Abstract
Nitrate transporter 1/peptide transporter family (NPF) proteins play pivotal roles in nitrogen uptake, translocation, and stress adaptation in plants. To comprehensively investigate this gene family in sorghum (Sorghum bicolor), we conducted the genome-wide identification and characterization of SbNPF genes. A total of 88 SbNPF members were identified and classified into eight subfamilies based on phylogenetic analysis, displaying diverse gene structures, conserved motifs, and evolutionary relationships. Gene duplication analysis revealed that tandem duplication was the primary driver of SbNPF family expansion, with most duplicated pairs undergoing purifying selection. Synteny analysis showed extensive collinearity between sorghum and rice, but limited conservation with Arabidopsis, highlighting the evolutionary divergence between monocots and dicots. Cis-regulatory element prediction suggested that SbNPF genes are widely involved in abiotic stress responses, hormone signaling, and light responsiveness. Expression profiling using RNA-seq data revealed distinct tissue-specific expression patterns, indicating functional specialization across roots, stems, leaves, and reproductive tissues. Furthermore, RT-qPCR analysis under low-nitrogen (LN) conditions demonstrated that several SbNPF genes, including SbNPF1.1, SbNPF2.6, SbNPF2.7, and SbNPF4.5, were significantly upregulated in shoots, whereas SbNPF1.2, SbNPF2.7, and SbNPF3.1 were downregulated in roots, suggesting differential regulatory roles in nitrogen acquisition and utilization under nutrient-limiting environments. Collectively, these findings provide novel insights into the evolutionary dynamics and potential functions of the SbNPF gene family and establish a foundation for future functional studies and molecular breeding aimed at improving nitrogen use efficiency in sorghum.
1. Introduction
The NPF gene family plays a crucial role in plant growth and development, as well as in plant responses to abiotic stresses such as low nitrogen stress. The amino acid sequences encoded by the NPF family exhibit strong homology across various species, with most members displaying similar conserved structures. Typically, NPF members in higher plants consist of 450–600 amino acids and possess 12 conserved transmembrane domains, among which there is an intracellular hydrophilic ring structure between the sixth and seventh transmembrane domains [1,2] and a conserved motif in the fifth transmembrane domain [3]. Currently, 53, 198, 93, and 150 NPF members have been identified in Arabidopsis thaliana, Brassica napus, Oryza sativa, and Gossypium hirsutum, respectively [4,5,6,7]. Based on phylogenetic relationships, the NPF family is categorized into eight subfamilies (NPF1-NPF8), each exhibiting distinct functions. To date, the biological functions of most NPF family genes in Arabidopsis thaliana have been characterized, while the functions of 16 NPF family genes in rice have also been investigated.
Under normal external nitrate supply conditions, the absorption and transport of nitrate in plants primarily rely on the regulation of NPF (Nitrate Transporter Family). AtNPF6.3/AtNRT1.1/CHL1 is the first identified NO3− transporter within the NPF family in Arabidopsis thaliana [8]. Most of the other members of the NPF are believed to encode low-affinity nitrate transporters. AtNPF6.2 (AtNRT1.4) is predominantly expressed in the vacuolar membrane and veins of the leaf, facilitating the transport of NO3− from the vein to the vacuole for storage, thereby maintaining the balance and stability of NO3− concentrations in the petiole and leaf [9]. AtNPF7.3 (AtNRT1.5) is situated on the cell membrane of the column sheath in Arabidopsis roots, where its encoded protein loads NO3− into the xylem and enables long-distance transport of NO3− to the shoot via transpiration pull [10]. AtNPF7.2 (AtNRT1.8) and AtNPF2.9 are expressed in the thin-walled cell membranes of the xylem and in phloem cells [11]. Additionally, AtNPF5.11, AtNPF5.12, and AtNPF5.16, which are localized on the vacuolar membrane, are thought to mediate the transport of nitrate from the vacuole to the cytoplasm and regulate the distribution of nitrate within the root and shoot of the plant [12]. In Oryza sativa, OsNRT1.1a and OsNRT1.1b are low-affinity nitrate transporters predominantly expressed in roots [13]. OsNPF2.2 is another low-affinity transporter that is induced by NO3− and is primarily expressed in xylem membrane cells [14].
Understanding the roles of NPF family genes in biological and physiological processes presents a valuable opportunity for analyzing and enhancing crop defenses against both biotic and abiotic stresses. Sorghum [Sorghum bicolor (L.) Moench] is the fifth most important cereal crop globally, and is cultivated on approximately 40 million hectares with an annual production of over 55 million tons. It serves as a staple food, feed, and bioenergy crop in semi-arid and arid regions. Sorghum exhibits significant stress tolerance, allowing it to thrive under abiotic conditions such as drought, salinity, and nutrient-deficient soils, thereby establishing it as an ideal model for investigating the mechanisms of nitrogen transport and utilization under challenging environmental conditions [15]. Improving nitrogen use efficiency in sorghum is of great importance for enhancing productivity and sustainability in stress-prone agricultural systems. While the DIR gene family has been extensively studied in model organisms such as Arabidopsis thaliana and Oryza sativa, a comprehensive analysis of this gene family in sorghum is still lacking. Consequently, in this study, we identified 88 SbNPF genes based on the sorghum genome. We then conducted a systematic bioinformatics analysis of the SbNPF gene family, which encompassed phylogenetic relationships, conserved protein motifs, gene structures, cis-acting elements, and tissue-specific expression levels under various abiotic stresses. This research will provide a theoretical foundation for future functional studies and enhance our understanding of the roles of SbNPF genes in the growth, development, and stress responses of sorghum.
2. Materials and Methods
2.1. Identification of Gene Family Members
In this study, HMMER 3.3.2 software was used to perform a homology search (E-value threshold of 1 × 10−5) based on the Hidden Markov Model (HMM) of the NPF domain in the Pfam database (https://pfam.xfam.org/)(accessed on 1 January 2025) for sorghum BTx623 protein sequences downloaded from the Phytozome v13 (https://phytozome-next.jgi.doe.gov/) (accessed on 1 January 2025). The gene sequences obtained from the initial screening were further validated for conserved structural domains using NCBI Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (accessed on 1 January 2025) to ensure that the identified sequences contained complete and typical NPF domains, thus accurately identifying members of the sorghum NPF gene family [16].
