Gene Editing for Sugar Perception Transport and Source–Sink Optimization in Soybean
Abstract
1. Introduction
2. Core Components of the Plant Sugar Sensing Network
2.1. The Glucose Sensor Hexokinase
2.2. Trehalose-6-Phosphate: A Key Signal of Sucrose Status
2.3. SnRK1: The Central Energy Sensor
2.4. TOR: The Master Regulator of Growth
3. Sugar Transport for Plant Growth and Development
3.1. Phloem Loading in Source Leaves
3.2. Phloem Unloading in Sink Tissues
4. The Source–Sink Regulatory Network: An Integrated View
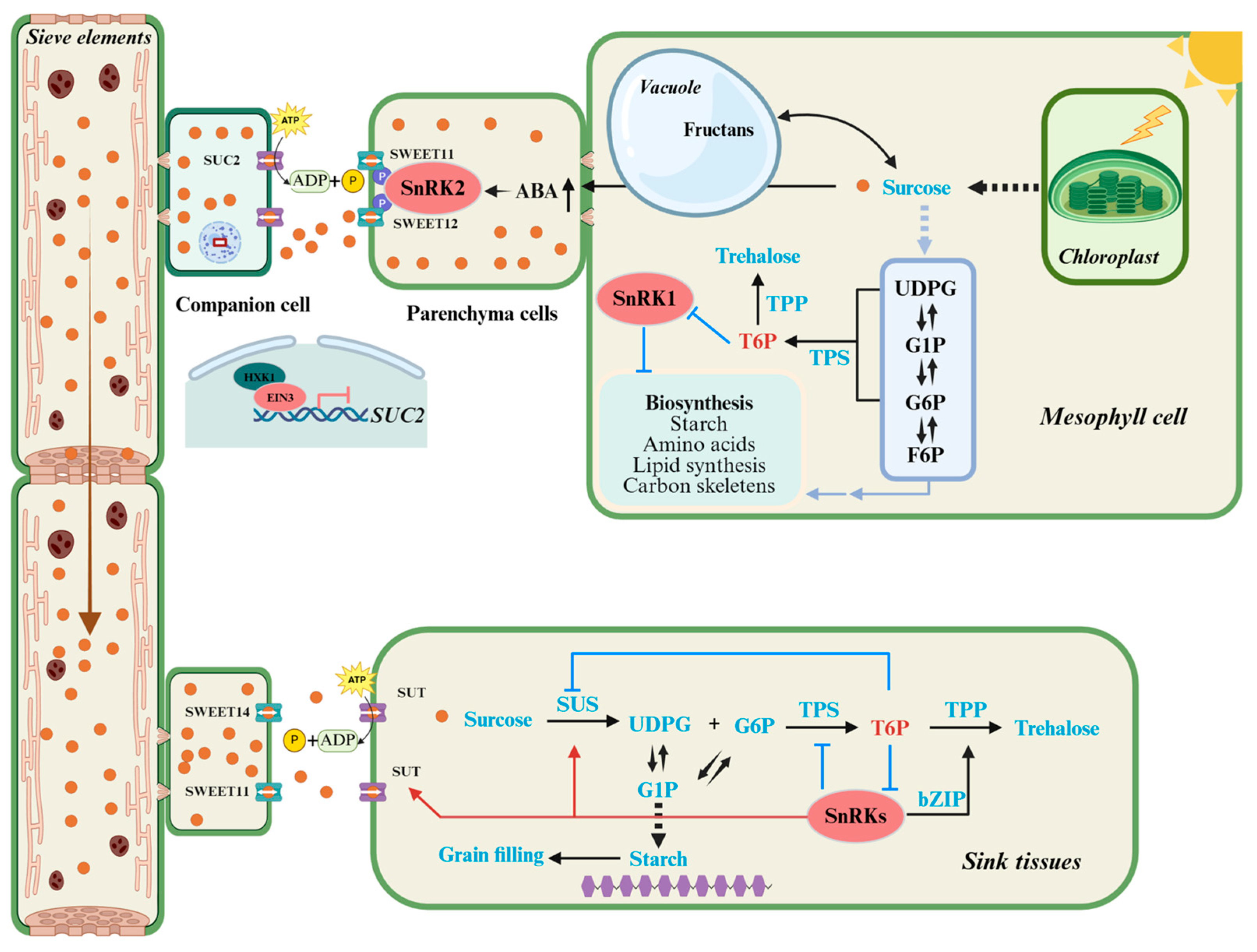
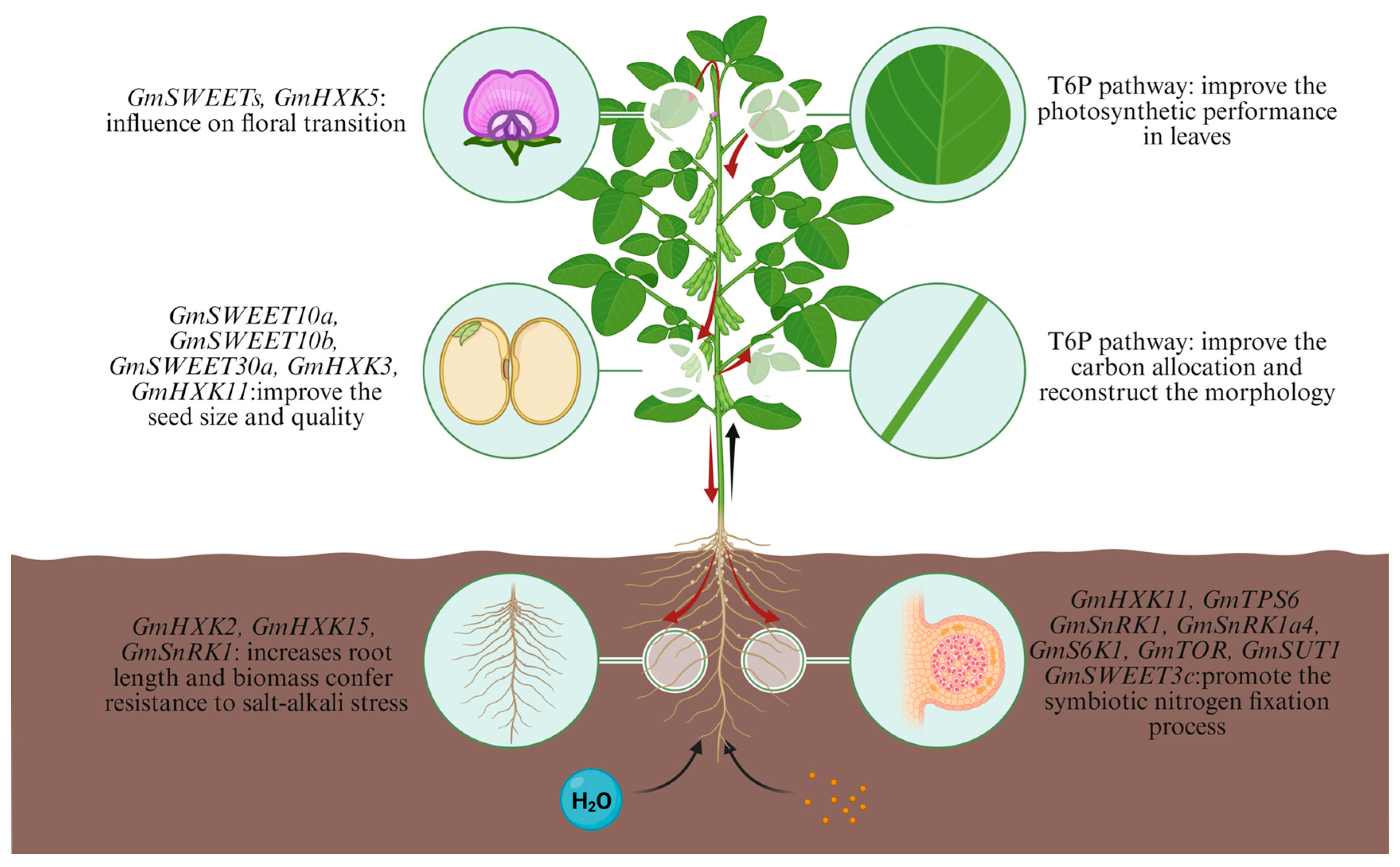
5. Targeted Editing to Optimize Source–Sink Relationships for Soybean Yield Enhancement
6. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar Sensing and Signaling in Plants: Conserved and Novel Mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef]
- Sonnewald, U.; Fernie, A.R. Next-Generation Strategies for Understanding and Influencing Source–Sink Relations in Crop Plants. Curr. Opin. Plant Biol. 2018, 43, 63–70. [Google Scholar] [CrossRef]
- Fernie, A.R.; Bachem, C.W.B.; Helariutta, Y.; Neuhaus, H.E.; Prat, S.; Ruan, Y.-L.; Stitt, M.; Sweetlove, L.J.; Tegeder, M.; Wahl, V.; et al. Synchronization of Developmental, Molecular and Metabolic Aspects of Source–Sink Interactions. Nat. Plants 2020, 6, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, J.; Li, G. Dynamic Epigenetic Modifications in Plant Sugar Signal Transduction. Trends Plant Sci. 2022, 27, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Rolland, F.; Moore, B.; Sheen, J. Sugar Sensing and Signaling in Plants. Plant Cell 2002, 14, S185–S205. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C.P. Legumes: Importance and Constraints to Greater Use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for Soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Z.; Li, H.; Yue, L.; Tan, C.; Liu, H.; Hu, Y.; Yang, Y.; Yao, X.; Kong, L.; et al. Dt1 Inhibits SWEET-Mediated Sucrose Transport to Regulate Photoperiod-Dependent Seed Weight in Soybean. Mol. Plant 2024, 17, 496–508. [Google Scholar] [CrossRef]
- Zhang, L.; Bai, M.; Kong, F.; Liu, B.; Guan, Y. Carbon-Nitrogen Bottlenecks: A Perfect Storm to Face in Soybean Yield Improvement. Mol. Plant 2025, 18, 1413–1416. [Google Scholar] [CrossRef]
- Kakiuchi, J.; Kobata, T. High Carbon Requirements for Seed Production in Soybeans [Glycine max (L.) Merr.]. Plant Prod. Sci. 2008, 11, 198–202. [Google Scholar] [CrossRef]
