Integrative Genomics and Precision Breeding for Stress-Resilient Cotton: Recent Advances and Prospects
Abstract
1. Introduction
2. Genomic Characterisation and Utilisation of Cotton Germplasm for Pre-Breeding and Genetic Improvement
3. Genome Sequencing Paving the Way for Molecular Marker Identification for Precision Breeding
4. Cotton Genome Sequencing Projects
5. Emerging Pangenomic Frameworks for Structural Variation Analysis
6. Progress in Molecular Marker Development for Genetic Linkage Mapping
| Population/Panel | Genotyping Assay | N (Mapping) | Mapped Markers | Map Distance (cm) | QTLs/Loci | Trait(s) | Reference |
|---|---|---|---|---|---|---|---|
| Multiple populations (meta-QTL synthesis) | Mixed (SSR/AFLP/SNP; published QTLs) | — | — | — | 300+ QTLs integrated | Abiotic & biotic stress (VW, FW, RKN, drought, salt) | [6,100] |
| GWAS panel (diverse Upland cotton) | CottonSNP63K array (Illumina) | 120 accessions | 63,000 SNPs (genotyped) | — | 5 QTLs/18 lead SNPs | Verticillium wilt resistance | [101,102] |
| RIL (G. hirsutum × G. barbadense) | High-density SNP (GBS + array) | not stated | high-density map | 2865 | 119 QTLs (DI & incidence) | Verticillium wilt resistance | [103] |
| BC1/BIL population | SSR (~175–400) | 94–200 | 175–400 | 2895–4403 | 10–17 QTLs | Verticillium wilt/heat tolerance | [97] |
| RIL (AS2 × MCU-13) | GBS + SSRs | 48 | 1898 | 2808.3 | 19 QTLs | Drought tolerance (chlorophyll stability index, proline, etc.) | [93] |
| GWAS panel (US Upland cotton) | CottonSNP63K | 376 | 26,301 polymorphic SNPs | — | 53 (drought), 78 (salt), 8 (thrips) | Seedling-stage drought, salt, thrips | [104] |
| F2:3 interspecific (G. tomentosum × G. hirsutum) | GBS (~93k SNPs) | 277 | 93,384 SNPs; 26 LGs | — | QTL clusters | Salt tolerance (seedling stage) | [105] |
| Introgression lines from wild donor (BC-derived) | GBS/resequencing | — | dense SNPs | — | 30 stable QTLs + 89 candidates | Drought-tolerance physiological traits | [106] |
| Multiple populations (NemX, SJ-2, Pima S-7, etc.) | SSRs linked to rkn1 | multiple | — | — | major-effect QTL(s) | Root-knot nematode resistance | [107,108] |
| Natural panel (200 accessions) | SLAF-seq + GWAS | 200 | ~10,000 SNPs | — | significant SNPs; GhSAD1 | Cold tolerance (ABA-mediated) | [94] |
| BSA-seq (extreme pools) | BSA-seq | pools | — | 6.27 Mb (physical) | 3 QTL intervals (A13, A10, A07) | Drought resistance (PH, SBW) | [109,110] |
7. Advances of Genomics Approach in Marker-Assisted Breeding
8. Biotic Stress Resistance
| Pathogen/Stress | Key QTLs/Candidate Genes | Population Type | Marker System/Platform | Main Finding/Significance | Reference |
|---|---|---|---|---|---|
| Verticillium wilt (VW) | QTL clusters on c16 and c19 | 376 upland cotton accessions (GWAS panel) | CottonSNP63K array (high density) | Identified 15 VW and 13 FOV4 QTLs; clusters on c16 and c19 were consistent across environments | [153,154,155] |
| Verticillium wilt (VW) | Major QTLs on c16 (D7) and c23 (D9) | Backcross Inbred Line (BIL) population | SSR-based linkage map (~2895 cM, 392 SSRs) | Ten VW QTLs and 28 clusters identified; hotspots correlated with NBS gene density | [143,156] |
| Verticillium wilt (VW) | SNPs on A01 and A10 (GhAMT2 candidate) | Multi-parent Advanced Generation Inter-cross (MAGIC) population | GWAS integrated with transcriptomics | Major QTLs identified; GhAMT2 implicated in VW response and resistance signalling | [157,158] |
| Cotton blue disease (CBD) | Cbd gene on chromosome 10 (0.75 cM) | ΔOpal F2 families | SSR marker (DC20027) + SNP haplotype analysis | Early example of MAS: trait tagging via haplotype-based SNP markers for CBD resistance | [159] |
| Cotton bacterial blight (CBB) | GhSWEET10 (TAL effector target) | Pathogen–cotton interaction study | Transcriptomics + functional validation | GhSWEET10 induced by TAL effector Avrb6; gene silencing reduced susceptibility | [160] |
9. Abiotic Stress
| Stress Type | Population/Material | Marker/Platform | Approach | Key Findings/Loci | Candidate Genes | Reference |
|---|---|---|---|---|---|---|
| Drought | F2/F3 (G. hirsutum × G. barbadense) | RFLP, SSR | QTL mapping | 33 QTLs for water-use efficiency, photosynthesis, productivity | WUE/photosynthesis pathways | [173] |
| Upland cotton (multiple populations) | SSR | QTL mapping | QTLs for physiology, yield, architecture | – | [174] | |
| Segregating breeding lines | SSR (NAU2715, NAU2954, BNL loci) | QTL mapping, MAS | NAU2715/2954 linked to RWC; BNL loci with osmotic adjustment | – | [93] | |
| F2:3 (G. tomentosum × G. hirsutum) | SSR | QTL mapping | 13 QTL clusters on 9 chromosomes; consistent loci | – | [175] | |
| Upland cotton diversity panel | SNP (array/GBS) | GWAS | Loci for yield under drought stress | Stress-response pathways | [176] | |
| Salinity | Chinese/U.S. panels (196–323 accessions) | CottonSNP70K | GWAS | Stage-specific salt-tolerance SNPs | Ion transporters, osmotic adjustment genes | [118] |
| 320 G. hirsutum accessions | SNP array + RNA-seq | GWAS × transcriptomics | 33 SNPs, 13 QTLs, 98 candidate genes | ROS scavengers, signalling genes, transporters | [105] | |
| 268 upland cotton accessions | CottonSNP70K | GWAS + transcriptomics | 27 SNPs (15 salt-tolerance index) | Gh_D01G0943, Gh_A01G0906 | [117] | |
| Upland panel (multi-environment) | GBS, SNP arrays | GWAS | Consistent drought & salinity QTLs | ABA signalling & ion transport | [177] | |
| Heat | Elite Pakistani breeding lines | SSR, KASP | MAS pyramiding | Improved cell membrane stability & RWC | Heat-shock proteins, membrane stability | [178] |
| Natural diversity panel | SNP array/GBS | Multi-locus GWAS | QTNs for heat tolerance across traits | ERF, HSF, HSP families | [179] | |
| Cold | 110 G. hirsutum genotypes | SSR (101 markers) | Assoc. mapping | 16 marker–trait associations; 10 major loci (PVE > 10%) | BNL0569, CIR081, CIR202 | [180] |
| Transgenic G. hirsutum lines | Overexpression/genome editing | Functional validation | Genes enhancing drought, cold, and heat tolerance | GhDREB1B, GhKCS13, AtSAP5, AmCBF1 | [181] |
10. Agronomic Traits
| Trait Focus | Population/Material | Marker/Platform | Approach | Key loci/Findings | Candidate Genes (if Reported) | Reference |
|---|---|---|---|---|---|---|
| Fibre quality (strength, length) | Segregating BC1F4 populations | RAPD → SCAR (SCAR1920) | MAS | Marker linked to QTL for fibre strength; improved genotype selection | – | [198] |
| Fibre strength (QTLFS1) | Segregating populations | SSRs | QTL mapping + MAS | DNA markers linked to QTLFS1 | – | [199] |
| Fibre yield & quality | RIL population (G. hirsutum) | SSRs | QTL mapping | Stable QTLs for lint % and fibre length | – | [200,201] |
| Fibre quality (multiple traits) | MAGIC population (G. hirsutum) | Whole-genome resequencing SNPs | GWAS | Seven major regions; 14 genes with non-synonymous SNPs | Structural & regulatory genes | [191] |
| Fibre quality (length, strength, fineness) | 334 G. hirsutum accessions | SSRs, EST-SSRs | LD mapping | 12–22 markers linked with fibre traits across environments | – | [186] |
| Fibre quality (yield & properties) | G. hirsutum × G. barbadense (F2) | SSRs | QTL mapping | QTLs for lint %, lint yield, fibre traits | – | [89] |
| Fibre traits (yield, architecture) | Upland cotton panels | SNP arrays (63K Illumina) | GWAS | ~10,500 SNPs; 46 significant loci across 15 chromosomes | 612 genes (163 for fibre length, 120 for strength) | [36,202] |
| Fibre quality (13 traits) | 419 accessions resequenced | WGS SNPs (~3.6M) | GWAS | Multiple novel associations | – | [203] |
| Agronomic + fibre quality traits | Diverse germplasm panels | SNPs + GS models | GWAS + Genomic prediction | SNPs strongly associated with yield + fibre traits | – | [129,204,205,206] |
11. Leveraging Markers for Cotton Breeding Using Genomic Selection Approach
| Trait Focus | Population/Material | Marker/Platform | Approach/Models Compared | Key Findings/Prediction Accuracy | Candidate Genes (if Reported) | Reference |
|---|---|---|---|---|---|---|
| Fibre length & strength | 215 upland cotton lines (Australia) | 13,330 SNPs (array) | GS with BayesB, GBLUP, rrBLUP, M × E models | Multi-environment GS improved accuracy: 0.71 (length), 0.59 (strength) | – | [202] |
| Fibre quality traits (6) | 550 RILs (MAGIC population) | 6292 SNPs (GBS) | GBLUP, rrBLUP, BayesB, Bayes LASSO, RKHS | Prediction accuracy 0.55–0.69; BayesB most effective | – | [192] |
| Fibre traits & yield | 80 elite upland cultivars (14 environments) | CottonSNP63K array | GBLUP, BayesB, GWAS-assisted GS | Accuracy 0.45–0.68 depending on trait; GWAS signals increased predictive power | – | [133] |
| Fibre length, uniformity, micronaire | 1385 CSIRO breeding lines | 12,296 SNPs (GBS) | Bayesian ridge regression, BayesB, ML models | Prediction accuracy up to 0.76 (length), 0.65 (strength), 0.64 (yield); combining pedigree + genomic data improved predictions | – | [218] |
| Fibre yield & quality | 300 upland cotton accessions | CottonSNP70K array | GBLUP, BayesCπ, Random Forest (ML + Bayesian) | Prediction accuracy 0.52–0.72; combined models gave stable predictions | – | [219] |
12. Harnessing Genomics and CRISPR/Cas Systems for Next-Generation Precision Breeding
12.1. Fibre Quality Enhancement
12.2. Cotton Oil Enhancement
12.3. Fibre Enhancement
| No. | Author (Year) | Target Gene(s) | Effect on Fibre Length |
|---|---|---|---|
| 1 | Wu et al. (2024) [222] | GhHDZ76 | Shortening |
| 2 | Wang et al. (2024) [256] | GhFAD3-4 | Shortening |
| 3 | Jia et al. (2024) [248] | GhFAD7A-1 | Shortening |
| 4 | Jia et al. (2024) [248] | GhKNOX6 | Increasing |
| 5 | Li et al. (2024) [249] | GhATL68b | Shortening |
| 6 | Jiao et al. (2025) [247] | GhMDHAR1A/GhDHAR2A | Increasing |
| 7 | Zhu et al. (2023) [250] | GhEXPA3-1 | Shortening |
| 8 | Tian et al. (2024) [227] | GhGRF4/GhARF2-GhGASA24 | Shortening |
| 9 | Zhu et al. (2021) [225] | GhAlaRP | Shortening |
13. Development of Stress-Resistant Cotton
14. Cross-Talk Between Genomics and Speed Breeding for Fast-Tracked Cotton Improvement
15. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kabish, A.K.; Degefu, D.T.; Gebregiorgis, Z.D. Cotton Value Chain and Economics. In Cotton Sector Development in Ethiopia: Challenges and Opportunities; Springer: Berlin/Heidelberg, Germany, 2024; pp. 441–463. [Google Scholar]
- Bitew, Y.; Abate, A. Cotton Agronomy and Production. In Cotton Sector Development in Ethiopia: Challenges and Opportunities; Springer: Berlin/Heidelberg, Germany, 2024; pp. 3–17. [Google Scholar]
- Juneja, R.; Gupta, A.; Gulati, A. Gene Revolution in Agriculture: Case of Cotton in India. In From Farm to Foreign; Indian Council for Research on International Economic Relations (ICRIER): New Delhi, India, 2025; p. 12. [Google Scholar]
- Poghosyan, A.; Isengildina-Massa, O.; Stewart, S.L. Futures-Based Forecasts of Cotton Prices: Beyond Historical Averages. J. Agric. Appl. Econ. 2025, 57, 114–134. [Google Scholar] [CrossRef]
- Dohlman, E.; Hansen, J.; Chambers, W.; Interagency Agricultural Projections Committee. USDA Agricultural Projections to 2034. 2025. Available online: https://ageconsearch.umn.edu/record/350164?v=pdf (accessed on 1 August 2025).