2.2. Analysis of Gene Structure and Conserved Motifs
Domain and positional information were determined utilizing the Pfam-Scan software (https://www.ebi.ac.uk/jdispatcher/pfa/pfamscan) (accessed on 1 January 2025) [17]. Conserved motifs were analyzed by MEME software 5.3.3 [18] with the maximun number of 10 and optimum widths of 10–50 amino acids, with subsequent extraction performed by Python 3.13.5 scripts. The exon and intron locations of SbNPF genes were derived from GFF3 annotation files. The visualization of domains, conserved motifs and gene structures was accomplished using the R package ggtree (https://github.com/YuLab-SMU/ggtree, accessed on 1 January 2025) and ggenes (https://github.com/wilkox/gggenes) (accessed on 1 January 2025).
2.3. Phylogenetic Tree Construction
Muscle software was used to perform multiple sequence alignment of the OsNPF, AtNPF and SbNPF proteins (https://www.ebi.ac.uk/jdispatcher/msa/muscle) (accessed on 1 January 2025) [19]. Subsequently, we constructed a phylogenetic tree using IQ-TREE (2.0.3) employing the maximum likelihood (ML) method with bootstrap values derived from 1000 replicates [20], and visualized the tree using the R package ggtree [21].
2.4. Analysis of Chromosome Distribution, Gene Dulication, and Selection Pressure
The chromosomal distribution of SbNPF genes was determined utilizing the GFF3 annotation file. Gene duplication and collinearity analyses were executed using the MCScanX software (https://github.com/wyp1125/MCScanX) (accessed on 1 January 2025) [22], which identified the duplication types, including tandem duplications (TP) and whole-genome duplications (WGD). For the analysis and visualization of inter-species collinearity, the JCVI software [23] was employed (https://github.com/tanghaibao/jcvi) (accessed on 1 January 2025). The alignment of protein sequences and coding sequences (CDS) of NPF genes with gene duplication was proformed using ClustalW software (https://www.genome.jp/tools-bin/clustalw) (accessed on 1 January 2025) [24]. Furthermore, the KaKs_Calculator software was utilized to compote the synonymous substitution rate (Ks), nonsynonymous substitution rate (Ka), and the evolutionary ratio (Ka/Ks) between duplicate gene pairs of NPF genes [25].
2.5. Gene Expression Analysis
Transcriptome data of different tissues of sorghum (including seedlings, leaves, roots, stems, inflorescences, and seeds, etc.) at different developmental stages were obtained from the Sorghum Genome and Mutant Bank (SGMD) database. The data are shown in a heatmap with gene expression in different tissues with row-scaled transcriptome atlas (TPM values). Red and blue boxes indicate high and low expression levels of NPF genes [26].
Hydroponic experiments were conducted in a controlled-environment growth chamber. The seedlings were treated with low nitrogen (0.05 mmol L−1 N) at two leaves stage and the control plants were maintained under normal nitrogen conditions. After seven days of treatment, shoot and root samples were collected for RNA extraction for qPCR analysis. Quantitative real-time PCR was performed to examine the expression levels of nine selected SbNPF genes (SbNPF1.1, SbNPF1.2, SbNPF2.6, SbNPF2.7, SbNPF3.1, SbNPF4.5, SbNPF6.2, SbNPF7.4, and SbNPF8.4) in both leaves and roots under LN and control conditions. Gene-specific primers for the SbNPF family were designed using Primer 5.0 software (Supplemental Table S1). Total RNA was extracted from sorghum shoots and roots with the Tiangen RNA Extraction Kit [Tiangen biotech(Beijing) Co., Ltd., Beijing, China], and first-strand cDNA was synthesized using the Tiangen RT Reagent Kit according to the manufacturer’s instructions [27].
3. Results
3.1. Genome-Wide Identification and Characterization of Sorghum NPF Genes
In this study, we identified 88 NPF genes in sorghum and named them SbNPF1 to SbNPF88 according to their locations on chromosomes. There are 1.7 times more SbNPF proteins than Arabidopsis proteins, similar to the levels of rice. This difference is attributed to gene expansion in sorghum during evolution. Supplemental Table S2 presented the details the gene IDs, genomic locations, coding sequence lengths, protein lengths, and other characteristics of the identified SbNPF genes. SbNPF protein lengths range from 188 to 688 amino acids, averaging 568 amino acids. Their molecular weights range from 20.5 to 72.33 kilodaltons (kDa), averaging 61.73 kDa. Additionally, the protein isoelectric points (pI) range widely (4.57–10.6), possibly due to significant differences in protein length (Figure 1).
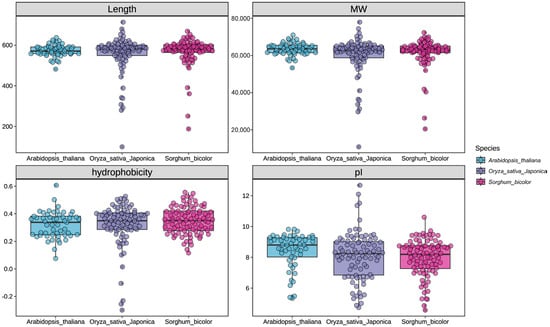
Figure 1.
Characteristics of NPF family members in Arabidopsis thaliana, Oryza sativa and Sorghum bicolor including protein length, molecular weight (MW), protein isoelectric point (pI) and hydrophilicity.
3.2. Phylogenetic Tree of SbNPF Gene Family in Sorghum
To clarify the evolutionary classification of the sorghum NPF gene family, this study used iqtree software 2.0.3 to perform a phylogenetic analysis of the amino acid sequences of 88 SbNPF members. The results showed that the NPF proteins can be divided into eight subfamilies: NPF1, NPF2, NPF3, NPF4, NPF5, NPF6, NPF7, and NPF8 (Figure 2). NPF1 comprises five members, NPF2 comprises eight members, NPF3 comprises seven members, NPF4 comprises 18 members, NPF5 comprises 19 members, NPF6 comprises five members, NPF7 comprises 10 members, and NPF8 comprises 16 members.