- Du, C.; Bai, H.; Chen, J.; Wang, J.; Wang, Z.; Zhang, Z. Alternative Splicing Regulation of Glycine-Rich Proteins via Target of Rapamycin-Reactive Oxygen Species Pathway in Arabidopsis Seedlings Upon Glucose Stress. Front. Plant Sci. 2022, 13, 830140. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-Based Methods for Genome Engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Li, W.; Wang, L.; Xue, H.; Zhang, M.; Song, H.; Qin, M.; Dong, Q. Molecular and Genetic Basis of Plant Architecture in Soybean. Front. Plant Sci. 2024, 15, 1477616. [Google Scholar] [CrossRef]
- Chiou, T.-J.; Bush, D.R. Sucrose Is a Signal Molecule in Assimilate Partitioning. Proc. Natl. Acad. Sci. USA 1998, 95, 4784–4788. [Google Scholar] [CrossRef]
- Goddijn, O.; Smeekens, S. Sensing Trehalose Biosynthesis in Plants. Plant J. 1998, 14, 143–146. [Google Scholar] [CrossRef]
- Roitsch, T. Source-Sink Regulation by Sugar and Stress. Curr. Opin. Plant Biol. 1999, 2, 198–206. [Google Scholar] [CrossRef]
- Li, L.; Liu, K.; Sheen, J. Dynamic Nutrient Signaling Networks in Plants. Annu. Rev. Cell Dev. Biol. 2021, 37, 341–367. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Kumar, A.; Kaur, N.; Kaur, H. Molecular Cues of Sugar Signaling in Plants. Physiol. Plant. 2022, 174, e13630. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J. Master Regulators in Plant Glucose Signaling Networks. J. Plant Biol. 2014, 57, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.-H.; Liu, Y.-X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis Glucose Sensor HXK1 in Nutrient, Light, and Hormonal Signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef]
- Barbier, F.F.; Cao, D.; Fichtner, F.; Weiste, C.; Perez-Garcia, M.-D.; Caradeuc, M.; Le Gourrierec, J.; Sakr, S.; Beveridge, C.A. HEXOKINASE1 Signalling Promotes Shoot Branching and Interacts with Cytokinin and Strigolactone Pathways. New Phytol. 2021, 231, 1088–1104. [Google Scholar] [CrossRef]
- Copeland, L.; Morell, M. Hexose Kinases from the Plant Cytosolic Fraction of Soybean Nodules. Plant Physiol. 1985, 79, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Chen, Y.; Zhang, D.; Wu, J. Genome-Wide Characterization of Soybean Hexokinase Genes Reveals a Positive Role of GmHXK15 in Alkali Stress Response. Plants 2023, 12, 3121. [Google Scholar] [CrossRef]
- Chen, S.; Tian, Z.; Guo, Y. Characterization of Hexokinase Gene Family Members in Glycine Max and Functional Analysis of GmHXK2 under Salt Stress. Front. Genet. 2023, 14, 1135290. [Google Scholar] [CrossRef]