- Kebede, M. Food and Nutrition (Cotton as a Feed and Food Crop). In Cotton Sector Development in Ethiopia: Challenges and Opportunities; Springer: Berlin/Heidelberg, Germany, 2024; pp. 379–412. [Google Scholar]
- Hussain, M.; Gao, X.; Qin, D.; Qin, X.; Wu, G. Role of biotic and abiotic factors for sustainable cotton production. In Best Crop Management and Processing Practices for Sustainable Cotton Production; IntechOpen: London, UK, 2023. [Google Scholar]
- Fu, J. Gene Stacking Strategies to Enhance the Durability of Bt Crops. Bt Res. 2024, 15, 96–109. [Google Scholar] [CrossRef]
- Kulwal, P.L.; Mir, R.R.; Varshney, R.K. Efficient Breeding of Crop Plants. In Fundamentals of Field Crop Breeding; Springer: Berlin/Heidelberg, Germany, 2022; pp. 745–777. [Google Scholar]
- Montalvo, N.; Requena, F.; Capriotti, E.; Rausell, A. Federated Learning for the pathogenicity annotation of genetic variants in multi-site clinical settings. Bioinformatics 2025, btaf523. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Anwar, S.; Abbas, M.; Aneeq, M.; de Jong, F.; Ayaz, M.; Wei, Y.; Zhang, R. Impacts of climate change on cotton production and advancements in genomic approaches for stress resilience enhancement. J. Cotton Res. 2025, 8, 17. [Google Scholar] [CrossRef]
- Tyagi, A.; Mir, Z.A.; Almalki, M.A.; Deshmukh, R.; Ali, S. Genomics-assisted breeding: A powerful breeding approach for improving plant growth and stress resilience. Agronomy 2024, 14, 1128. [Google Scholar] [CrossRef]
- Alemu, A.; Åstrand, J.; Montesinos-Lopez, O.A.; y Sanchez, J.I.; Fernandez-Gonzalez, J.; Tadesse, W.; Vetukuri, R.R.; Carlsson, A.S.; Ceplitis, A.; Crossa, J. Genomic selection in plant breeding: Key factors shaping two decades of progress. Mol. Plant 2024, 17, 552–578. [Google Scholar] [CrossRef]
- Vikram, P.; Shokat, S.; Mohan, A.; Sehgal, D.; Kashyap, M. Genomics assisted improvement of crop plants for adaptation to marginal environments. Front. Genet. 2024, 15, 1461709. [Google Scholar] [CrossRef]
- De Santiago, L.M. Identifying, Mapping and Overcoming Genomic Impediments to Intraspecific Genetic Improvement of Upland Cotton Through Interspecific Hybridization and Introgression. 2020. Available online: https://hdl.handle.net/1969.1/192244 (accessed on 1 August 2025).
- Mangal, V.; Verma, L.K.; Singh, S.K.; Saxena, K.; Roy, A.; Karn, A.; Rohit, R.; Kashyap, S.; Bhatt, A.; Sood, S. Triumphs of genomic-assisted breeding in crop improvement. Heliyon 2024, 10, e35513. [Google Scholar] [CrossRef]
- Chavhan, R.L.; Hinge, V.R.; Wankhade, D.J.; Deshmukh, A.S.; Mahajan, N.; Kadam, U.S. Bioinformatics for molecular breeding and enhanced crop performance: Applications and perspectives. In Bioinformatics for Plant Research and Crop Breeding; Wiley: Hoboken, NJ, USA, 2024; pp. 21–74. [Google Scholar]
- Kumar, R.; Das, J.; Puttaswamy, R.K.; Kumar, M.; Balasubramani, G.; Prasad, Y.G. Targeted genome editing for cotton improvement: Prospects and challenges. Nucleus 2024, 67, 181–203. [Google Scholar] [CrossRef]
- Hui, F.; Tang, X.; Li, B.; Alariqi, M.; Xu, Z.; Meng, Q.; Hu, Y.; Wang, G.; Zhang, Y.; Zhang, X. Robust CRISPR/Mb2Cas12a genome editing tools in cotton plants. Imeta 2024, 3, e209. [Google Scholar] [CrossRef]
- Li, C.; Tuerxun, Z.; Yang, Y.; Li, X.; Hui, F.; Li, J.; Liu, Z.; Chen, G.; Cai, D.; Zhang, H. Application of an endogenous pGhαGloA promoter in CRISPR/Cas12a system for efficient genome editing to creat glandless cotton germplasm. J. Integr. Agric. 2024, in press. [CrossRef]
- Samantara, K.; Bohra, A.; Mohapatra, S.R.; Prihatini, R.; Asibe, F.; Singh, L.; Reyes, V.P.; Tiwari, A.; Maurya, A.K.; Croser, J.S. Breeding more crops in less time: A perspective on speed breeding. Biology 2022, 11, 275. [Google Scholar] [CrossRef]
- Yunus, M.H.; Firdaus, A.; Khan, Z.; Ansari, M.Y.K. Genomics-Assisted Breeding (GAB) for Trait Improvement: Unveiling Genomic Strategies for Accelerated Crop Enhancement. In Plant Breeding Technology: Future Trends and Challenges; CABI GB: Oxfordshire, UK, 2025; pp. 138–165. [Google Scholar]
- Amin, A.; Zaman, W.; Park, S. Harnessing Multi-Omics and Predictive Modeling for Climate-Resilient Crop Breeding: From Genomes to Fields. Genes 2025, 16, 809. [Google Scholar] [CrossRef]
- Kun, W.; Shoupu, H.; Yuxian, Z. Cotton2035: From genomics research to optimized breeding. Mol. Plant 2025, 18, 298–312. [Google Scholar] [CrossRef] [PubMed]
- Conaty, W.C.; Broughton, K.J.; Egan, L.M.; Li, X.; Li, Z.; Liu, S.; Llewellyn, D.J.; MacMillan, C.P.; Moncuquet, P.; Rolland, V. Cotton breeding in Australia: Meeting the challenges of the 21st century. Front. Plant Sci. 2022, 13, 904131. [Google Scholar] [CrossRef] [PubMed]
- Robert, P.; Goudemand, E.; Auzanneau, J.; Oury, F.-X.; Rolland, B.; Heumez, E.; Bouchet, S.; Caillebotte, A.; Mary-Huard, T.; Le Gouis, J. Phenomic selection in wheat breeding: Prediction of the genotype-by-environment interaction in multi-environment breeding trials. Theor. Appl. Genet. 2022, 135, 3337–3356. [Google Scholar] [CrossRef]
- Bakala, H.S.; Mandahal, K.S.; Sarao, L.K.; Srivastava, P. Breeding Wheat for Biotic Stress Resistance: Achievements, Challenges and Prospects; IntechOpen: London, UK, 2021. [Google Scholar]
- Ruperao, P. Development of a core set from large germplasm collections in genebank. In Bioinformatics for Plant Research and Crop Breeding; Wiley: Hoboken, NJ, USA, 2024; pp. 269–282. [Google Scholar]
- Panahi, B.; Hosseinzadeh Gharajeh, N.; Mohammadzadeh Jalaly, H. Advances in barley germplasm diversity characterization through next-generation sequencing approach. Genet. Resour. Crop Evol. 2025, 72, 3829–3843. [Google Scholar] [CrossRef]
- Nguyen, G.N.; Norton, S.L. Genebank phenomics: A strategic approach to enhance value and utilization of crop germplasm. Plants 2020, 9, 817. [Google Scholar] [CrossRef]
- Hinze, L.L.; Hulse-Kemp, A.M.; Wilson, I.W.; Zhu, Q.-H.; Llewellyn, D.J.; Taylor, J.M.; Spriggs, A.; Fang, D.D.; Ulloa, M.; Burke, J.J. Diversity analysis of cotton (Gossypium hirsutum L.) germplasm using the CottonSNP63K Array. BMC Plant Biol. 2017, 17, 37. [Google Scholar] [CrossRef]
- Orken, A.; Manabayeva, S.; Makhmadjanov, S.; Ramazanova, M.; Kali, B.; Rakhimzhanova, A.; Tussipkan, D. Cotton (Gossypium L.) global distribution and adaptation to different geographic region. J. Biol. Res. 2025, 1, 43–55. [Google Scholar] [CrossRef]
- Ali, Z.; Maryam, H.; Saddique, M.A.B.; Ikram, R.M. Exploiting genetic diversity in enhancing phenotypic plasticity to develop climate-resilient cotton. Genet. Resour. Crop Evol. 2023, 70, 1305–1320. [Google Scholar] [CrossRef]
- Zhu, D.; Li, X.; Wang, Z.; You, C.; Nie, X.; Sun, J.; Zhang, X.; Zhang, D.; Lin, Z. Genetic dissection of an allotetraploid interspecific CSSLs guides interspecific genetics and breeding in cotton. BMC Genom. 2020, 21, 431. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; De Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Joshi, B.; Singh, S.; Tiwari, G.J.; Kumar, H.; Boopathi, N.M.; Jaiswal, S.; Adhikari, D.; Kumar, D.; Sawant, S.V.; Iquebal, M.A. Genome-wide association study of fiber yield-related traits uncovers the novel genomic regions and candidate genes in Indian upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 2023, 14, 1252746. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhang, Y.; Wu, L.; Zhang, G.; Sun, Z.; Li, Z.; Jiang, Y.; Ke, H.; Chen, B.; Liu, Z. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement. Nat. Genet. 2021, 53, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Xie, P.; Xu, Z.; Tang, J.; Hui, L.; Gu, J.; Gu, X.; Jiang, S.; Rong, Y.; Zhang, J. Pangenome analysis reveals yield-and fiber-related diversity and interspecific gene flow in Gossypium barbadense L. Nat. Commun. 2025, 16, 4995. [Google Scholar] [CrossRef]
- Hitzelberger, J.C. Development and Characterization of Chromosome Substitution and Chromosome Segment Substitution Lines in Cotton (Gossypium spp.). 2022. Available online: https://hdl.handle.net/1969.1/198676 (accessed on 10 September 2025).
- Zhang, F.; Wang, J.; Chen, Y.; Huang, J.; Liang, W. Genome-Wide Identification of MKK Gene Family and Response to Hormone and Abiotic Stress in Rice. Plants 2024, 13, 2922. [Google Scholar] [CrossRef]
- Shrestha, A. Utilizing the Potential of Landraces as Novel Sources of Genetic Variation for the Agronomic Improvement of Upland Cotton (Gossypium hirsutum). 2025. Available online: https://hdl.handle.net/2346/103216 (accessed on 1 September 2025).