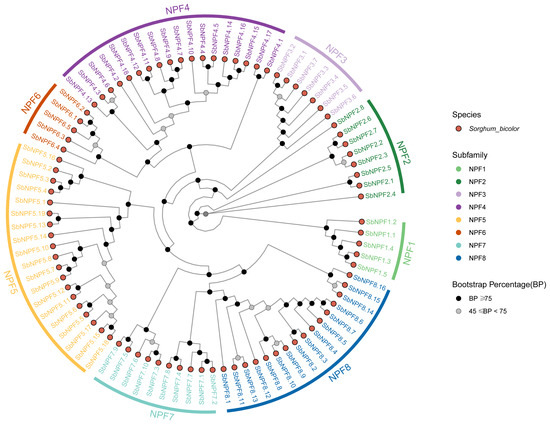
Figure 2.
The evolutionary examination of the NPF protein sequences in sorghum. These 88 NPF proteins were categorized into eight distinct subgroups, labeled NPF1–NPF8.
To clarify the phylogenetic position of the sorghum NPF gene further, this study integrated NPF protein sequences from sorghum, A. thaliana, and O. sativa, then performed a multiple comparison analysis. The phylogenetic tree constructed using the neighbor-joining method revealed that the NPF genes of all species fall into eight distinct branches (Figure 3). NPF1 includes five SbNPFs, three OsNPFs, and three AtNPFs. NPF2 is mainly composed of eight SbNPFs, eight OsNPFs, and 14 AtNPFs. NPF3 primarily includes seven SbNPFs, five OsNPFs, and one AtNPF. NPF4 primarily includes 16 SbNPFs, 12 OsNPFs, and eight AtNPFs. NPF5 primarily includes 19 SbNPFs, 23 OsNPFs, and 16 AtNPFs. NPF6 includes five SbNPFs, five OsNPFs, and three AtNPFs. NPF7 includes 10 SbNPFs, 11 OsNPFs, and three AtNPFs. NPF8 includes 16 SbNPFs, 20 OsNPFs, and five AtNPFs. Phylogenetic analysis results indicate that the NPF gene family of sorghum has established evolutionary associations with NPF genes from other plants, yet it has retained its own specificity.
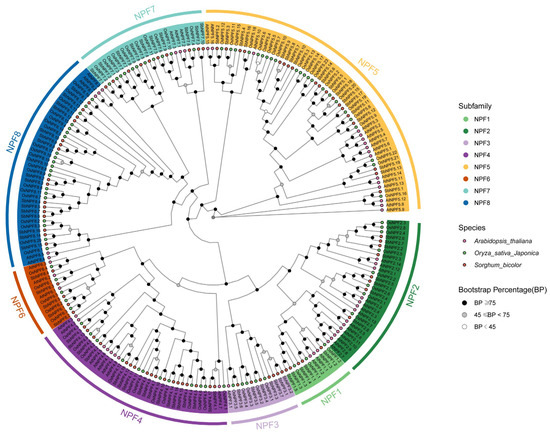
Figure 3.
The evolutionary examination of the NPF protein sequences in Arabidopsis thaliana, Oryza sativa and Sorghum bicolor. The NPF proteins were categorized into eight distinct subgroups, labeled NPF1–NPF8.
3.3. Structure and Conserved Motif Analysis of SbNPF Genes
This study analyzed the exon/intron structure of NPF genes to investigate the structural characteristics of sorghum NPF genes throughout evolution. Significant structural differences were observed among the SbNPF genes. Specifically, the number of exons in the 88 SbNPF genes ranged from one to eight, with SbNPF2.4 exhibiting the highest count of eight exons. Most NPF1 and NPF3 subgroup genes contained two or three introns, while the majority of NPF2 subgroup genes exhibited one to seven introns. NPF4 subgroup genes typically had three or six introns, NPF5 subgroup genes had two to four introns, NPF7 subgroup genes had one to three introns, and NPF8 subgroup genes had zero to four introns (Figure 4). These findings suggest that introns may have undergone changes through gain and loss during the evolutionary process of the SbNPF gene family, thereby contributing to the functional diversity of SbNPF gene.
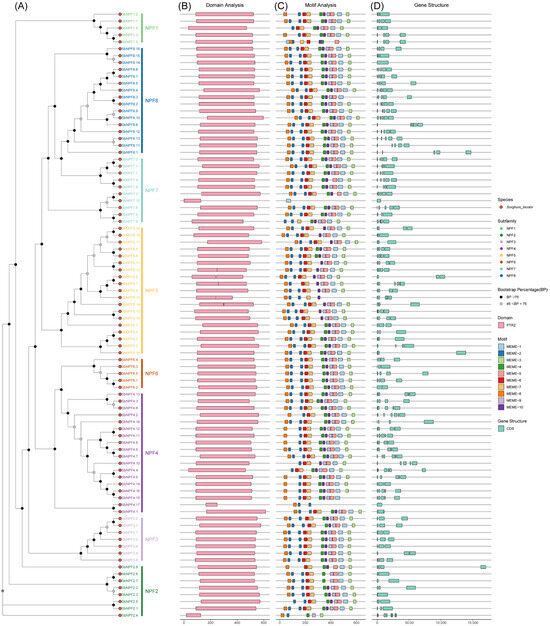
Figure 4.
Analysis of phylogenetic relationships, conserved domains, motifs, and gene structures related to NPF genes in sorghum. (A) Phylogenetic tree of NPF proteins; (B) conserved domains in NPF protein sequences; (C) distribution of 15 conserved motifs in NPF proteins; (D) gene structure of NPF genes.
3.4. Gene Duplication Analysis of SbNPF Genes
This study used the MCScan tool to analyze gene duplication events and elucidate amplification patterns of the SbNPF gene family members in sorghum to reveal evolutionary duplication events. Figure 5 shows the results, which reveal that a total of 22 pairs of tandemly repeated genes were identified. Additionally, ten pairs of segmental duplication genes were identified in the sorghum SbNPF gene family (Supplemental Table S3). These results suggest that tandem duplication plays a dominant role in the evolution of the SbNPF gene family.
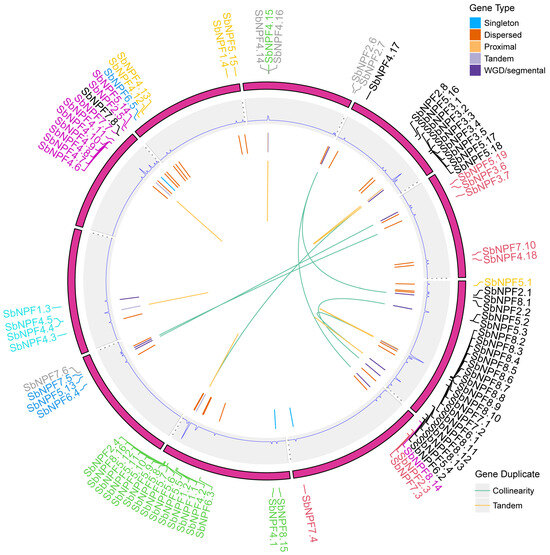
Figure 5.