- Griffiths, C.A.; Sagar, R.; Geng, Y.; Primavesi, L.F.; Patel, M.K.; Passarelli, M.K.; Gilmore, I.S.; Steven, R.T.; Bunch, J.; Paul, M.J.; et al. Chemical Intervention in Plant Sugar Signalling Increases Yield and Resilience. Nature 2016, 540, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Yadav, U.P.; Ivakov, A.; Feil, R.; Duan, G.Y.; Walther, D.; Giavalisco, P.; Piques, M.; Carillo, P.; Hubberten, H.-M.; Stitt, M.; et al. The Sucrose–Trehalose 6-Phosphate (Tre6P) Nexus: Specificity and Mechanisms of Sucrose Signalling by Tre6P. J. Exp. Bot. 2014, 65, 1051–1068. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.M.; Lunn, J.E. A Tale of Two Sugars: Trehalose 6-Phosphate and Sucrose. Plant Physiol. 2016, 172, 7–27. [Google Scholar] [CrossRef]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose Metabolism in Plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef]
- Pilon-Smits, E.A.H.; Terry, N.; Sears, T.; Kim, H.; Zayed, A.; Hwang, S.; van Dun, K.; Voogd, E.; Verwoerd, T.C.; Krutwagen, R.W.H.H.; et al. Trehalose-Producing Transgenic Tobacco Plants Show Improved Growth Performance under Drought Stress. J. Plant Physiol. 1998, 152, 525–532. [Google Scholar] [CrossRef]
- Zhu, X.; Fang, D.; Li, D.; Zhang, J.; Jiang, H.; Guo, L.; He, Q.; Zhang, T.; Macho, A.P.; Wang, E.; et al. Phytophthora Sojae Boosts Host Trehalose Accumulation to Acquire Carbon and Initiate Infection. Nat. Microbiol. 2023, 8, 1561–1573. [Google Scholar] [CrossRef]
- He, W.; Chai, Q.; Zhao, C.; Yin, W.; Fan, H.; Yu, A.; Fan, Z.; Hu, F.; Sun, Y.; Wang, F. Soybean Plant Growth and Tre6P Metabolism Under Red/Far-Red and Blue Light. J. Plant Growth Regul. 2024, 43, 473–485. [Google Scholar] [CrossRef]
- Li, G.; Zhao, Y. The Critical Roles of Three Sugar-Related Proteins (HXK, SnRK1, TOR) in Regulating Plant Growth and Stress Responses. Hortic. Res. 2024, 11, uhae099. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Primavesi, L.F.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.A.C.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-Related Protein Kinase1 Activity and Regulation of Metabolic Pathways by Trehalose-6-Phosphate. Plant Physiol. 2009, 149, 1860–1871. [Google Scholar] [CrossRef] [PubMed]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The Plant Energy Sensor: Evolutionary Conservation and Divergence of SnRK1 Structure, Regulation, and Function. J. Exp. Bot. 2016, 67, 6215–6252. [Google Scholar] [CrossRef]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A Central Integrator of Transcription Networks in Plant Stress and Energy Signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Han, C.; Wang, J.-G.; Chu, X.; Shi, W.; Yao, L.; Chen, J.; Hao, W.; Deng, Z.; Fan, M.; et al. Regulatory Functions of Cellular Energy Sensor SnRK1 for Nitrate Signalling through NLP7 Repression. Nat. Plants 2022, 8, 1094–1107. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, H.; You, H.; Liu, Y.; Chen, C.; Feng, X.; Yu, X.; Wu, S.; Wang, L.; Zhong, S.; et al. Identification of Novel Interactors and Potential Phosphorylation Substrates of GsSnRK1 from Wild Soybean (Glycine soja). Plant Cell Environ. 2019, 42, 145–157. [Google Scholar] [CrossRef]
- Li, H.Q.; Chen, C.; Chen, R.R.; Song, X.W.; Li, J.N.; Zhu, Y.M.; Ding, X.D. Preliminary Analysis of the Role of GmSnRK1.1 and GmSnRK1.2 in the ABA and Alkaline Stress Response of the Soybean Using the CRISPR/Cas9-Based Gene Double-Knockout System. Yi Chuan 2018, 40, 496–507. [Google Scholar]
- Liu, Y.; Cao, L.; Wu, X.; Wang, S.; Zhang, P.; Li, M.; Jiang, J.; Ding, X.; Cao, X. Functional Characterization of Wild Soybean (Glycine soja) GsSnRK1.1 Protein Kinase in Plant Resistance to Abiotic Stresses. J. Plant Physiol. 2023, 280, 153881. [Google Scholar] [CrossRef]
- Guo, D.; Liu, P.; Liu, Q.; Zheng, L.; Liu, S.; Shen, C.; Liu, L.; Fan, S.; Li, N.; Dong, J.; et al. Legume-Specific SnRK1 Promotes Malate Supply to Bacteroids for Symbiotic Nitrogen Fixation. Mol. Plant 2023, 16, 1396–1412. [Google Scholar] [CrossRef]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR Signaling and Nutrient Sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef]
- Ren, M.; Qiu, S.; Venglat, P.; Xiang, D.; Feng, L.; Selvaraj, G.; Datla, R. Target of Rapamycin Regulates Development and Ribosomal RNA Expression through Kinase Domain in Arabidopsis. Plant Physiol. 2011, 155, 1367–1382. [Google Scholar] [CrossRef]
- Zhang, N.; Meng, Y.; Li, X.; Zhou, Y.; Ma, L.; Fu, L.; Schwarzländer, M.; Liu, H.; Xiong, Y. Metabolite-Mediated TOR Signaling Regulates the Circadian Clock in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 25395–25397. [Google Scholar] [CrossRef]
- Li, X.; Cai, W.; Liu, Y.; Li, H.; Fu, L.; Liu, Z.; Xu, L.; Liu, H.; Xu, T.; Xiong, Y. Differential TOR Activation and Cell Proliferation in Arabidopsis Root and Shoot Apexes. Proc. Natl. Acad. Sci. USA 2017, 114, 2765–2770. [Google Scholar] [CrossRef]
- Um, J.-H.; Kim, S.; Kim, Y.-K.; Song, S.-B.; Lee, S.-H.; Verma, D.P.S.; Cheon, C.-I. RNA Interference-Mediated Repression of S6 Kinase 1 Impairs Root Nodule Development in Soybean. Mol. Cells 2013, 35, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar Transporters for Intercellular Exchange and Nutrition of Pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Ayre, B.G. Membrane-Transport Systems for Sucrose in Relation to Whole-Plant Carbon Partitioning. Mol. Plant 2011, 4, 377–394. [Google Scholar] [CrossRef]