- Meshram, P. Plant Breeding for Resistance to Pests and Diseases; Academic Guru Publishing House: Bhopal, India, 2025; ISBN 978-93-49028-90-6. [Google Scholar]
- Ćeran, M.; Miladinović, D.; Đorđević, V.; Trkulja, D.; Radanović, A.; Glogovac, S.; Kondić-Špika, A. Genomics-assisted speed breeding for crop improvement: Present and future. Front. Sustain. Food Syst. 2024, 8, 1383302. [Google Scholar] [CrossRef]
- Sinha, D.; Maurya, A.K.; Abdi, G.; Majeed, M.; Agarwal, R.; Mukherjee, R.; Ganguly, S.; Aziz, R.; Bhatia, M.; Majgaonkar, A. Integrated genomic selection for accelerating breeding programs of climate-smart cereals. Genes 2023, 14, 1484. [Google Scholar] [CrossRef]
- Paux, E.; Lafarge, S.; Balfourier, F.; Derory, J.; Charmet, G.; Alaux, M.; Perchet, G.; Bondoux, M.; Baret, F.; Barillot, R. Breeding for economically and environmentally sustainable wheat varieties: An integrated approach from genomics to selection. Biology 2022, 11, 149. [Google Scholar] [CrossRef]
- Chaney, L.; Sharp, A.R.; Evans, C.R.; Udall, J.A. Genome mapping in plant comparative genomics. Trends Plant Sci. 2016, 21, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; van Wersch, R.; Zhang, Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant-Microbe Interact. 2018, 31, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, J.; Lin, W.; Li, S.; Li, H.; Zhou, J.; Ni, P.; Dong, W.; Hu, S.; Zeng, C. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3, e38. [Google Scholar] [CrossRef]
- Hu, J.; Huang, L.; Chen, G.; Liu, H.; Zhang, Y.; Zhang, R.; Zhang, S.; Liu, J.; Hu, Q.; Hu, F. The elite alleles of OsSPL4 regulate grain size and increase grain yield in rice. Rice 2021, 14, 1–18. [Google Scholar] [CrossRef]
- Hu, G.; Grover, C.E.; Jareczek, J.; Yuan, D.; Dong, Y.; Miller, E.; Conover, J.L.; Wendel, J.F. Evolution and diversity of the cotton genome. In Cotton Precision Breeding; Springer: Berlin/Heidelberg, Germany, 2021; pp. 25–78. [Google Scholar]
- Fang, L.; Gong, H.; Hu, Y.; Liu, C.; Zhou, B.; Huang, T.; Wang, Y.; Chen, S.; Fang, D.D.; Du, X. Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol. 2017, 18, 33. [Google Scholar] [CrossRef]
- Huang, G.; Huang, J.-Q.; Chen, X.-Y.; Zhu, Y.-X. Recent advances and future perspectives in cotton research. Annu. Rev. Plant Biol. 2021, 72, 437–462. [Google Scholar] [CrossRef]
- Udall, J.A.; Long, E.; Hanson, C.; Yuan, D.; Ramaraj, T.; Conover, J.L.; Gong, L.; Arick, M.A.; Grover, C.E.; Peterson, D.G. De novo genome sequence assemblies of Gossypium raimondii and Gossypium turneri. G3 Genes Genomes Genet. 2019, 9, 3079–3085. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Deschamps, S.; Llaca, V. Strategies for sequence assembly of plant genomes. In Plant Genomics; IntechOpen: London, UK, 2016. [Google Scholar][Green Version]
- Hulse-Kemp, A.M. Development of Genomic Markers and Mapping Tools for Assembling the Allotetraploid Gossypium hirsutum L. Draft Genome Sequence. 2015. Available online: https://hdl.handle.net/1969.1/155055 (accessed on 1 September 2025).[Green Version]
- Liu, X.; Zhao, B.; Zheng, H.-J.; Hu, Y.; Lu, G.; Yang, C.-Q.; Chen, J.-D.; Chen, J.-J.; Chen, D.-Y.; Zhang, L. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 2015, 5, 14139. [Google Scholar] [CrossRef]
- Chang, X.; He, X.; Li, J.; Liu, Z.; Pi, R.; Luo, X.; Wang, R.; Hu, X.; Lu, S.; Zhang, X. High-quality Gossypium hirsutum and Gossypium barbadense genome assemblies reveal the landscape and evolution of centromeres. Plant Commun. 2024, 5, 100722. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Sreedasyam, A.; Lovell, J.T.; Mamidi, S.; Khanal, S.; Jenkins, J.W.; Plott, C.; Bryan, K.B.; Li, Z.; Shu, S.; Carlson, J. Genome resources for three modern cotton lines guide future breeding efforts. Nat. Plants 2024, 10, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Sreedasyam, A.; Ando, A.; Song, Q.; De Santiago, L.M.; Hulse-Kemp, A.M.; Ding, M.; Ye, W.; Kirkbride, R.C.; Jenkins, J. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020, 52, 525–533. [Google Scholar] [CrossRef]
- Yu, J.; Jung, S.; Cheng, C.-H.; Ficklin, S.P.; Lee, T.; Zheng, P.; Jones, D.; Percy, R.G.; Main, D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014, 42, D1229–D1236. [Google Scholar] [CrossRef]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Zaynab, M.; Sharif, Y.; Al-Yahyai, R.; Hussain, A.; Sadder, M.; Perveen, K.; Bukhari, N.A.; Li, S. Genome-wide and expression analysis to understand the DUF789 gene family during development of Arabidopsis thaliana. J. King Saud Univ. Sci. 2024, 36, 103478. [Google Scholar] [CrossRef]
- Shan, S.; Spoelhof, J.P.; Blischak, P.D.; Batley, J.; Soltis, P.S.; Soltis, D.E.; Edwards, D. Pangenomes provide new insights into polyploidy in plants. Evol. J. Linn. Soc. 2025, 4, kzaf010. [Google Scholar] [CrossRef]
- He, W.; Li, X.; Qian, Q.; Shang, L. The developments and prospects of plant super-pangenomes: Demands, approaches, and applications. Plant Commun. 2025, 6, 101230. [Google Scholar] [CrossRef]
- Li, J.; Yuan, D.; Wang, P.; Wang, Q.; Sun, M.; Liu, Z.; Si, H.; Xu, Z.; Ma, Y.; Zhang, B. Cotton pan-genome retrieves the lost sequences and genes during domestication and selection. Genome Biol. 2021, 22, 119. [Google Scholar] [CrossRef]
- Mendu, L.; Ghose, K.; Mendu, V. Population Genomics of Cotton. In Population Genomics: Crop Plants; Springer: Berlin/Heidelberg, Germany, 2022; pp. 691–740. [Google Scholar]
- Secomandi, S.; Gallo, G.R.; Rossi, R.; Rodríguez Fernandes, C.; Jarvis, E.D.; Bonisoli-Alquati, A.; Gianfranceschi, L.; Formenti, G. Pangenome graphs and their applications in biodiversity genomics. Nat. Genet. 2025, 57, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Han, Z.; Hu, Y.; Si, Z.; Dai, F.; He, L.; Cheng, Y.; Li, Y.; Zhao, T.; Fang, L. Structural variation (SV)-based pan-genome and GWAS reveal the impacts of SVs on the speciation and diversification of allotetraploid cottons. Mol. Plant 2023, 16, 678–693. [Google Scholar] [CrossRef]
- Li, X.; Jin, X.; Wang, H.; Zhang, X.; Lin, Z. Structure, evolution, and comparative genomics of tetraploid cotton based on a high-density genetic linkage map. DNA Res. 2016, 23, 283–293. [Google Scholar] [CrossRef]
- Green, E.L. Linkage, recombination and mapping. In Genetics and Probability in Animal Breeding Experiments; Springer: Berlin/Heidelberg, Germany, 1981; pp. 77–113. [Google Scholar]
- Amom, T.; Nongdam, P. The use of molecular marker methods in plants: A review. Int. J. Curr. Res. Rev. 2017, 9, 1–7. [Google Scholar]
- Reinisch, A.J.; Dong, J.-M.; Brubaker, C.L.; Stelly, D.M.; Wendel, J.F.; Paterson, A.H. A detailed RFLP map of cotton, Gossypium hirsutum x Gossypium barbadense: Chromosome organization and evolution in a disomic polyploid genome. Genetics 1994, 138, 829–847. [Google Scholar] [CrossRef] [PubMed]
- Mei, M.; Syed, N.; Gao, W.; Thaxton, P.; Smith, C.; Stelly, D.; Chen, Z. Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theor. Appl. Genet. 2004, 108, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.I.; Awan, F.S.; Sadia, B.; Rana, R.M.; Khan, I.A. Genetic diversity studies among coloured cotton genotypes by using RAPD markers. Pak. J. Bot 2010, 42, 71–77. [Google Scholar]
- Lin, Z.-X.; He, D.; Zhang, X.-L.; Nie, Y.; Guo, X.; Feng, C.; Stewart, J.M. Linkage map construction and mapping QTL for cotton fibre quality using SRAP, SSR and RAPD. Plant Breed. 2005, 124, 180–187. [Google Scholar] [CrossRef]
- Sheeja, T.E.; Kumar, I.P.V.; Giridhari, A.; Minoo, D.; Rajesh, M.K.; Babu, K.N. Amplified fragment length polymorphism: Applications and recent developments. In Molecular Plant Taxonomy: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2020; pp. 187–218. [Google Scholar]
- Liu, Z.J. Amplified fragment length polymorphism (AFLP). In Stock Identification Methods; Elsevier: Amsterdam, The Netherlands, 2005; pp. 389–411. [Google Scholar]
- Badigannavar, A.; Myers, G.O.; Jones, D.C. Molecular diversity revealed by AFLP markers in upland cotton genotypes. J. Crop Improv. 2012, 26, 627–640. [Google Scholar] [CrossRef]
- Malik, W.; Ashraf, J.; Iqbal, M.Z.; Ali Khan, A.; Qayyum, A.; Ali Abid, M.; Noor, E.; Qadir Ahmad, M.; Hasan Abbasi, G. Molecular markers and cotton genetic improvement: Current status and future prospects. Sci. World J. 2014, 2014, 607091. [Google Scholar] [CrossRef]
- Hamid, R.; Tomar, R.S.; Marashi, H.; Shafaroudi, S.M.; Golakiya, B.A.; Mohsenpour, M. Transcriptome profiling and cataloging differential gene expression in floral buds of fertile and sterile lines of cotton (Gossypium hirsutum L.). Gene 2018, 660, 80–91. [Google Scholar] [CrossRef]
- Khan, M.K.; Chen, H.; Zhou, Z.; Ilyas, M.K.; Wang, X.; Cai, X.; Wang, C.; Liu, F.; Wang, K. Genome-wide SSR high density genetic map construction from an interspecific cross of Gossypium hirsutum × Gossypium tomentosum. Front. Plant Sci. 2016, 7, 436. [Google Scholar] [CrossRef]
- Dwivedi, A.; Suthar, K.P.; Hamid, R.; Lakhani, K.G.; Singh, D.; Kumar, S.; BK, R.; Vekariya, V.; Prajapat, P. Exploitation of novel drought responsive EST-SSR markers in tetraploid cotton (Gossypium hirsutum L.). Gene Rep. 2025, 38, 102097. [Google Scholar] [CrossRef]
- Nie, X.; Huang, C.; You, C.; Li, W.; Zhao, W.; Shen, C.; Zhang, B.; Wang, H.; Yan, Z.; Dai, B. Genome-wide SSR-based association mapping for fiber quality in nation-wide upland cotton inbreed cultivars in China. BMC Genom. 2016, 17, 352. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Chen, M.; Yi, X.; Bie, S.; Zhang, C.; Zhang, Y.; Lan, J.; Meng, Y.; Yuan, Y.; Jiao, C. Identification of associated SSR markers for yield component and fiber quality traits based on frame map and upland cotton collections. PLoS ONE 2015, 10, e0118073. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Z.; Xiao, C.; Ilyas, M.K.; Ibrar, D.; Khan, S.; Guo, L.; Wang, W.; Wang, B.; Huang, H.; Li, Y. Use of SSR markers for the exploration of genetic diversity and DNA finger-printing in early-maturing upland cotton (Gossypium hirsutum L.) for future breeding program. Agronomy 2022, 12, 1513. [Google Scholar] [CrossRef]