The chromosome location and duplicated gene pair of NPF genes in sorghum. The duplicate gene types were displayed in different colors. WGD and TD events are shown in orange and blue, respectively.
To better understand the evolutionary constraints controlling the functional diversification of the SbNPF gene family, we calculated the non-synonymous substitution rate (Ka), the synonymous substitution rate (Ks), and their ratio (Ka/Ks). Most repeated SbNPF gene pairs showed a Ka/Ks ratio less than 1, suggesting that the SbNPF gene family experienced selective pressure during evolution (Figure 6, Supplemental Table S4).
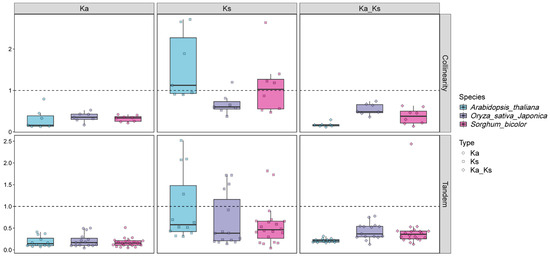
Figure 6.
The value of Ka, Ks and Ka/Ks of Arabidopsis thaliana, Oryza sativa and Sorghum bicolor. MCScanX was used to calculate the value of Ka, Ks, and Ka/Ks. Ka/Ks value of >1 signifies the gene is subjected to positive selection; Ka/Ks value of 1 indicates neutral selection; Ka/Ks value of <1 represents negative purifying selection.
3.5. Synteny Analysis of SbNPF Genes
To analyze the evolutionary characteristics of the NPF gene family in sorghum, we created a synteny map of homologous genes in Sorghum bicolor, Oryza sativa, and Arabidopsis thaliana using whole-genome alignment. Our analysis revealed that sorghum and rice share 42 pairs of highly conserved NPF collinear genes, whereas sorghum and Arabidopsis share only four. This significant difference in number (a 10.5-fold difference) strongly supports the notion that NPF genes in monocotyledonous plants are more closely related evolutionarily. These findings suggest that the SbNPF gene family has maintained core physiological functions and that its molecular mechanisms may be related to the unique nitrogen utilization strategies of monocotyledonous plants (Figure 7).
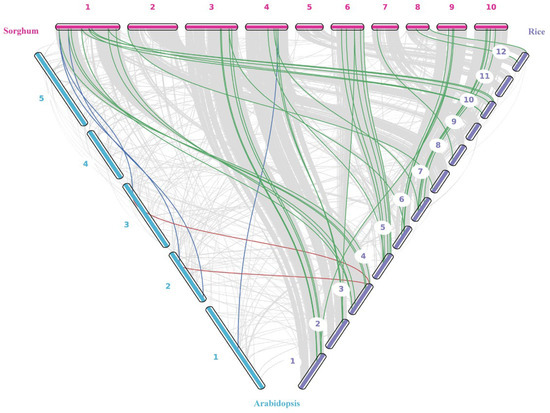
Figure 7.
Syntenic analysis of NPF genes between Oryza sativa, Arabidopsis thaliana, Sorghum bicolor, respectively. The collinear blocks and NPF homologous genes pairs were shown by gray, red and green lines, respectively. The numbers indicated the chromosomes of each species.
3.6. Analysis of Cis-Acting Elements of the SbNPF Gene Family
To better understand the biological functions of the SbNPF gene family, we analyzed the cis-regulatory elements in the 2000-bp upstream region of 88 SbNPF genes, revealing their potential response mechanisms. We identified 21 cis-regulatory elements associated with stress response, tissue-specific expression, response to plant hormones, and response to light. Five of these elements are related to the stress response: AS-1, ARE, LTR, MBS, and A-box. These elements are involved in responses to stresses such as low temperature, salt stress, defense, and drought. Five elements related to growth and development were also identified: TATA-box, CAT-box, AAGAA-motif, CCGTCC-motif, and CCGTCC-box. Additionally, six hormone-related elements were identified: ABREs, CGTCA-motifs, TGACG-motifs, TCA elements, ABRE3s, and ABRE4s. This category includes abscisic acid response elements (ABREs) and methyl jasmonate response elements (CGTCA-motif and TGACG-motif). Five cis-acting elements related to light response were also identified: CAAT-box, G-box, Box 4, G-box, and GT1-motif. The Box 4 element and G-box are relatively abundant among light-responsive cis-acting regulatory elements. These results collectively suggest that the SbNPF gene plays an important role in non-biotic stress responses, defense-related signal transduction, and plant hormone responses. Furthermore, these genes may be involved in various light responses during sorghum growth (Figure 8).
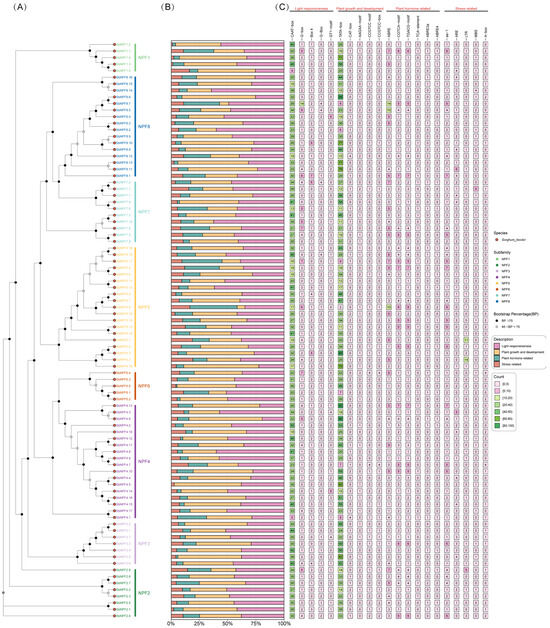
Figure 8.
Cis-elements analysis of NPF genes in sorghum. (A) Phylogenetic tree of NPF proteins; (B) bar chart illustrating the summary of cis-acting elements, with colors denoting various functional categories; (C) distribution of cis-acting element counts, with colors indicating different count ranges.
