber PI 197085 Against Pseudoperonospora cubensis. Agronomy, 15(11), 2633. https://doi.org/10.3390/agronomy15112633