- Kushanov, F.N.; Turaev, O.S.; Ernazarova, D.K.; Gapparov, B.M.; Oripova, B.B.; Kudratova, M.K.; Rafieva, F.U.; Khalikov, K.K.; Erjigitov, D.S.; Khidirov, M.T. Genetic diversity, QTL mapping, and marker-assisted selection technology in cotton (Gossypium spp.). Front. Plant Sci. 2021, 12, 779386. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Fu, J. Improving Cotton Yield and Fiber Quality Based on QTL Mapping and Genomic Selection. Cotton Genom. Genet. 2025, 16, 117–125. [Google Scholar] [CrossRef]
- Hulse-Kemp, A.M.; Lemm, J.; Plieske, J.; Ashrafi, H.; Buyyarapu, R.; Fang, D.D.; Frelichowski, J.; Giband, M.; Hague, S.; Hinze, L.L. Development of a 63K SNP array for cotton and high-density mapping of intraspecific and interspecific populations of Gossypium spp. G3 Genes Genomes Genet. 2015, 5, 1187–1209. [Google Scholar] [CrossRef]
- Hou, S.; Zhu, G.; Li, Y.; Li, W.; Fu, J.; Niu, E.; Li, L.; Zhang, D.; Guo, W. Genome-wide association studies reveal genetic variation and candidate genes of drought stress related traits in cotton (Gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 1276. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z.; Gong, Z.; Liang, Y.; Ai, X.; Sang, Z.; Guo, J.; Li, X.; Zheng, J. Analysis of the genetic structure and diversity of upland cotton groups in different planting areas based on SNP markers. Gene 2022, 809, 146042. [Google Scholar] [CrossRef]
- Shukla, R.P.; Tiwari, G.J.; Joshi, B.; Song-Beng, K.; Tamta, S.; Boopathi, N.M.; Jena, S.N. GBS-SNP and SSR based genetic mapping and QTL analysis for drought tolerance in upland cotton. Physiol. Mol. Biol. Plants 2021, 27, 1731–1745. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Wang, L.; Yang, Y.; Liu, R.; Liu, S.; Chen, J.; Shen, Q.; Ma, H.; Li, Y.; Zhang, S. Genome-wide association study identifies variants of GhSAD1 conferring cold tolerance in cotton. J. Exp. Bot. 2022, 73, 2222–2237. [Google Scholar] [CrossRef] [PubMed]
- Purkaystha, S.; Das, P.; Rashmi, K.; Rout, S.; Nanda, S. Advances in Genetic Mapping of Loci Governing Disease Resistance in Plants. In Biotechnological Advances for Disease Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2024; pp. 1–27. [Google Scholar]
- Sinha, S.; Kushwaha, B.K.; Deshmukh, R.K. QTL Mapping Using Advanced Mapping Populations and High-throughput Genotyping. In Genotyping by Sequencing for Crop Improvement; Wiley: Hoboken, NJ, USA, 2022; pp. 52–79. [Google Scholar]
- Ayyaz, M.; Chang, Z.; Ding, S.; Han, P.; Xu, L.; Abudukeyoumu, A.; Siddho, I.A.; Li, Z.; Lin, H.; Xu, J. QTL mapping associated with Verticillium wilt resistance in cotton based on MAGIC population. J. Cotton Res. 2025, 8, 1–15. [Google Scholar] [CrossRef]
- Singh, M.; Nara, U.; Kumar, A.; Thapa, S.; Jaswal, C.; Singh, H. Enhancing genetic gains through marker-assisted recurrent selection: From phenotyping to genotyping. Cereal Res. Commun. 2022, 50, 523–538. [Google Scholar] [CrossRef]
- Panahi, B.; Jalaly, H.M.; Hamid, R. Using next-generation sequencing approach for discovery and characterization of plant molecular markers. Curr. Plant Biol. 2024, 40, 100412. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Liu, F.; Song, M.; Zhang, J.F. A meta-analysis of quantitative trait loci for abiotic and biotic stress resistance in tetraploid cotton. Mol. Genet. Genom. 2017, 292, 1221–1235. [Google Scholar] [CrossRef]
- Zhu, Y.; Thyssen, G.N.; Abdelraheem, A.; Teng, Z.; Fang, D.D.; Jenkins, J.N.; McCarty, J.C.; Wedegaertner, T.; Hake, K.; Zhang, J. A GWAS identified a major QTL for resistance to Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) race 4 in a MAGIC population of Upland cotton and a meta-analysis of QTLs for Fusarium wilt resistance. Theor. Appl. Genet. 2022, 135, 2297–2312. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, W.; Cui, Y.; Sang, X.; Lu, J.; Jing, H.; Wang, W.; Zhao, P.; Wang, H. Detection of candidate genes and development of KASP markers for Verticillium wilt resistance by combining genome-wide association study, QTL-seq and transcriptome sequencing in cotton. Theor. Appl. Genet. 2021, 134, 1063–1081. [Google Scholar] [CrossRef]
- Bardak, A.; Çelik, S.; Erdoğan, O.; Ekinci, R.; Dumlupinar, Z. Association mapping of Verticillium wilt disease in a worldwide collection of cotton (Gossypium hirsutum L.). Plants 2021, 10, 306. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Thyssen, G.N.; Fang, D.D.; Jenkins, J.N.; McCarty, J.C.; Wedegaertner, T.; Zhang, J. GWAS reveals consistent QTL for drought and salt tolerance in a MAGIC population of 550 lines derived from intermating of 11 Upland cotton (Gossypium hirsutum) parents. Mol. Genet. Genom. 2021, 296, 119–129. [Google Scholar] [CrossRef]
- Yuan, Y.; Xing, H.; Zeng, W.; Xu, J.; Mao, L.; Wang, L.; Feng, W.; Tao, J.; Wang, H.; Zhang, H. Genome-wide association and differential expression analysis of salt tolerance in Gossypium hirsutum L. at the germination stage. BMC Plant Biol. 2019, 19, 394. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Cai, X.; Zhou, Z.; Agong, S.G.; Wang, K.; Liu, F. Identification of QTLs and candidate genes for physiological traits associated with drought tolerance in cotton. J. Cotton Res. 2020, 3, 3. [Google Scholar] [CrossRef]
- Wang, C.; Ulloa, M.; Duong, T.T.; Roberts, P.A. QTL analysis of transgressive nematode resistance in tetraploid cotton reveals complex interactions in chromosome 11 regions. Front. Plant Sci. 2017, 8, 1979. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, M.; Wang, C.; Roberts, P. Gene action analysis by inheritance and quantitative trait loci mapping of resistance to root-knot nematodes in cotton. Plant Breed. 2010, 129, 541–550. [Google Scholar] [CrossRef]
- Geng, S.; Gao, W.; Li, S.; Chen, Q.; Jiao, Y.; Zhao, J.; Wang, Y.; Wang, T.; Qu, Y.; Chen, Q. Rapidly mining candidate cotton drought resistance genes based on key indicators of drought resistance. BMC Plant Biol. 2024, 24, 129. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chen, Q.; Fu, J.; Jiang, H.; Sun, F.; Geng, S.; Wang, Y.; Zhao, J.; Xie, Y.; Zhou, M. Using association mapping and local interval haplotype association analysis to improve the cotton drought stress response. Plant Sci. 2023, 335, 111813. [Google Scholar] [CrossRef]
- Thottappilly, G.; Magonouna, H.; Omitogun, O. The use of DNA markers for rapid improvement of crops in Africa. Afr. Crop Sci. J. 2000, 8, 99–108. [Google Scholar] [CrossRef]
- Markert, C.L.; Whitt, G. Molecular varieties of isozymes. Experientia 1968, 24, 977–991. [Google Scholar] [CrossRef]
- Jonah, P.; Bello, L.; Lucky, O.; Midau, A.; Moruppa, S. The importance of molecular markers in plant breeding programmes. Glob. J. Sci. Front. Res. 2011, 11, 5–12. [Google Scholar]
- Wang, M.; Li, J.; Qi, Z.; Long, Y.; Pei, L.; Huang, X.; Grover, C.E.; Du, X.; Xia, C.; Wang, P. Genomic innovation and regulatory rewiring during evolution of the cotton genus Gossypium. Nat. Genet. 2022, 54, 1959–1971. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Li, D.; Li, Z.; Feng, H.; Feng, Z.; Wei, F.; Zhou, J.; Ma, Z.; Yang, J. A comprehensive review on elucidating the host disease resistance mechanism from the perspective of the interaction between cotton and Verticillium dahliae. J. Cotton Res. 2025, 8, 5. [Google Scholar] [CrossRef]
- Bag, S.K.; Rai, K.; Singh, S.K.; Spriggs, A. Development of a 63K SNP array for cotton and high-density mapping of intra-and inter-specific populations of Gossypium spp. G3 Genes Genomes Genet. 2013, 5, 1187–1209. [Google Scholar]
- Xu, P.; Guo, Q.; Meng, S.; Zhang, X.; Xu, Z.; Guo, W.; Shen, X. Genome-wide association analysis reveals genetic variations and candidate genes associated with salt tolerance related traits in Gossypium hirsutum. BMC Genom. 2021, 22, 26. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; He, S.; Sun, G.; Geng, X.; Pan, Z.; Gong, W.; Jia, Y.; Du, X. A genome-wide association study revealed key SNPs/genes associated with salinity stress tolerance in upland cotton. Genes 2019, 10, 829. [Google Scholar] [CrossRef]
- Panahi, B.; Hamid, R. Decoding core molecular mechanisms related to multiple abiotic stress adaptation in cotton: Insights from RNA-seq data meta-analysis in combination with machine learning approach. Curr. Plant Biol. 2025, 43, 100503. [Google Scholar] [CrossRef]
- Darmanov, M.M.; Makamov, A.K.; Ayubov, M.S.; Khusenov, N.N.; Buriev, Z.T.; Shermatov, S.E.; Salakhutdinov, I.B.; Ubaydullaeva, K.A.; Norbekov, J.K.; Kholmuradova, M.M. Development of superior fibre quality upland cotton cultivar series ‘Ravnaq’using marker-assisted selection. Front. Plant Sci. 2022, 13, 906472. [Google Scholar] [CrossRef]
- Razzaq, A.; Zafar, M.M.; Ali, A.; Hafeez, A.; Batool, W.; Shi, Y.; Gong, W.; Yuan, Y. Cotton germplasm improvement and progress in Pakistan. J. Cotton Res. 2021, 4, 1. [Google Scholar] [CrossRef]
- Scott, M.F.; Ladejobi, O.; Amer, S.; Bentley, A.R.; Biernaskie, J.; Boden, S.A.; Clark, M.; Dell’Acqua, M.; Dixon, L.E.; Filippi, C.V. Multi-parent populations in crops: A toolbox integrating genomics and genetic mapping with breeding. Heredity 2020, 125, 396–416. [Google Scholar] [CrossRef]
- Sakthipriya, M.; Subramanian, A.; Premalatha, N.; Marimuthu, S.; Chitra, N. Weaving the wild: Harnessing the potential of cotton relatives for superior fibre quality. Genet. Resour. Crop Evol. 2025, 72, 9147–9164. [Google Scholar] [CrossRef]
- Kennedy, H.D. Selection and Response of Yield and Fiber Traits in Upland Cotton. 2018. Available online: https://hdl.handle.net/1969.1/173991 (accessed on 12 September 2025).