3.7. Expression Patterns of SbNPF Genes in Different Tissues
To gain a deeper understanding of the functional differentiation of the SbNPF gene family in sorghum, we analyzed the transcriptional expression characteristics of its members in various organs and tissues, such as roots, stems, leaves, flowers, seeds, and seedlings. Alignment analysis of multi-tissue RNA sequencing data revealed significant tissue-specific expression patterns among the gene family members, suggesting their potential involvement in fine-tuning organ development or physiological processes.
In leaves, the core organ of photosynthesis, we observed obvious leaf tissue preference for the following genes: SbNPF5.19, SbNPF6.3, SbNPF4.15, SbNPF4.17, SbNPF5.7, and SbNPF8.16. This suggests that these genes may be involved in nitrogen allocation or photosynthetic product transport networks in leaves. SbNPF4.9, SbNPF5.11, SbNPF6.2, SbNPF5.17, SbNPF7.2, SbNPF3.4, SbNPF3.7, SbNPF5.5, SbNPF3.8, SbNPF4.7, SbNPF4.18, SbNPF8.9, SbNPF5.18, SbNPF8.11, and SbNPF4.8 were primarily expressed in roots. This suggests that they may constitute core components of nitrogen absorption and signal transduction in the root system.13, 8.12, 8.13, and 5.14 are primarily expressed in roots. This suggests that they may be core components of nitrogen absorption and signal transduction in the root system.
Genes related to vascular bundle development show a preference for expression in the stem. Specifically, SbNPF4.14, SbNPF5.1, SbNPF8.8, SbNPF4.3, SbNPF2.8, and SbNPF2.2 are primarily expressed in the stem. SbNPF4.16, SbNPF8.5, SbNPF8.7, SbNPF8.3, SbNPF5.2, SbNPF2.1, and SbNPF2.5 are specifically expressed during the seedling stage. These genes may play a key role during the nutritional transition period after seed germination. Their dynamic expression patterns provide important clues for elucidating the environmental adaptation mechanisms of sorghum seedlings. The SbNPF family’s multi-level expression pattern indicates that it achieves coordinated regulation of multiple physiological processes, including nitrogen metabolism, hormone transport, and stress responses, through the functional specialization of its gene members (Figure 9).
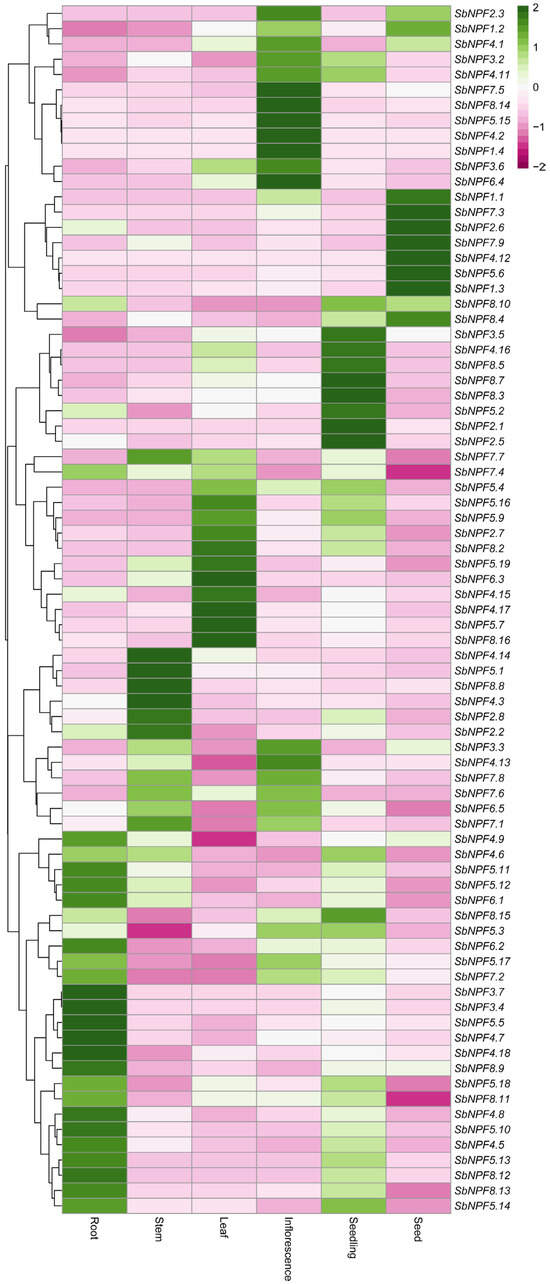
Figure 9.
Analysis of the expression patterns of SbNPF genes across various tissues by RNA-Seq.
3.8. Expression of SbNPF Genes in Response to Low Nitrogen Stress by RT-qPCR
To further explore the potential functions of SbNPF genes, we selected four genes that were responsive to low nitrogen stress based on RT-qPCR analysis. As illustrated in Figure 10, these candidate genes displayed distinct expression patterns in response to low nitrogen (LN) treatment, indicating their differential regulatory mechanisms under nutrient-limiting conditions. Notably, SbNPF1.1, SbNPF2.6, SbNPF2.7, and SbNPF4.5 were significantly upregulated in response to LN stress in the shoot. This induction suggests that these paralogs may play active roles in enhancing nitrogen uptake or remobilization under limiting conditions by potentially facilitating nitrate transport across cellular membranes.
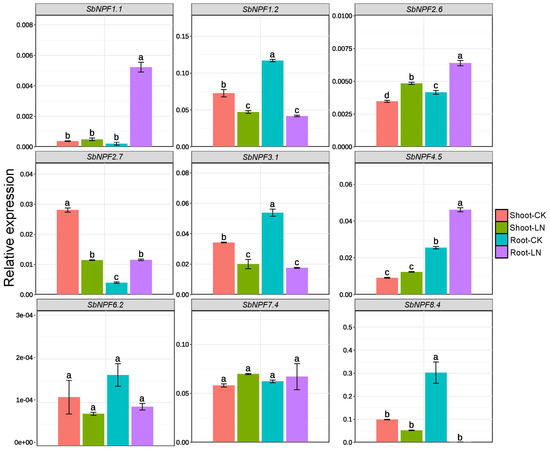
Figure 10.
NPF genes’ expression level under low nitrogen stress in shoot and root of sorghum. Different lowercase letters indicate significant differences (p < 0.05).