- Ludewig, F.; Flügge, U.-I. Role of Metabolite Transporters in Source-Sink Carbon Allocation. Front. Plant Sci. 2013, 4, 231. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Hu, T.; Li, X.; Song, C.-P.; Zhu, J.-K.; Chen, L.; Zhao, Y. Phosphorylation of SWEET Sucrose Transporters Regulates Plant Root:Shoot Ratio under Drought. Nat. Plants 2022, 8, 68–77. [Google Scholar] [CrossRef]
- Aubry, E.; Clément, G.; Gilbault, E.; Dinant, S.; Le Hir, R. Changes in SWEET-Mediated Sugar Partitioning Affect Photosynthesis Performance and Plant Response to Drought. Physiol. Plant. 2024, 176, e14623. [Google Scholar] [CrossRef]
- Bezrutczyk, M.; Hartwig, T.; Horschman, M.; Char, S.N.; Yang, J.; Yang, B.; Frommer, W.B.; Sosso, D. Impaired Phloem Loading in Zmsweet13a,b,c Sucrose Transporter Triple Knock-out Mutants in Zea mays. New Phytol. 2018, 218, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, E.A.; Bush, D.R. Carbohydrate Export from the Leaf: A Highly Regulated Process and Target to Enhance Photosynthesis and Productivity. Plant Physiol. 2011, 155, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Das, S.; Jagadis Gupta, K.; Ranjan, A.; Foyer, C.H.; Thakur, J.K. Physiological Implications of SWEETs in Plants and Their Potential Applications in Improving Source–Sink Relationships for Enhanced Yield. Plant Biotechnol. J. 2023, 21, 1528–1541. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, Y.; Liu, S.; Wang, S.; Li, J.; Fang, C.; Liu, Y.; Yang, X.; Tian, D.; Song, S.; et al. Genomic Atlas of 8,105 Accessions Reveals Stepwise Domestication, Global Dissemination, and Improvement Trajectories in Soybean. Cell 2025, 188, 6519–6535. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.-C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.-Q.; et al. Simultaneous Changes in Seed Size, Oil Content and Protein Content Driven by Selection of SWEET Homologues during Soybean Domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]
- Fei, H.; Yang, Z.; Lu, Q.; Wen, X.; Zhang, Y.; Zhang, A.; Lu, C. OsSWEET14 Cooperates with OsSWEET11 to Contribute to Grain Filling in Rice. Plant Sci. 2021, 306, 110851. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, D.; Miao, Q.; Yang, J.; Xuan, Y.; Hu, Y. Essential Role of Sugar Transporter OsSWEET11 During the Early Stage of Rice Grain Filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef]
- Deng, L.; Zhao, S.; Yang, G.; Zhu, S.; Tian, J.; Wang, X. Soybean GmSUT1 Transporter Participates in Sucrose Transport to Nodules during Rhizobial Symbiosis. Plant Growth Regul. 2022, 96, 119–129. [Google Scholar] [CrossRef]