- Slater, A.T.; Cogan, N.O.; Rodoni, B.C.; Daetwyler, H.D.; Hayes, B.J.; Caruana, B.; Badenhorst, P.E.; Spangenberg, G.C.; Forster, J.W. Breeding differently—The digital revolution: High-throughput phenotyping and genotyping. Potato Res. 2017, 60, 337–352. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; De Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Ramzan, M.T.; Razaq, L.; Zhang, X.; Zia, M.S.; Yaseen, U.; Chaudhary, M.U.M.; Ali, M.J. Integrating genomic tools and traditional breeding for climate-resilient cotton: A comprehensive review. Int. J. Cotton Res. Technol. 2025, 7, 1–8. [Google Scholar] [CrossRef]
- Luo, M. AI-Assisted Genomic Prediction Models in Cotton Breeding. Cotton Genom. Genet. 2025, 16, 137–147. [Google Scholar] [CrossRef]
- Li, Z.; Liu, S.; Conaty, W.; Zhu, Q.-H.; Moncuquet, P.; Stiller, W.; Wilson, I. Genomic prediction of cotton fibre quality and yield traits using Bayesian regression methods. Heredity 2022, 129, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Delvento, C.; Ricciardi, L.; Lotti, C.; Ciani, E.; D’Agostino, N. Recommendations for choosing the genotyping method and best practices for quality control in crop genome-wide association studies. Front. Genet. 2020, 11, 447. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Yang, X.; Peng, Z.; Xu, L.; Wang, J. Development and applications of a high throughput genotyping tool for polyploid crops: Single-nucleotide polymorphism (SNP) array. Front. Plant Sci. 2018, 9, 104. [Google Scholar] [CrossRef]
- Geibel, J.; Reimer, C.; Weigend, S.; Weigend, A.; Pook, T.; Simianer, H. How array design creates SNP ascertainment bias. PLoS ONE 2021, 16, e0245178. [Google Scholar] [CrossRef]
- Islam, M.S.; Fang, D.D.; Jenkins, J.N.; Guo, J.; McCarty, J.C.; Jones, D.C. Evaluation of genomic selection methods for predicting fiber quality traits in Upland cotton. Mol. Genet. Genom. 2020, 295, 67–79. [Google Scholar] [CrossRef]
- Kamburova, V.; Salakhutdinov, I.; Abdurakhmonov, I.Y. Cotton Breeding in the View of Abiotic and Biotic Stresses: Challenges and Perspectives; IntechOpen: London, UK, 2022. [Google Scholar]
- Degefu, D.T.; Gebregiorgis, Z.D. Cotton biotechnology. In Cotton Sector Development in Ethiopia: Challenges and Opportunities; Springer: Berlin/Heidelberg, Germany, 2024; pp. 65–88. [Google Scholar]
- Chohan, S.; Perveen, R.; Abid, M.; Tahir, M.N.; Sajid, M. Cotton diseases and their management. In Cotton Production and Uses: Agronomy, Crop Protection, and Postharvest Technologies; Springer: Berlin/Heidelberg, Germany, 2020; pp. 239–270. [Google Scholar]
- Razzaq, A.; Zafar, M.M.; Ali, A.; Li, P.; Qadir, F.; Zahra, L.T.; Shaukat, F.; Laghari, A.H.; Yuan, Y.; Gong, W. Biotechnology and solutions: Insect-pest-resistance management for improvement and development of Bt cotton (Gossypium hirsutum L.). Plants 2023, 12, 4071. [Google Scholar] [CrossRef]
- Huo, W.-Q.; Zhang, Z.-Q.; Ren, Z.-Y.; Zhao, J.-J.; Song, C.-X.; Wang, X.-X.; Pei, X.-Y.; Liu, Y.-G.; He, K.-L.; Zhang, F. Unraveling genomic regions and candidate genes for multiple disease resistance in upland cotton using meta-QTL analysis. Heliyon 2023, 9, e18731. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Elassbli, H.; Zhu, Y.; Kuraparthy, V.; Hinze, L.; Stelly, D.; Wedegaertner, T.; Zhang, J. A genome-wide association study uncovers consistent quantitative trait loci for resistance to Verticillium wilt and Fusarium wilt race 4 in the US Upland cotton. Theor. Appl. Genet. 2020, 133, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Abdelraheem, A.; Zhu, Y.; Zeng, L.; Stetina, S.; Zhang, J. A genome-wide association study for resistance to Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) race 4 in diploid cotton (Gossypium arboreum) and resistance transfer to tetraploid Gossypium hirsutum. Mol. Genet. Genom. 2024, 299, 30. [Google Scholar] [CrossRef] [PubMed]
- Said, J.I.; Song, M.; Wang, H.; Lin, Z.; Zhang, X.; Fang, D.D.; Zhang, J. A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol. Genet. Genom. 2015, 290, 1003–1025. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ma, X.; Li, N.; Zhou, L.; Liu, Z.; Han, H.; Gui, Y.; Bao, Y.; Chen, J.; Dai, X. Genome-wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2017, 15, 1520–1532. [Google Scholar] [CrossRef]
- Zhang, J.; Manikanda Boopathi, N. Disease resistance in cotton. In Genomic Designing for Biotic Stress Resistant Technical Crops; Springer: Berlin/Heidelberg, Germany, 2022; pp. 191–225. [Google Scholar]
- Zhao, J.; Liu, J.; Xu, J.; Zhao, L.; Wu, Q.; Xiao, S. Quantitative trait locus mapping and candidate gene analysis for Verticillium wilt resistance using Gossypium barbadense chromosomal segment introgressed line. Front. Plant Sci. 2018, 9, 682. [Google Scholar] [CrossRef]
- Shahbazi, S.; Ghaffarian, S.; Razinataj, M.; Zangi, M.R.; Hamid, R.; Panahi, B. SSR-based molecular characterization of Verticillium wilt resistance in Iranian cotton cultivars. Biochem. Biophys. Rep. 2025, 42, 102059. [Google Scholar] [CrossRef]
- Pathak, D.; Rathore, P.; Kaur, H.; Singh, B.; Kumar, H.; Ali, A.; Punia, S.; Sekhon, P.S.; Singh, K. Introgression and Mapping of Cotton Leaf Curl Disease Resistance from Wild Gossypium armourianum Kearney into Upland Cotton (G. hirsutum). Plant Dis. 2025, 109, 554–557. [Google Scholar] [CrossRef]
- Ullah, R.; Akhtar, K.P.; Moffett, P.; Mansoor, S.; Briddon, R.W.; Saeed, M. An analysis of the resistance of Gossypium arboreum to cotton leaf curl disease by grafting. Eur. J. Plant Pathol. 2014, 139, 837–847. [Google Scholar] [CrossRef]
- Abbas, A.; Iqbal, M.A.; Rahman, M.-u.; Paterson, A.H. Estimating genetic diversity among selected cotton genotypes and the identificationof DNA markers associated with resistance to cotton leaf curl disease. Turk. J. Bot. 2015, 39, 1033–1041. [Google Scholar] [CrossRef]
- Hashim, H.O.; Al-Shuhaib, M.B.S. Exploring the potential and limitations of PCR-RFLP and PCR-SSCP for SNP detection: A review. J. Appl. Biotechnol. Rep. 2019, 6, 137–144. [Google Scholar] [CrossRef]
- Sarwar, M.; Hamed, M.; Yousaf, M.; Hussain, M. Identification of resistance to insect pests infestations in cotton (Gossypium hirsutum L.) varieties evaluated in the field experiment. Int. J. Sci. Res. Environ. Sci. 2013, 1, 317. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhu, Z.; Jiao, Y.; Zhang, G. Pan-genome analysis of GT64 gene family and expression response to Verticillium wilt in cotton. BMC Plant Biol. 2024, 24, 893. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, R.; Taoutaou, A.; Hakeem, K.R.; Gawwad, M.R.A.; Tah, J. Molecular marker-assisted technologies for crop improvement. In Crop Improvement in the Era of Climate Change; I.K. International Publishing House: New Delhi, India, 2014; pp. 241–258. [Google Scholar]
- Khalid, M.; Rehman, H.M.; Cheung, T.Y.; Ahmed, S.; Chan, T.F.; Lam, H.M. A Repertoire of Major Genes From Crop Wild Relatives for Breeding Disease-Resistant Wheat, Rice, Maize, Soybean and Cotton Crops. Plant Breed. 2025. [CrossRef]
- Biswas, P.; Kumar, N. Application of molecular markers for the assessment of genetic fidelity of in vitro raised plants: Current status and future prospects. In Molecular Marker Techniques: A Potential Approach of Crop Improvement; Springer: Berlin/Heidelberg, Germany, 2023; pp. 233–256. [Google Scholar]
- Zhao, Y.; Wang, H.; Chen, W.; Zhao, P.; Gong, H.; Sang, X.; Cui, Y. Regional association analysis-based fine mapping of three clustered QTL for verticillium wilt resistance in cotton (G. hirsutum. L). BMC Genom. 2017, 18, 661. [Google Scholar] [CrossRef]
- Aini, N.; Jibril, A.N.; Liu, S.; Han, P.; Pan, Z.; Zhu, L.; Nie, X. Advances and prospects of genetic mapping of Verticillium wilt resistance in cotton. J. Cotton Res. 2022, 5, 5. [Google Scholar] [CrossRef]
- Palanga, K.K.; Jamshed, M.; Rashid, M.H.o.; Gong, J.; Li, J.; Iqbal, M.S.; Liu, A.; Shang, H.; Shi, Y.; Chen, T. Quantitative trait locus mapping for Verticillium wilt resistance in an upland cotton recombinant inbred line using SNP-based high density genetic map. Front. Plant Sci. 2017, 8, 382. [Google Scholar] [CrossRef]
- Ynturi, P.; Jenkins, J.N.; McCarty Jr, J.C.; Gutierrez, O.A.; Saha, S. Association of root-knot nematode resistance genes with simple sequence repeat markers on two chromosomes in cotton. Crop Sci. 2006, 46, 2670–2674. [Google Scholar] [CrossRef][Green Version]
- Fang, D.D.; Xiao, J.; Canci, P.C.; Cantrell, R.G. A new SNP haplotype associated with blue disease resistance gene in cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2010, 120, 943–953. [Google Scholar] [CrossRef]
- Cox, K.L.; Meng, F.; Wilkins, K.E.; Li, F.; Wang, P.; Booher, N.J.; Carpenter, S.C.; Chen, L.-Q.; Zheng, H.; Gao, X. TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat. Commun. 2017, 8, 15588. [Google Scholar] [CrossRef]
- Mahmood, T.; Khalid, S.; Abdullah, M.; Ahmed, Z.; Shah, M.K.N.; Ghafoor, A.; Du, X. Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 2019, 9, 105. [Google Scholar] [CrossRef]
- Younis, A.; Ramzan, F.; Ramzan, Y.; Zulfiqar, F.; Ahsan, M.; Lim, K.B. Molecular markers improve abiotic stress tolerance in crops: A review. Plants 2020, 9, 1374. [Google Scholar] [CrossRef]
- Oluoch, G.; Zheng, J.; Wang, X.; Khan, M.K.R.; Zhou, Z.; Cai, X.; Wang, C.; Wang, Y.; Li, X.; Wang, H. QTL mapping for salt tolerance at seedling stage in the interspecific cross of Gossypium tomentosum with Gossypium hirsutum. Euphytica 2016, 209, 223–235. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Jamshed, M.; Shi, Y.; Liu, A.; Gong, J.; Wang, S.; Zhang, J.; Sun, F.; Jia, F. Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population. Plant Biotechnol. J. 2020, 18, 239–253. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Fang, D.D.; Zhang, J. Quantitative trait locus mapping of drought and salt tolerance in an introgressed recombinant inbred line population of Upland cotton under the greenhouse and field conditions. Euphytica 2018, 214, 8. [Google Scholar] [CrossRef]