In contrast, another subset of genes—SbNPF1.2, SbNPF2.7, and SbNPF3.1—displayed marked down-regulation upon LN treatment in roots. Such repression could indicate a regulatory strategy to conserve energy by downregulating nonessential transporters under nutrient stress. Alternatively, it could reflect their involvement in distinct signaling pathways that are repressed under LN conditions.
These expression patterns collectively indicate that SbNPF genes exhibit intricate and specific transcriptional regulatory behaviors in response to low nitrogen stress. This implies their potential involvement in coordinating plant adaptation to this nutrient-stress condition.
4. Discussion
4.1. Identification and Evolutionary Characteristics of the Sorghum NPF Gene Family: A Balance Between Conservation and Species Specificity
This study identified 88 NPF family members (SbNPFs) in the sorghum genome. This number falls between the 53 identified in Arabidopsis and the 93 identified in rice [4,6]. Phylogenetic analysis revealed that the SbNPFs can be divided into eight subfamilies (NPF1–NPF8). This classification is highly consistent with those of Arabidopsis and rice [1,7]. This indicates that the core evolutionary branch of the NPF gene family formed before the diversification of angiosperms. Further cross-species synteny analysis revealed that sorghum and rice share 42 pairs of homologous genes, whereas sorghum and Arabidopsis share only four pairs. This finding suggests that the NPF family exhibits higher functional conservation among monocots [5] and may be closely related to monocot-specific nitrogen utilization strategies, such as the efficient uptake of nitrate by fibrous root systems. Comparable patterns are evident across other monocots: hexaploid wheat carries 331 NPF genes grouped into 113 homoeologous sets, and maize contains 78 NPF members partitioned into the eight canonical subfamilies, underscoring a monocot tendency toward repertoire expansion with subfamily-level conservation and functional diversification [28,29].
Analysis of gene expansion mechanisms revealed that 22 pairs of tandem repeat genes and 10 pairs of fragments repeat genes drove the evolution of the SbNPF family. Most repeat gene pairs had Ka/Ks ratios less than 1, indicating that purifying selection primarily maintains function. This pattern is similar to that observed in the Arabidopsis NPF family through tandem repeats [3]. However, the role of tandem repeats is more pronounced in sorghum, potentially due to the enrichment of chromosomal regions in its genome. This enrichment may have facilitated the refinement of functional specialization within subfamilies. For example, the NPF5 subfamily contains 19 members that are hypothesized to be involved in complex nitrogen allocation regulation.
4.2. Gene Structure and Cis-Elements: The Molecular Basis of Functional Diversity
The number of exons in SbNPF genes varies significantly (1–8), and structural preferences are observed within subfamilies (e.g., the NPF4 subgroup often contains 3 or 6 introns). This structural variation may drive functional diversification through exon shuffling mechanisms. For example, AtNPF6.3 achieves both nitrate transport and IAA signaling through a specific exon combination [30], while members of the NPF6 subfamily in sorghum (5 genes) have exon numbers concentrated at 3–4, suggesting they may retain similar multifunctionality.
Promoter cis-element analysis revealed the potential of SbNPFs to broadly participate in stress responses and hormone regulation: the enrichment of elements such as ABRE (abscisic acid response) and CGTCA-motif (jasmonic acid response) aligns with the hormone-regulated characteristics of NPF genes in Arabidopsis [8]. Additionally, the presence of MBS (drought response) and LTR (cold response) elements provides molecular evidence for SbNPFs’ involvement in abiotic stress responses, which aligns with the upregulation pattern of NRT1.7 in Salicornia under salt stress [13]. Consistent with these promoter signatures, multiple Arabidopsis NPFs have been experimentally verified as hormone carriers: NPF4.6/AIT1 imports ABA, and NPF3 transports GA (and ABA in vitro). Post-translational regulation fine-tunes NPF4.6 function, with CEPR2 phosphorylation stabilizing ABA transport activity and SOS2 phosphorylation modulating Na+ influx under salt stress, thereby linking nutrient transport to stress adaptation [31,32,33,34]. It is suggesting that sorghum NPF genes may coordinate the balance between nitrogen utilization and stress resistance by integrating hormone signals with environmental stress signals.
4.3. Tissue-Specific Expression and Stress Response: The Physiological Significance of Functional Division
Members such as SbNPF4.9 and SbNPF5.11 are highly expressed in roots and may be similar to rice OsNRT1.1a/b in their role in nitrate absorption in the root system [13]. SbNPF6.3, which is enriched in leaves, may function similarly to AtNPF6.2 by maintaining the nitrogen balance in leaves and regulating nitrate storage in vacuoles [9]. SbNPF4.14 and other members that are specifically expressed in stems are speculated to participate in long-distance nitrogen transport, similar to the role of AtNPF7.3 in nitrate loading in the xylem [10]. These tissue-biased patterns scale to agronomic outcomes in cereals: natural variation at OsNRT1.1B/OsNPF6.5 underlies indica–japonica divergence in nitrate use and improves grain yield and NUE in field trials; overexpressing OsNPF3.1 enhances NUE and growth; notably, OsNRT1.1B also shapes root-associated microbiota that support nitrogen acquisition, emphasizing how NPF activity links organ-level distribution to whole-plant performance [35,36,37]. Furthermore, we took a RT-qPCR under LN stress to evaluation the gene expression pattern of SbNPF. The reason to perform RT-qPCR under LN conditions because members of the NPF gene family are primarily involved in nitrate uptake, transport, and redistribution in plants. Nitrogen deficiency is one of the most common environmental stresses affecting crop growth and productivity, and it strongly induces the expression of nitrate transporters. By applying LN treatment, we aimed to reveal the transcriptional responsiveness of SbNPF genes to nitrogen limitation, thereby providing functional clues about their potential roles in regulating nitrogen transport and utilization under stress conditions. This approach also allowed us to identify candidate SbNPF genes that may contribute to improving nitrogen use efficiency in sorghum, which is of great importance for cultivation in nutrient-poor soils. The results indicated that the upregulation of SbNPF1.1, SbNPF2.6, and others in the aboveground parts may adapt to nutrient deficiency by enhancing nitrate redistribution. This is similar to the function of AtNPF7.2, which promotes nitrogen transport from old leaves to new leaves under low-nitrogen conditions [11]; whereas the downregulation of SbNPF1.2 in roots may reflect an adaptive resource allocation strategy that prioritizes critical metabolic processes in the aboveground parts. Notably, the differential expression of SbNPF2.7 between roots and aboveground parts (down- vs. up-regulated) suggests that it may switch functions through tissue-specific splicing. This phenomenon has not been reported previously in the Arabidopsis NPF family and warrants further functional validation. The functions of certain NPF genes in rice (such as OsNPF6.5 (OsNRT1.1A)) have been demonstrated to be significantly correlated with nitrogen fertilizer use efficiency. We compared homologs of these functionally important genes in sorghum and identified differences in their coding sequences and promoter cis-elements. For example, the response patterns and chromatin accessibility regulation of the rice OsNRT2 gene family under low-nitrogen stress differ from those in wheat, suggesting that the sorghum NPF gene family may also possess unique regulatory mechanisms [38]. In future work, we plan to utilize plant transgenic systems to validate the substrate selectivity and transport functions of these key sorghum NPF genes, thereby directly confirming their functional divergence from rice and Arabidopsis homologs.