- Chen, K.; Wang, X.; Pang, R.; Chen, L.; Chen, J.; Ren, Z.; Wang, S.; Wang, Y.; Li, X.; Su, C. The Sucrose Transporter GmSWEET3c Drives Soybean Nodulation by Regulating Root Sucrose Allocation. Curr. Biol. 2025, 35, 4121–4134.e4. [Google Scholar] [CrossRef]
- Andrés, F.; Kinoshita, A.; Kalluri, N.; Fernández, V.; Falavigna, V.S.; Cruz, T.M.D.; Jang, S.; Chiba, Y.; Seo, M.; Mettler-Altmann, T.; et al. The Sugar Transporter SWEET10 Acts Downstream of FLOWERING LOCUS T during Floral Transition of Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 53. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Wu, K.; Ye, Y.; Yu, J.; Zhang, J.; Liu, Q.; Hu, M.; Li, H.; Tong, Y.; et al. Modulating Plant Growth–Metabolism Coordination for Sustainable Agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef]
- O’Hara, L.E.; Paul, M.J.; Wingler, A. How Do Sugars Regulate Plant Growth and Development? New Insight into the Role of Trehalose-6-Phosphate. Mol. Plant 2013, 6, 261–274. [Google Scholar] [CrossRef]
- Tong, C.; Li, C.; Cao, X.-Y.; Sun, X.-D.; Bao, Q.-X.; Mu, X.-R.; Liu, C.-Y.; Loake, G.J.; Chen, H.; Meng, L.-S. Long-Distance Transport of Sucrose in Source Leaves Promotes Sink Root Growth by the EIN3-SUC2 Module. PLoS Genet. 2022, 18, e1010424. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Watson, A.; Griffiths, C.A. Trehalose 6-Phosphate Signalling and Impact on Crop Yield. Biochem. Soc. Trans. 2020, 48, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- Oszvald, M.; Primavesi, L.F.; Griffiths, C.A.; Cohn, J.; Basu, S.S.; Nuccio, M.L.; Paul, M.J. Trehalose 6-Phosphate Regulates Photosynthesis and Assimilate Partitioning in Reproductive Tissue. Plant Physiol. 2018, 176, 2623–2638. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR Technologies in Research and Beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, L.; Chen, L.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; Yao, W.; Jiang, B.; Yuan, S.; et al. Mutagenesis of GmFT2a and GmFT5a Mediated by CRISPR/Cas9 Contributes for Expanding the Regional Adaptability of Soybean. Plant Biotechnol. J. 2020, 18, 298–309. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Hou, W. Genome Editing Technologies Accelerate Innovation in Soybean Breeding. Agronomy 2023, 13, 2045. [Google Scholar] [CrossRef]
- Kawall, K.; Cotter, J.; Then, C. Broadening the GMO Risk Assessment in the EU for Genome Editing Technologies in Agriculture. Environ. Sci. Eur. 2020, 32, 106. [Google Scholar] [CrossRef]