- Feng, L.; Chen, Y.; Ma, T.; Zhou, C.; Sang, S.; Li, J.; Ji, S. Integrative physiology and transcriptome sequencing reveal differences between G. hirsutum and G. barbadense in response to salt stress and the identification of key salt tolerance genes. BMC Plant Biol. 2024, 24, 787. [Google Scholar] [CrossRef] [PubMed]
- Muthuramalingam, P.; Jeyasri, R.; Rakkammal, K.; Satish, L.; Shamili, S.; Karthikeyan, A.; Valliammai, A.; Priya, A.; Selvaraj, A.; Gowri, P. Multi-Omics and integrative approach towards understanding salinity tolerance in rice: A review. Biology 2022, 11, 1022. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, X.; Wang, J.; Wang, D.; Yin, Z.; Fan, W.; Wang, S.; Ye, W. Mining and analysis of SNP in response to salinity stress in upland cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0158142. [Google Scholar] [CrossRef]
- Saleem, M.; Malik, T.; Shakeel, A.; Ashraf, M. QTL mapping for some important drought tolerant traits in upland cotton. JAPS J. Anim. Plant Sci. 2015, 25, 502–509. [Google Scholar]
- Saleem, M.A.; Amjid, M.W.; Ahmad, M.Q.; Riaz, H.; Arshad, S.F.; Zia, Z.U. EST-SSR based analysis revealed narrow genetic base of in-use cotton varieties of Pakistan. Pak. J. Bot 2020, 52, 1667–1672. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, Z.; Liang, Y.; Gong, Z.; Zhang, N.; Ditta, A.; Sang, Z.; Wang, J.; Li, X. Whole transcriptome sequencing reveals drought resistance-related genes in upland cotton. Genes 2022, 13, 1159. [Google Scholar] [CrossRef]
- Ndudzo, A.; Makuvise, A.S.; Moyo, S.; Bobo, E.D. CRISPR-Cas9 genome editing in crop breeding for climate change resilience: Implications for smallholder farmers in Africa. J. Agric. Food Res. 2024, 16, 101132. [Google Scholar] [CrossRef]
- Saranga, Y.; Jiang, C.X.; Wright, R.; Yakir, D.; Paterson, A. Genetic dissection of cotton physiological responses to arid conditions and their inter-relationships with productivity. Plant Cell Environ. 2004, 27, 263–277. [Google Scholar] [CrossRef]
- Wang, H.; Huang, C.; Guo, H.; Li, X.; Zhao, W.; Dai, B.; Yan, Z.; Lin, Z. QTL mapping for fiber and yield traits in upland cotton under multiple environments. PLoS ONE 2015, 10, e0130742. [Google Scholar] [CrossRef]
- Zheng, J.; Oluoch, G.; Riaz Khan, M.; Wang, X.; Cai, X.; Zhou, Z.; Wang, C.; Wang, Y.; Li, X.; Liu, F. Mapping QTLs for drought tolerance in an F2: 3 population from an inter-specific cross between Gossypium tomentosum and Gossypium hirsutum. Genet. Mol. Res. GMR 2016, 15, gmr.15038477. [Google Scholar] [CrossRef]
- Baytar, A.A.; Peynircioğlu, C.; Sezener, V.; Basal, H.; Frary, A.; Frary, A.; Doğanlar, S. Genome-wide association mapping of yield components and drought tolerance-related traits in cotton. Mol. Breed. 2018, 38, 74. [Google Scholar] [CrossRef]
- Zhu, G.; Hou, S.; Song, X.; Wang, X.; Wang, W.; Chen, Q.; Guo, W. Genome-wide association analysis reveals quantitative trait loci and candidate genes involved in yield components under multiple field environments in cotton (Gossypium hirsutum). BMC Plant Biol. 2021, 21, 250. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.A.; Malik, W.; Ahmad, M.Q.; Arshad, S.F.; Baig, M.M.A.; Asif, M.; Nauman, M.; Anwar, M. Gene pyramiding improved cell membrane stability under heat stress in cotton (Gossypium hirsutum L.). BMC Plant Biol. 2024, 24, 886. [Google Scholar] [CrossRef] [PubMed]
- Luqman, T.; Hussain, M.; Ahmed, S.R.; Ijaz, I.; Maryum, Z.; Nadeem, S.; Khan, Z.; Khan, S.M.U.D.; Aslam, M.; Liu, Y. Cotton under heat stress: A comprehensive review of molecular breeding, genomics, and multi-omics strategies. Front. Genet. 2025, 16, 1553406. [Google Scholar] [CrossRef]
- Baytar, A.A.; Peynircioğlu, C.; Sezener, V.; Frary, A.; Doğanlar, S. Association analysis of germination level cold stress tolerance and candidate gene identification in Upland cotton (Gossypium hirsutum L.). Physiol. Mol. Biol. Plants 2022, 28, 1049–1060. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; Yu, X.; Zhao, J.; Tian, Z.; Liu, X.; Wang, G.; Zhang, L.; Guo, X. Enhancing cold and drought tolerance in cotton: A protective role of SikCOR413PM1. BMC Plant Biol. 2023, 23, 577. [Google Scholar] [CrossRef]
- Ullah, K.; Waheed, A. Genetic Improvement in Livestock: A Journey from Conventional Breeding to Genomic Precision. Vet. Biomed. Clin. J. 2025, 7, 30–44. [Google Scholar] [CrossRef]
- Somegowda, V.K.; Reddy, S.D.; Gaddameedi, A.; Kiranmayee, K.U.; Naravula, J.; Kishor, P.K.; Penna, S. Genomics breeding approaches for developing Sorghum bicolor lines with stress resilience and other agronomic traits. Curr. Plant Biol. 2024, 37, 100314. [Google Scholar] [CrossRef]
- Rong, J.; Feltus, F.A.; Waghmare, V.N.; Pierce, G.J.; Chee, P.W.; Draye, X.; Saranga, Y.; Wright, R.J.; Wilkins, T.A.; May, O.L. Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics 2007, 176, 2577–2588. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Guo, W.; Zhu, X.; Wu, Y.; Huang, N.; Zhang, T. QTL mapping of yield and yield components for elite hybrid derived-RILs in upland cotton. J. Genet. Genom. 2007, 34, 35–45. [Google Scholar] [CrossRef]
- Abdurakhmonov, I.Y.; Kohel, R.J.; Yu, J.; Pepper, A.; Abdullaev, A.; Kushanov, F.; Salakhutdinov, I.; Buriev, Z.; Saha, S.; Scheffler, B. Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics 2008, 92, 478–487. [Google Scholar] [CrossRef]
- Saeed, M.; Song, X.; Iqbal, M.A.; Sun, X. Genomics-Assisted Breeding for Fiber Quality Traits in Cotton. In Cotton Precision Breeding; Springer: Berlin/Heidelberg, Germany, 2021; pp. 157–172. [Google Scholar]
- Cao, Z.; Wang, P.; Zhu, X.; Chen, H.; Zhang, T. SSR marker-assisted improvement of fiber qualities in Gossypium hirsutum using G. barbadense introgression lines. Theor. Appl. Genet. 2014, 127, 587–594. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, X.; Liu, Z.; Gu, Q.; Zhang, Y.; Li, Z.; Ke, H.; Yang, J.; Wu, J.; Wu, L. A genome-wide association study uncovers novel genomic regions and candidate genes of yield-related traits in upland cotton. Theor. Appl. Genet. 2018, 131, 2413–2425. [Google Scholar] [CrossRef]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef]
- Liu, W.; Song, C.; Ren, Z.; Zhang, Z.; Pei, X.; Liu, Y.; He, K.; Zhang, F.; Zhao, J.; Zhang, J. Genome-wide association study reveals the genetic basis of fiber quality traits in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 2020, 20, 395. [Google Scholar] [CrossRef]
- Thyssen, G.N.; Jenkins, J.N.; McCarty, J.C.; Zeng, L.; Campbell, B.T.; Delhom, C.D.; Islam, M.S.; Li, P.; Jones, D.C.; Condon, B.D. Whole genome sequencing of a MAGIC population identified genomic loci and candidate genes for major fiber quality traits in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2019, 132, 989–999. [Google Scholar] [CrossRef]
- Sarfraz, Z.; Iqbal, M.S.; Geng, X.; Iqbal, M.S.; Nazir, M.F.; Ahmed, H.; He, S.; Jia, Y.; Pan, Z.; Sun, G. GWAS mediated elucidation of heterosis for metric traits in cotton (Gossypium hirsutum L.) across multiple environments. Front. Plant Sci. 2021, 12, 565552. [Google Scholar] [CrossRef]
- Abdurakhmonov, I.; Buriev, Z.; Saha, S.; Pepper, A.; Musaev, J.; Almatov, A.; Shermatov, S.; Kushanov, F.; Mavlonov, G.; Reddy, U. Microsatellite markers associated with lint percentage trait in cotton, Gossypium hirsutum. Euphytica 2007, 156, 141–156. [Google Scholar] [CrossRef]
- Abdurakhmonov, I.Y.; Saha, S.; Jenkins, J.N.; Buriev, Z.T.; Shermatov, S.E.; Scheffler, B.E.; Pepper, A.E.; Yu, J.Z.; Kohel, R.J.; Abdukarimov, A. Linkage disequilibrium based association mapping of fiber quality traits in G. hirsutum L. variety germplasm. Genetica 2009, 136, 401–417. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Cooper, M.; Hayes, B.J. Accelerating crop genetic gains with genomic selection. Theor. Appl. Genet. 2019, 132, 669–686. [Google Scholar] [CrossRef]
- Watson, A.; Ghosh, S.; Williams, M.; Cuddy, W.; Simmonds, J.; Rey, M.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, T.; Shen, X.; Yu, J.Z.; Kohel, R.J. Development of SCAR marker linked to a major QTL for high fiber strength and its usage in molecular-marker assisted selection in upland cotton. Crop Sci. 2003, 43, 2252–2256. [Google Scholar] [CrossRef]
- Zhang, T.; Yuan, Y.; Yu, J.; Guo, W.; Kohel, R.J. Molecular tagging of a major QTL for fiber strength in Upland cotton and its marker-assisted selection. Theor. Appl. Genet. 2003, 106, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Lacape, J.-M.; Llewellyn, D.; Jacobs, J.; Arioli, T.; Becker, D.; Calhoun, S.; Al-Ghazi, Y.; Liu, S.; Palaï, O.; Georges, S. Meta-analysis of cotton fiber quality QTLs across diverse environments in a Gossypium hirsutum × G. barbadense RIL population. BMC Plant Biol. 2010, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Kuraparthy, V.; Fang, H.; Zhu, L.; Sood, S.; Jones, D.C. High-density linkage map construction and QTL analyses for fiber quality, yield and morphological traits using CottonSNP63K array in upland cotton (Gossypium hirsutum L.). BMC Genom. 2019, 20, 889. [Google Scholar] [CrossRef]
- Gapare, W.; Conaty, W.; Zhu, Q.-H.; Liu, S.; Stiller, W.; Llewellyn, D.; Wilson, I. Genome-wide association study of yield components and fibre quality traits in a cotton germplasm diversity panel. Euphytica 2017, 213, 66. [Google Scholar] [CrossRef]
- Yu, J.; Hui, Y.; Chen, J.; Yu, H.; Gao, X.; Zhang, Z.; Li, Q.; Zhu, S.; Zhao, T. Whole-genome resequencing of 240 Gossypium barbadense accessions reveals genetic variation and genes associated with fiber strength and lint percentage. Theor. Appl. Genet. 2021, 134, 3249–3261. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. GWAS Revealed the Key Genetic Factors Affecting Cotton Fiber Quality. Cotton Genom. Genet. 2024, 15, 1–8. [Google Scholar] [CrossRef]