4.4. Research Limitations and Future Prospects
This study systematically analyzed the evolutionary and expression characteristics of the SbNPF family. However, several limitations remain. First, the substrate specificity of SbNPFs remains unclear e.g., do they transport nitrate and ABA simultaneously, as AtNPF4.1 does? Second, the functional redundancy and differentiation mechanisms of tandem repeat genes require validation through in vitro experiments. Future studies could use heterologous expression systems, such as Xenopus oocytes, to measure transport activity. They could also use CRISPR technology to knockout candidate genes, such as SbNPF6.3 and SbNPF4.5, to investigate their roles in nitrogen use efficiency and stress tolerance.
In summary, the sorghum NPF family has formed a complex functional network through the synergy of evolutionary conservation and species specificity. This study provides key targets for elucidating the molecular mechanisms underlying efficient nitrogen utilization in monocotyledonous crops and establishes a theoretical basis for the molecular breeding of low-nitrogen-tolerant sorghum varieties.
5. Conclusions
This study identified 88 NPF genes in sorghum and classified them into eight subfamilies. Tandem duplication was found to be the major force driving family expansion, while purifying selection maintained functional conservation. Promoter analysis revealed stress- and hormone-related cis-elements, suggesting involvement in abiotic stress responses and signaling pathways. Expression profiling demonstrated clear tissue specificity: some SbNPFs function in nitrate uptake in roots, whereas others participate in transport and allocation in leaves, stems, and reproductive organs. Under low-nitrogen stress, genes such as SbNPF1.1, SbNPF2.6, SbNPF2.7, and SbNPF4.5 were strongly induced in shoots, highlighting their potential roles in nitrogen redistribution. In contrast, repression of SbNPF1.2 and SbNPF3.1 in roots suggests adaptive resource allocation strategies. Collectively, these findings provide valuable insights into the molecular basis of nitrogen utilization in sorghum and identify promising candidate genes for improving nitrogen use efficiency through molecular breeding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15102402/s1, Supplemental Table S1. qPCR primer of SbNPF genes. Supplemental Table S2. The gene IDs, genomic locations, coding sequence lengths, protein lengths, and other characteristics of the identified SbNPF genes. Supplemental Table S3. The NPF duplicate gene pairs in Sorghum bicolor. Supplemental Table S4. The Ka, Ks, and Ka/Ks of NPF duplicate gene pairs in Sorghum bicolor.
Author Contributions
Z.L. and Z.J. wrote the manuscript. Y.L., D.L., S.Z., S.L. and S.S. analyzed the data. C.L. supervised the research and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Major Project of Food Crop Production Based on Technological Application of Liaoning Province (2023JH1/10200001-030-01), China Agriculture Research System of MOF and MARA (CARS-06-14.5-A17) and Phd Starting Foundation of Shenyang Agricultural University (X2021022).
Data Availability Statement
Data are contained within the article or Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Forde, B.G. Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta (BBA) Biomembr. 2000, 1465, 219–235. [Google Scholar] [CrossRef]
- Jargeat, P.; Rekangalt, D.; Verner, M.C.; Gay, G.; Debaud, J.C.; Marmeisse, R.; Fraissinet-Tachet, L. Characterisation and expression analysis of a nitrate transporter and nitrite reductase genes, two members of a gene cluster for nitrate assimilation from the symbiotic basidiomycete Hebeloma cylindrosporum. Curr. Genet. 2003, 43, 199–205. [Google Scholar] [CrossRef]
- Crawford, N.M. Nitrate: Nutrient and signal for plant growth. Plant Cell 1995, 7, 859–868. [Google Scholar] [CrossRef][Green Version]
- Chiba, Y.; Shimizu, T.; Miyakawa, S.; Kanno, Y.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 2015, 128, 679–686. [Google Scholar] [CrossRef]
- Drechsler, N.; Courty, P.-E.; Brulé, D.; Kunze, R. Identification of arbuscular mycorrhiza-inducible Nitrate Transporter 1/Peptide Transporter Family (NPF) genes in rice. Mycorrhiza 2018, 28, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hu, J.; Ma, X.; Li, C.; Yang, Q.; Feng, S.; Li, M.; Li, N.; Song, X. Identification, evolution and expression analyses of whole genome-wide TLP gene family in Brassica napus. BMC Genom. 2020, 21, 264. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Wang, G.; Iqbal, A.; Muhammad, N.; Wang, X.; Gui, H.; Zhang, H.; Kayoumu, M.; Li, X.; Zhang, X.; et al. Identification and Expression Analysis of the NPF Genes in Cotton. Int. J. Mol. Sci. 2022, 23, 14262. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.-Q.; Wang, R.; Chen, M.; Crawford, N.M. The Arabidopsis Dual-Affinity Nitrate Transporter Gene AtNRT1.1 (CHL1) Is Activated and Functions in Nascent Organ Development during Vegetative and Reproductive Growth. Plant Cell 2001, 13, 1761–1777. [Google Scholar] [CrossRef]
- Tsay, Y.-F.; Chiu, C.-C.; Tsai, C.-B.; Ho, C.-H.; Hsu, P.-K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef]