| Gene Family | Key Gene | Expression Tissue | Phenotype | Research Approach | Reference |
|---|---|---|---|---|---|
| HXK | GmHXK2 | Roots | Reduce expression exacerbates salt stress phenotype. | Homology inference | [24] |
| HXK | GmHXK3 | Pods | Unknown | Homology inference | [23] |
| HXK | GmHXK5 | Flowers | Unknown | Homology inference | [23] |
| HXK | GmHXK11 | Seeds, Root nodules | Unknown | Homology inference | [23] |
| HXK | GmHXK15 | Flowers | Overexpression promotes root growth and alkali tolerance. | Homology inference | [23] |
| T6P | GmTPS6 | Broadly expressed | Triggered by Phytophthora sojae effector to promote infection. | Experimental identification | [30] |
| SnRK1 | GmSnRK1.1/GmSnRK1.2 | Roots | Double knockout impairs root growth and reduces abiotic stress tolerance. | Homology inference | [38] |
| SnRK1 | SnRK1a4 | Roots | Enhance expression increases nodule size and nitrogenase activity. | Homology inference | [40] |
| TOR | GmTOR/GmS6K1 | Root nodules | Phenotype not studied | Experimental identification | [45] |
| SUT | GmSUT1 | Root nodules | Overexpression of GmSUT1 increases nodule number and dry weight and improves plant dry weight and nitrogen content. RNAi plants impaired sucrose transport and increased shoot sucrose concentration. | Homology inference | [60] |
| SWEET | GmSWEET3c | Root nodules | CRISPR knockdown significantly reduces infection threads and nodules. | Experimental identification | [61] |
| SWEET | GmSWEET10a/b | Seed coat | Knockout mutants have smaller seeds and reduced oil content. | Experimental identification | [57] |
| SWEET | GmSWEET30a/b | Seed coat | Double knockout mutants have reduced oil and protein content. | Experimental identification | [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, S.; Chen, L.; Hou, W.; Cai, Y. Gene Editing for Sugar Perception Transport and Source–Sink Optimization in Soybean. Agronomy 2025, 15, 2621. https://doi.org/10.3390/agronomy15112621
Ding S, Chen L, Hou W, Cai Y. Gene Editing for Sugar Perception Transport and Source–Sink Optimization in Soybean. Agronomy. 2025; 15(11):2621. https://doi.org/10.3390/agronomy15112621
Chicago/Turabian StyleDing, Shuqi, Li Chen, Wensheng Hou, and Yupeng Cai. 2025. "Gene Editing for Sugar Perception Transport and Source–Sink Optimization in Soybean" Agronomy 15, no. 11: 2621. https://doi.org/10.3390/agronomy15112621
APA StyleDing, S., Chen, L., Hou, W., & Cai, Y. (2025). Gene Editing for Sugar Perception Transport and Source–Sink Optimization in Soybean. Agronomy, 15(11), 2621. https://doi.org/10.3390/agronomy15112621