- Khalilisamani, N.; Li, Z.; Pettolino, F.A.; Moncuquet, P.; Reverter, A.; MacMillan, C.P. Leveraging transcriptomics-based approaches to enhance genomic prediction: Integrating SNPs and gene networks for cotton fibre quality improvement. Front. Plant Sci. 2024, 15, 1420837. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Pi, R.; Wu, Y.; You, J.; Qi, Z.; Liu, Z.; Chang, X.; Ding, S.; Zhang, Q.; Han, P. GWAS and eQTL analyses reveal genetic components influencing the key fiber yield trait lint percentage in upland cotton. Plant J. 2025, 121, e70036. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Shan, B.; Xiong, W.; Zhang, S. Dyeing method and properties of a novel blue azo-anthraquinone reactive dye on cotton. Molecules 2019, 24, 1334. [Google Scholar] [CrossRef]
- Gapare, W.; Liu, S.; Conaty, W.; Zhu, Q.-H.; Gillespie, V.; Llewellyn, D.; Stiller, W.; Wilson, I. Historical datasets support genomic selection models for the prediction of cotton fiber quality phenotypes across multiple environments. G3 Genes Genomes Genet. 2018, 8, 1721–1732. [Google Scholar] [CrossRef]
- Billings, G.T.; Jones, M.A.; Rustgi, S.; Bridges Jr, W.C.; Holland, J.B.; Hulse-Kemp, A.M.; Campbell, B.T. Outlook for implementation of genomics-based selection in public cotton breeding programs. Plants 2022, 11, 1446. [Google Scholar] [CrossRef]
- Souaibou, M.; Yan, H.; Dai, P.; Pan, J.; Li, Y.; Shi, Y.; Gong, W.; Shang, H.; Gong, J.; Yuan, Y. Machine Learning-Driven Identification of Key Environmental Factors Influencing Fiber Yield and Quality Traits in Upland Cotton. Plants 2025, 14, 2053. [Google Scholar] [CrossRef]
- Huang, W. The current situation and future of using GWAS strategies to accelerate the improvement of crop stress resistance traits. Mol. Plant Breed. 2024, 15, 52–62. [Google Scholar] [CrossRef]
- Khan, M.; Hu, D.; Dai, S.; Li, H.; Peng, Z.; He, S.; Awais, M.; Du, X.; Geng, X. Unraveling key genes and pathways involved in Verticillium wilt resistance by integrative GWAS and transcriptomic approaches in Upland cotton. Funct. Integr. Genom. 2025, 25, 1–31. [Google Scholar] [CrossRef]
- Sadohara, R.; Long, Y.; Izquierdo, P.; Urrea, C.A.; Morris, D.; Cichy, K. Seed coat color genetics and genotype× environment effects in yellow beans via machine-learning and genome-wide association. Plant Genome 2022, 15, e20173. [Google Scholar] [CrossRef]
- Hickey, J.M.; Chiurugwi, T.; Mackay, I.; Powell, W. Genomic prediction unifies animal and plant breeding programs to form platforms for biological discovery. Nat. Genet. 2017, 49, 1297–1303. [Google Scholar] [CrossRef]
- Crossa, J.; Martini, J.W.; Vitale, P.; Pérez-Rodríguez, P.; Costa-Neto, G.; Fritsche-Neto, R.; Runcie, D.; Cuevas, J.; Toledo, F.; Li, H. Expanding genomic prediction in plant breeding: Harnessing big data, machine learning, and advanced software. Trends Plant Sci. 2025, 30, 756–774. [Google Scholar] [CrossRef]
- Sharma, R.; Yang, C.J.; Rossi, N.; Irving, E.; Tuffin, A.; Aliki, H.; Powell, W.; Dawson, I.K. Integrating molecular genetics with plant breeding to deliver impact. Plant Physiol. 2025, 198, kiaf087. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Chen, H.; Cao, Y.; He, L.; Si, Z.; Hu, Y.; Lin, H.; Ning, X.; Li, J.; Ma, Q. Genomic insights into genetic improvement of upland cotton in the world’s largest growing region. Ind. Crops Prod. 2022, 183, 114929. [Google Scholar] [CrossRef]
- Zhao, L.; Um, D.; Nowka, K.; Landivar-Scott, J.L.; Landivar, J.; Bhandari, M. Cotton yield prediction utilizing unmanned aerial vehicles (UAV) and Bayesian neural networks. Comput. Electron. Agric. 2024, 226, 109415. [Google Scholar] [CrossRef]
- Salunkhe, S.R.; Ramasamy, S.P.; Rathnasamy, S.A.; Rajagopalan, V.R.; Muthurajan, R.; Manickam, S. Applications and Potential of Genome Editing in Industrial Crop Improvement. In Industrial Crops Improvement: Biotechnological Approaches for Sustainable Agricultural Development; Springer: Berlin/Heidelberg, Germany, 2025; pp. 1–19. [Google Scholar]
- Lee, J.J.; Woodward, A.W.; Chen, Z.J. Gene expression changes and early events in cotton fibre development. Ann. Bot. 2007, 100, 1391–1401. [Google Scholar] [CrossRef]
- Wu, C.; Xiao, S.; Zhang, X.; Ren, W.; Shangguan, X.; Li, S.; Zuo, D.; Cheng, H.; Zhang, Y.; Lv, L. GhHDZ76, a cotton HD-Zip transcription factor, involved in regulating the initiation and early elongation of cotton fiber development in G. hirsutum. Plant Sci. 2024, 345, 112132. [Google Scholar] [CrossRef]
- Verma, V.; Kumar, A.; Partap, M.; Thakur, M.; Bhargava, B. CRISPR-Cas: A robust technology for enhancing consumer-preferred commercial traits in crops. Front. Plant Sci. 2023, 14, 1122940. [Google Scholar] [CrossRef]
- Zhu, S.; Yu, X.; Li, Y.; Sun, Y.; Zhu, Q.; Sun, J. Highly efficient targeted gene editing in upland cotton using the CRISPR/Cas9 system. Int. J. Mol. Sci. 2018, 19, 3000. [Google Scholar] [CrossRef]
- Zhu, S.; Li, Y.; Zhang, X.; Liu, F.; Xue, F.; Zhang, Y.; Kong, Z.; Zhu, Q.-H.; Sun, J. GhAlaRP, a cotton alanine rich protein gene, involves in fiber elongation process. Crop J. 2021, 9, 313–324. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, C.; Wang, H.; Niu, E.; Zhao, P.; Fang, S.; Zhu, G.; Shang, X.; Guo, W. Cotton fiber development requires the pentatricopeptide repeat protein GhIm for splicing of mitochondrial Nad7 mRNA. Genetics 2021, 217, iyaa017. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Chen, B.; Sun, Y.; Sun, G.; Gao, X.; Pan, Z.; Song, G.; Du, X.; He, S. GhGRF4/GhARF2-GhGASA24 module regulates fiber cell wall thickness by modulating cellulose biosynthesis in upland cotton (Gossypium hirsutum). Plant J. 2024, 120, 1842–1856. [Google Scholar] [CrossRef] [PubMed]
- Vijay, S.; Harikrishnan, M.; Phanikanth, J.; Anshu, A.; Khan, R.G.; Baohong, Z. CRISPR/Cas genome editing for cotton precision breeding: Mechanisms, advances, and prospects. J. Cotton Res. 2025, 8, 4. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, J.; Tian, J.; Du, M.; Gou, L.; Zhang, Y.; Zhang, W. Different concentrations of chemical topping agents affect cotton yield and quality by regulating plant architecture. Agronomy 2023, 13, 1741. [Google Scholar] [CrossRef]
- Shang, X.; Cheng, C.; Ding, J.; Guo, W. Identification of candidate genes from the SAD gene family in cotton for determination of cottonseed oil composition. Mol. Genet. Genom. 2017, 292, 173–186. [Google Scholar] [CrossRef]
- Li, Z.; Shi, L.; Liang, D.; Li, F.; Wei, L.; Li, W.; Zha, X. Study on the hydrocarbon-rich bio-oil from catalytic fast co-pyrolysis cotton stalk and polypropylene over alkali-modified HZSM-5. Ind. Crops Prod. 2025, 224, 120352. [Google Scholar] [CrossRef]
- Kaupbayeva, B.; Tsoy, A.; Safarova, Y.; Nurmagambetova, A.; Murata, H.; Matyjaszewski, K.; Askarova, S. Unlocking genome editing: Advances and obstacles in CRISPR/Cas delivery technologies. J. Funct. Biomater. 2024, 15, 324. [Google Scholar] [CrossRef]
- Ramakrishnan, P.; Sundaram, T.; Lahiri, D.; Nag, M.; Bhattacharya, D. Genetic Engineering and Modulation of Metabolic Pathways. In Introduction to Metabolic Engineering and Application; Springer: Berlin/Heidelberg, Germany, 2025; pp. 295–330. [Google Scholar]
- Chen, Y.; Fu, M.; Li, H.; Wang, L.; Liu, R.; Liu, Z.; Zhang, X.; Jin, S. High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system. Plant Biotechnol. J. 2020, 19, 424. [Google Scholar] [CrossRef]
- Li, L.; Zhang, D.; Zhang, Z.; Zhang, B. CRISPR/Cas: A powerful tool for designing and improving oil crops. Trends Biotechnol. 2024, 43, 773–789. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Pei, W.; Wedegaertner, T.; Zhang, J.; Yu, J. Genetics, breeding and genetic engineering to improve cottonseed oil and protein: A review. Front. Plant Sci. 2022, 13, 864850. [Google Scholar] [CrossRef] [PubMed]
- APHIS, U. Movement of certain genetically engineered organisms. Fed. Regist 2020, 85, 96. [Google Scholar]
- Molinari, H.; Vieira, L.; Freitas, N.; Justen, F.; de Jesus, V.; de Oliveira, B. Regulatory framework of genome editing in Brazil and worldwide. In CRISPR Technology in Plant Genome Editing: Biotechnology Applied to Agriculture; Embrapa: Brasília, Brazil, 2021; pp. 169–195. [Google Scholar]
- da Cunha, N.B.; Silva Junior, J.J.d.; Araújo, A.M.; de Souza, L.R.; Leite, M.L.; Medina, G.d.S.; Rodriguez, G.R.; Dos Anjos, R.M.; Rodrigues, J.C.; Costa, F.F. Updates on the Regulatory Framework of Edited Organisms in Brazil: A Molecular Revolution in Brazilian Agribusiness. Genes 2025, 16, 553. [Google Scholar] [CrossRef]
- Mundorf, J.; Simon, S.; Engelhard, M. The European Commission’s Regulatory Proposal on New Genomic Techniques in Plants: A Spotlight on Equivalence, Complexity, and Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2025. [Google Scholar]
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome edited crops touch the market: A view on the global development and regulatory environment. Front. Plant Sci. 2020, 11, 586027. [Google Scholar] [CrossRef]
- Subedi, U.; Jayawardhane, K.N.; Pan, X.; Ozga, J.; Chen, G.; Foroud, N.A.; Singer, S.D. The potential of genome editing for improving seed oil content and fatty acid composition in oilseed crops. Lipids 2020, 55, 495–512. [Google Scholar] [CrossRef]
- Pandeya, D.; Campbell, L.M.; Puckhaber, L.; Suh, C.; Rathore, K.S. Gossypol and related compounds are produced and accumulate in the aboveground parts of the cotton plant, independent of roots as the source. Planta 2023, 257, 21. [Google Scholar] [CrossRef]
- Geneste, T. Regulation of Fatty Acid Desaturation and Lipid Engineering; Université Paris-Saclay: Paris, France, 2022. [Google Scholar]
- Tesema, G.B. Cotton Quality Requirements for Spinning. In Cotton Sector Development in Ethiopia: Challenges and Opportunities; Springer: Berlin/Heidelberg, Germany, 2024; pp. 241–284. [Google Scholar]
- Constable, G.; Llewellyn, D.; Walford, S.A.; Clement, J.D. Cotton breeding for fiber quality improvement. In Industrial Crops: Breeding for Bioenergy and Bioproducts; Springer: Berlin/Heidelberg, Germany, 2014; pp. 191–232. [Google Scholar]