- Cui, Y.-N.; Li, X.-T.; Yuan, J.-Z.; Wang, F.-Z.; Wang, S.-M.; Ma, Q. Nitrate transporter NPF7.3/NRT1.5 plays an essential role in regulating phosphate deficiency responses in Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 508, 314–319. [Google Scholar] [CrossRef]
- Huang, N.-C.; Liu, K.-H.; Lo, H.-J.; Tsay, Y.-F. Cloning and Functional Characterization of an Arabidopsis Nitrate Transporter Gene That Encodes a Constitutive Component of Low-Affinity Uptake. Plant Cell 1999, 11, 1381–1392. [Google Scholar] [CrossRef]
- Dölfors, F.; Ilbäck, J.; Bejai, S.; Fogelqvist, J.; Dixelius, C. Nitrate transporter protein NPF5.12 and major latex-like protein MLP6 are important defense factors against Verticillium longisporum. J. Exp. Bot. 2024, 75, 4148–4164. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, B.; Yuan, D.; Liu, Y.; Che, R.; Hu, Y.; Ou, S.; Liu, Y.; Zhang, Z.; Wang, H.; et al. Expression of the Nitrate Transporter Gene OsNRT1.1A/OsNPF6.3 Confers High Yield and Early Maturation in Rice. Plant Cell 2018, 30, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ouyang, J.; Wang, Y.-Y.; Hu, R.; Xia, K.; Duan, J.; Wang, Y.; Tsay, Y.-F.; Zhang, M. Disruption of the rice nitrate transporter OsNPF2.2 hinders root-to-shoot nitrate transport and vascular development. Sci. Rep. 2015, 5, 9635. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wang, Y.; Hu, L.; Xu, J.; Yan, J.; Cao, P.; Liu, C.; Shi, X.; Liu, C.; Jiang, Y.; et al. The Identification of Kabatiella zeae as a Causal Agent of Northern Anthracnose of Sorghum in China and Estimation of Host Resistance. Plants 2024, 13, 1857. [Google Scholar] [CrossRef]
- Xue, B.; Liang, Z.; Liu, Y.; Li, D.; Cao, P.; Liu, C. Comparative Analysis of Casparian Strip Membrane Domain Protein Family in Oryza sativa (L.) and Arabidopsis thaliana (L.). Int. J. Mol. Sci. 2024, 25, 9858. [Google Scholar] [CrossRef]
- Bateman, A.; Birney, E.; Durbin, R.; Eddy, S.R.; Howe, K.L.; Sonnhammer, E.L. The Pfam Protein Families Database. Nucleic Acids Res. 2002, 30, 276–280. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Liang, Z.; Shi, S.; Xue, B.; Li, D.; Liu, Y.; Liu, C. Comparative Analysis of TALE Gene Family in Gramineae. Agronomy 2025, 15, 1460. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H.; et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom. 2014, 15, 312. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2, 2.3. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Liang, Z.; Liu, Y.; Li, D.; Liu, C. Genome-Wide Identification of the RALF Gene Family and Expression Pattern Analysis in Zea mays (L.) under Abiotic Stresses. Plants 2024, 13, 2883. [Google Scholar] [CrossRef]
- Xue, B.; Liang, Z.; Li, D.; Liu, Y.; Liu, C. Genome-wide identification and expression analysis of CASPL gene family in Zea mays (L.). Front. Plant Sci. 2024, 15, 1477383. [Google Scholar] [CrossRef]
- Liu, C.; Tian, L.; Yu, W.; Wang, Y.; Yao, Z.; Liu, Y.; Yang, L.; Liu, C.; Shi, X.; Liu, T.; et al. Natural variation in SbTEF1 contributes to salt tolerance in sorghum seedlings. J. Integr. Agric. 2024, in press. [Google Scholar] [CrossRef]
- Wang, H.; Wan, Y.; Buchner, P.; King, R.; Ma, H.; Hawkesford, M.J. Phylogeny and gene expression of the complete NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY in Triticum aestivum. J. Exp. Bot. 2020, 71, 4531–4546. [Google Scholar] [CrossRef]
- Jia, L.; Hu, D.; Wang, J.; Liang, Y.; Li, F.; Wang, Y.; Han, Y. Genome-Wide Identification and Functional Analysis of Nitrate Transporter Genes (NPF, NRT2 and NRT3) in Maize. Int. J. Mol. Sci. 2023, 24, 12941. [Google Scholar] [CrossRef]
- Hsu, P.-K.; Tsay, Y.-F. Two Phloem Nitrate Transporters, NRT1.11 and NRT1.12, are Important for Redistributing Xylem-Borne Nitrate to Enhance Plant Growth. Plant Physiol. 2013, 163, 844–856. [Google Scholar] [CrossRef]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [PubMed]
- Tal, I.; Zhang, Y.; Jørgensen, M.E.; Pisanty, O.; Barbosa, I.C.R.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Olsen, C.E.; Weinstain, R.; et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, Z.; Xu, Y.; Yu, M.; Ren, Y.; Zhang, S.; Yang, G.; Huang, J.; Yan, K.; Zheng, C.; et al. Regulation of the stability and ABA import activity of NRT1.2/NPF4.6 by CEPR2-mediated phosphorylation in Arabidopsis. Mol. Plant 2021, 14, 633–646. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, L.; Zhao, Z.; Zheng, Y.; Ren, Y.; Zhao, X.; Zhang, S.; Yang, G.; Huang, J.; Yan, K.; et al. Regulation of the non-selective Na+ importer capacity of NRT1.2/NPF4.6/AIT1 by SOS2-mediated phosphorylation in Arabidopsis. Cell Rep. 2025, 44, 115729. [Google Scholar] [CrossRef]
- Hu, B.; Wang, W.; Doug, S.J.B.; Tang, J.Y.; Li, H.; Che, R.H.; Zhang, Z.H.; Chai, X.Y.; Wang, H.R.; Wang, Y.Q.; et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834–838. [Google Scholar] [CrossRef]
- Yang, X.; Nong, B.; Chen, C.; Wang, J.; Xia, X.; Zhang, Z.; Wei, Y.; Zeng, Y.; Feng, R.; Wu, Y.; et al. OsNPF3.1, a member of the NRT1/PTR family, increases nitrogen use efficiency and biomass production in rice. Crop. J. 2023, 11, 108–118. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Cui, F.; Han, X.; He, Y.; Zhao, L.; Zhang, N.; Zhang, H.; Zhu, H.; Liu, Z.; Ma, B.; et al. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204. Mol. Plant 2022, 15, 1440–1456. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).