- Jiao, J.; Chang, S.; Wang, F.; Yang, J.; Ismayil, A.; Wu, P.; Wang, L.; Li, H. Genes Affecting Cotton Fiber Length: A Systematic Review and Meta-Analysis. Plants 2025, 14, 1203. [Google Scholar] [CrossRef]
- Jia, T.; Wang, H.; Cui, S.; Li, Z.; Shen, Y.; Li, H.; Xiao, G. Cotton BLH1 and KNOX6 antagonistically modulate fiber elongation via regulation of linolenic acid biosynthesis. Plant Commun. 2024, 5, 100887. [Google Scholar] [CrossRef]
- Li, X.; Huang, G.; Zhou, Y.; Wang, K.; Zhu, Y. GhATL68b regulates cotton fiber cell development by ubiquitinating the enzyme required for β-oxidation of polyunsaturated fatty acids. Plant Commun. 2024, 5, 101003. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, H.; Zhu, J.; Wang, X.; Jiang, B.; Hou, L.; Xiao, G. A conserved brassinosteroid-mediated BES1-CERP-EXPA3 signaling cascade controls plant cell elongation. Cell Rep. 2023, 42, 112301. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Ji, M.; You, J.; Zhang, Y.; Lindsey, K.; Zhang, X.; Tu, L.; Wang, M. Synergistic interplay of redox homeostasis and polysaccharide synthesis promotes cotton fiber elongation. Plant J. 2024, 118, 405–422. [Google Scholar] [CrossRef] [PubMed]
- Liú, R.; Xiāo, X.; Gōng, J.; Lǐ, J.; Yán, H.; Gě, Q.; Lú, Q.; Lǐ, P.; Pān, J.; Shāng, H. Genetic linkage analysis of stable QTLs in Gossypium hirsutum RIL population revealed function of GhCesA4 in fiber development. J. Adv. Res. 2024, 65, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Khan, S.H.; Ahmed, A.; Iqbal, M.U.; Mubarik, M.S.; Ghouri, M.Z.; Ahmad, F.; Yaseen, S.; Ali, Z.; Khan, A.A. Genome editing in cotton: Challenges and opportunities. J. Cotton Res. 2023, 6, 3. [Google Scholar] [CrossRef]
- Gutierrez-Reinoso, M.; Aponte, P.; Garcia-Herreros, M. Genomic analysis, progress and future perspectives in dairy cattle selection: A review. Animals 2021, 11, 599. [Google Scholar] [CrossRef]
- He, X.; Qi, Z.; Liu, Z.; Chang, X.; Zhang, X.; Li, J.; Wang, M. Pangenome analysis reveals transposon-driven genome evolution in cotton. BMC Biol. 2024, 22, 92. [Google Scholar] [CrossRef]
- Sheri, V.; Kumar, M.; Jaconis, S.; Zhang, B. Antioxidant defense in cotton under environmental stresses: Unraveling the crucial role of a universal defense regulator for enhanced cotton sustainability. Plant Physiol. Biochem. 2023, 204, 108141. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Das, S.P.; Gupta, A.; Parihar, P.; Chandrasekhar, K.; Sarker, U.; Kumar, A.; Ramrao, D.P.; Sudhakar, C. Multi-omics pipeline and omics-integration approach to decipher plant’s abiotic stress tolerance responses. Genes 2023, 14, 1281. [Google Scholar] [CrossRef]
- Rahman, M.-u.; Zulfiqar, S.; Raza, M.A.; Ahmad, N.; Zhang, B. Engineering abiotic stress tolerance in crop plants through CRISPR genome editing. Cells 2022, 11, 3590. [Google Scholar] [CrossRef]
- Ahmed, M.Z.; De Barro, P.J.; Greeff, J.M.; Ren, S.X.; Naveed, M.; Qiu, B.L. Genetic identity of the Bemisia tabaci species complex and association with high cotton leaf curl disease (CLCuD) incidence in Pakistan. Pest Manag. Sci. 2011, 67, 307–317. [Google Scholar] [CrossRef]
- Binyameen, B.; Khan, Z.; Khan, S.H.; Ahmad, A.; Munawar, N.; Mubarik, M.S.; Riaz, H.; Ali, Z.; Khan, A.A.; Qusmani, A.T. Using multiplexed CRISPR/Cas9 for suppression of cotton leaf curl virus. Int. J. Mol. Sci. 2021, 22, 12543. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fu, C.; Zhou, J.; Hui, F.; Wang, Q.; Wang, F.; Wang, G.; Xu, Z.; Che, L.; Yuan, D. Highly efficient genome editing using geminivirus-based CRISPR/Cas9 system in cotton plant. Cells 2022, 11, 2902. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.J.; Zheng, J.; Yang, M.; Batool, R.; Abro, A.A.; Hou, Y.; Xu, Y.; Gebremeskel, H.; Wang, Y.; Zhou, Z. Insights to Gossypium defense response against Verticillium dahliae: The cotton cancer. Funct. Integr. Genom. 2023, 23, 142. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhu, L.; Xu, L.; Yuan, D.; Min, L.; Zhang, X. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 2014, 5, 5372. [Google Scholar] [CrossRef]
- Hu, Q.; Min, L.; Yang, X.; Jin, S.; Zhang, L.; Li, Y.; Ma, Y.; Qi, X.; Li, D.; Liu, H. Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 2018, 176, 1808–1823. [Google Scholar] [CrossRef]
- Nadeem, S.; Riaz Ahmed, S.; Luqman, T.; Tan, D.K.; Maryum, Z.; Akhtar, K.P.; Muhy Ud Din Khan, S.; Tariq, M.S.; Muhammad, N.; Khan, M.K.R. A comprehensive review on Gossypium hirsutum resistance against cotton leaf curl virus. Front. Genet. 2024, 15, 1306469. [Google Scholar] [CrossRef]
- Giband, M.; Kranthi, K.R. Climate-smart breeding of cotton: Enhancing resilience in the face of climate change. ICAC Rec. 2023, 41, 17–22. [Google Scholar]
- Miedaner, T.; Juroszek, P. Climate change will influence disease resistance breeding in wheat in Northwestern Europe. Theor. Appl. Genet. 2021, 134, 1771–1785. [Google Scholar] [CrossRef]
- Chiurugwi, T.; Kemp, S.; Powell, W.; Hickey, L.T. Speed breeding orphan crops. Theor. Appl. Genet. 2019, 132, 607–616. [Google Scholar] [CrossRef]
- Wang, G.; Sun, Z.; Yang, J.; Ma, Q.; Wang, X.; Ke, H.; Huang, X.; Zhang, L.; Wang, G.; Gu, Q. The speed breeding technology of five generations per year in cotton. Theor. Appl. Genet. 2025, 138, 79. [Google Scholar] [CrossRef]
- Caradus, J.R. Processes for regulating genetically modified and gene edited plants. GM Crops Food 2023, 14, 1–41. [Google Scholar]
- Nouman Tahir, M.; Zahra, S. Regulatory, Biosafety, and Ethical Perspectives of Plant Genome Editing. In Genome Editing for Crop Improvement: Theory and Methodology; CAB International: Wallingford, UK, 2025; pp. 262–273. [Google Scholar]
- Muleta, K.T.; Pressoir, G.; Morris, G.P. Optimizing genomic selection for a sorghum breeding program in Haiti: A simulation study. G3 Genes Genomes Genet. 2019, 9, 391–401. [Google Scholar] [CrossRef]
- Dipta, B.; Sood, S.; Mangal, V.; Bhardwaj, V.; Thakur, A.K.; Kumar, V.; Singh, B. KASP: A high-throughput genotyping system and its applications in major crop plants for biotic and abiotic stress tolerance. Mol. Biol. Rep. 2024, 51, 508. [Google Scholar] [CrossRef]
- Smith, D.T.; Potgieter, A.B.; Chapman, S.C. Scaling up high-throughput phenotyping for abiotic stress selection in the field. Theor. Appl. Genet. 2021, 134, 1845–1866. [Google Scholar] [CrossRef]
- Malhotra, N. Harnessing genomics for sustainable food systems with orphan crops. Discov. Agric. 2025, 3, 1–17. [Google Scholar] [CrossRef]
- Westengen, O.T.; Dalle, S.P.; Mulesa, T.H. Navigating toward resilient and inclusive seed systems. Proc. Natl. Acad. Sci. USA 2023, 120, e2218777120. [Google Scholar] [CrossRef]
- Fadda, C.; Mengistu, D.K.; Kidane, Y.G.; Dell’Acqua, M.; Pè, M.E.; Van Etten, J. Integrating conventional and participatory crop improvement for smallholder agriculture using the seeds for needs approach: A review. Front. Plant Sci. 2020, 11, 559515. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, M.; Spies, M.; Nielsen, J.Ø. Is there a future for smallholder farmers in bioeconomy? The case of ‘improved’seeds in South Punjab, Pakistan. For. Policy Econ. 2024, 158, 103100. [Google Scholar] [CrossRef]
- Yu, J.; Jung, S.; Cheng, C.-H.; Lee, T.; Zheng, P.; Buble, K.; Crabb, J.; Humann, J.; Hough, H.; Jones, D. CottonGen: The community database for cotton genomics, genetics, and breeding research. Plants 2021, 10, 2805. [Google Scholar] [CrossRef] [PubMed]
- Hamid, R.; Panahi, B.; Jacob, F. Identification of key pathways and associated transcription factor-miRNA-gene regulatory networks driving heterosis in cotton (Gossypium spp.). Funct. Plant Biol. 2025, 52, FP25041. [Google Scholar]
- Hamid, R.; Jacob, F.; Ghorbanzadeh, Z.; Jafari, L.; Alishah, O. Dynamic roles of small RNAs and DNA methylation associated with heterosis in allotetraploid cotton (Gossypium hirsutum L.). BMC Plant Biol. 2023, 23, 488. [Google Scholar] [CrossRef]
- Hamid, R.; Jacob, F.; Ghorbanzadeh, Z.; Nekouei, M.K.; Zeinalabedini, M.; Mardi, M.; Sadeghi, A.; Kumar, S.; Ghaffari, M.R. Genomic insights into CKX genes: Key players in cotton fibre development and abiotic stress responses. PeerJ 2024, 12, e17462. [Google Scholar] [CrossRef]
- Hamid, R.; Jacob, F.; Ghorbanzadeh, Z.; Mardi, M.; Ariaeenejad, S.; Zeinalabedini, M.; Ghaffari, M.R. Genome-wide identification and characterization of FORMIN genes in cotton: Implications for abiotic stress tolerance. Plant Gene 2024, 40, 100474. [Google Scholar] [CrossRef]
- Hamid, R.; Ghorbanzadeh, Z.; Jacob, F.; Nekouei, M.K.; Zeinalabedini, M.; Mardi, M.; Sadeghi, A.; Ghaffari, M.R. Decoding drought resilience: A comprehensive exploration of the cotton Eceriferum (CER) gene family and its role in stress adaptation. BMC Plant Biol. 2024, 24, 468. [Google Scholar] [CrossRef]
- Hamid, R.; Panahi, B.; Ghorbanzadeh, Z.; Jacob, F.; Zeinalabedini, M.; Ghaffari, M.R. Genome-wide identification and characterization of DUF789 genes in cotton: Implications for fibre development. BMC Plant Biol. 2025, 25, 1192. [Google Scholar] [CrossRef]
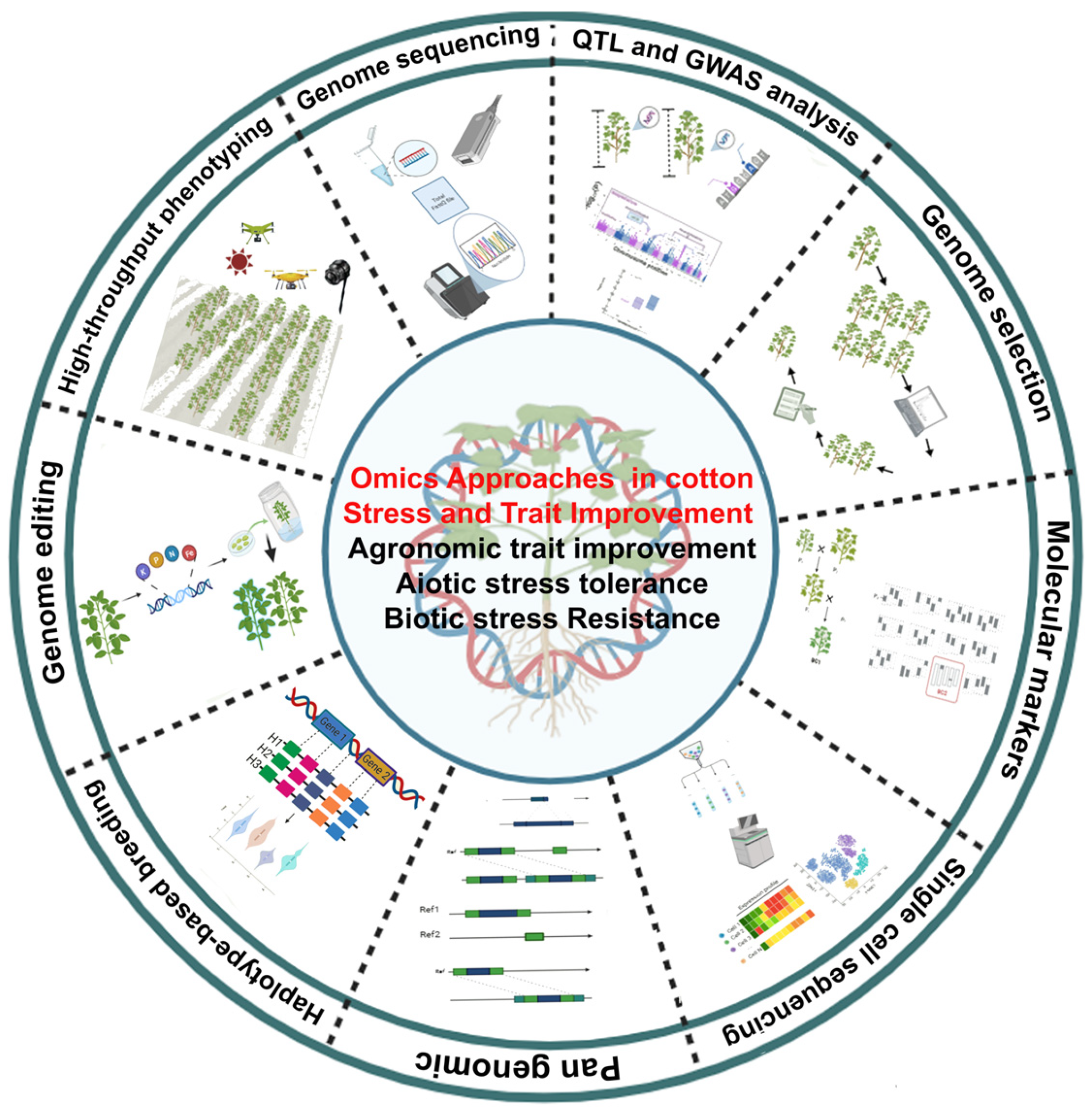
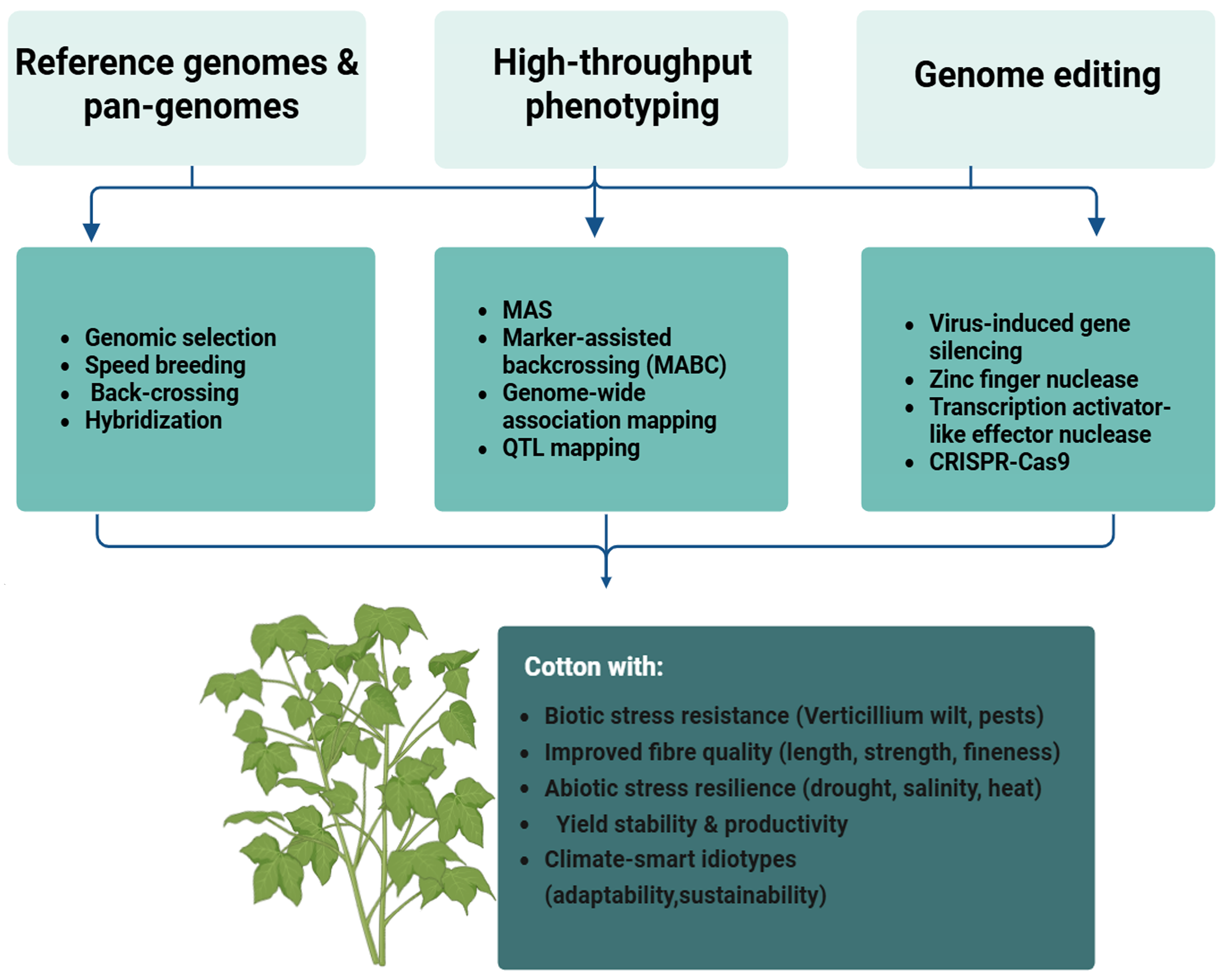
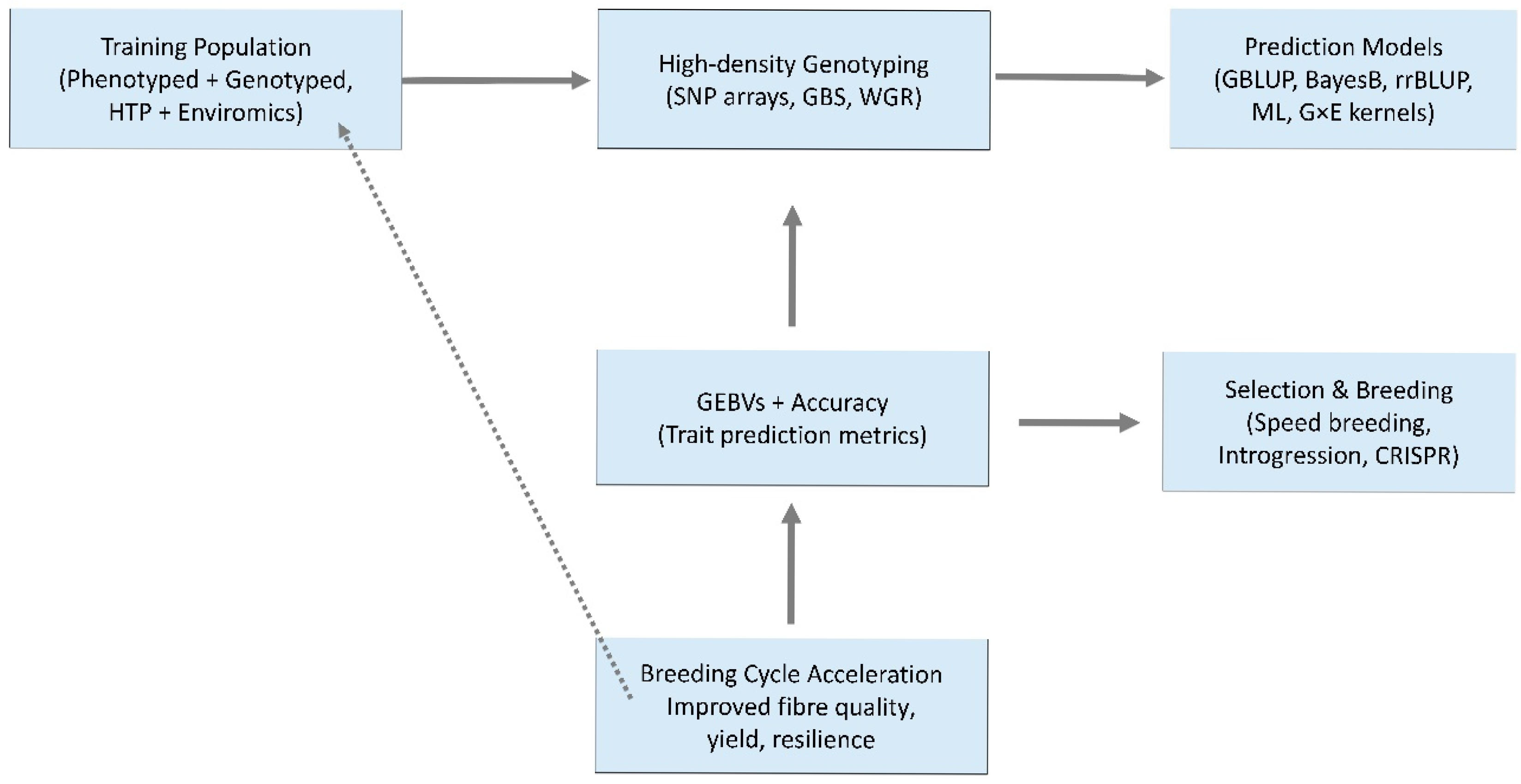

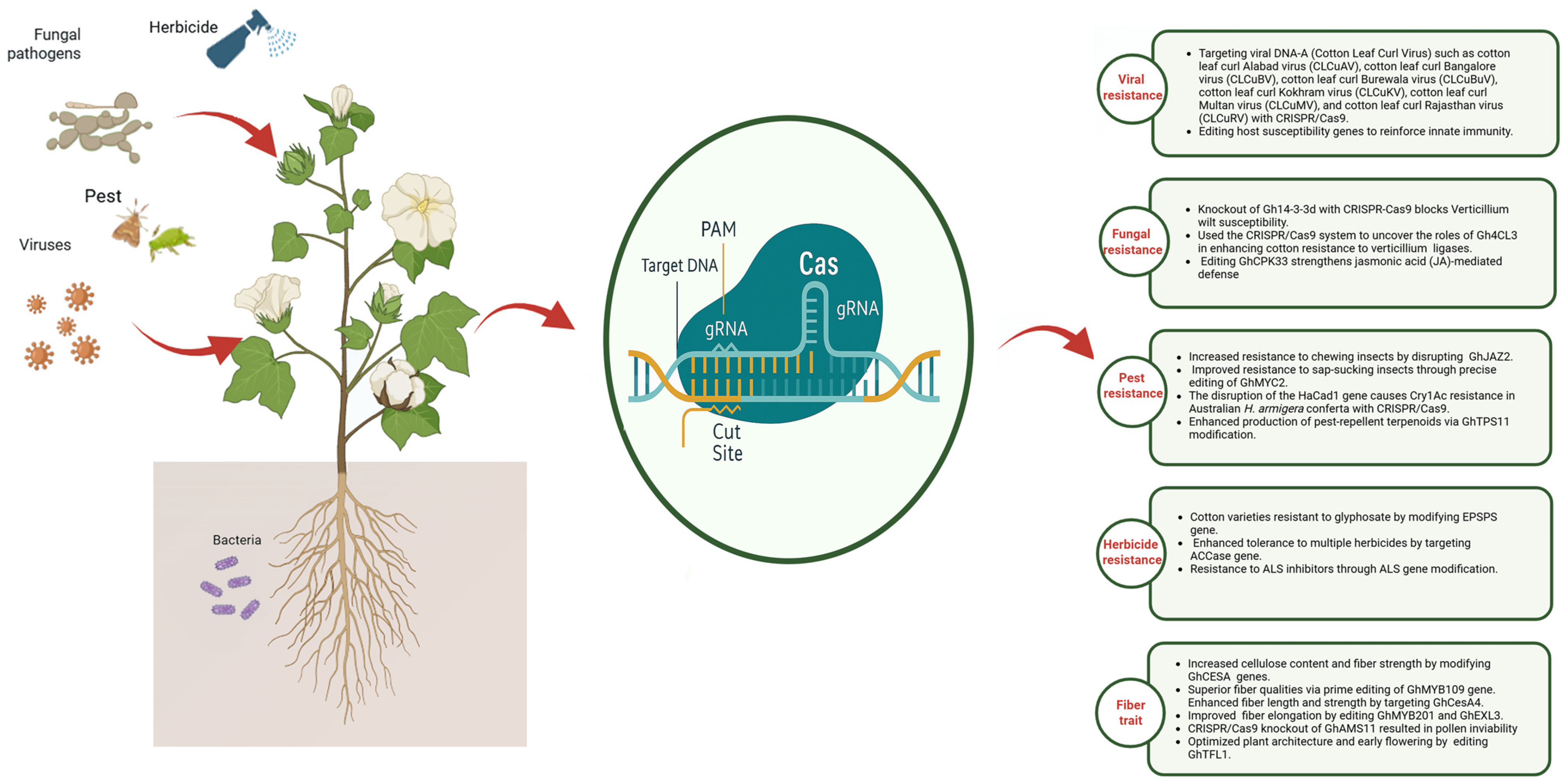
| Group | Species/ Accession | Genome Type | Year | Assembly Size (Gb) | N50 (Contig/Scaffold) | Technology Used | Key Contributions |
|---|---|---|---|---|---|---|---|
| Diploid references | G. raimondii (D5) | Diploid | 2012 | ~0.74 | 44.9–135.5 kb | Illumina short reads | First cotton genome; basis for comparative genomics |
| G. arboreum (A2) | Diploid | 2016 | ~1.7 | 72 kb | Illumina short reads | Insights into A-genome evolution and fibre traits | |
| Tetraploid references | G. hirsutum (TM-1) | Tetraploid | 2015 | ~2.3 | <1 Mb (fragmented) | Illumina short reads | First tetraploid draft; confirmed At–Dt subgenome collinearity |
| G. barbadense | Tetraploid | 2015 | ~2.3 | <1 Mb (fragmented) | Illumina short reads | Genomic basis for superior fibre quality | |
| G. hirsutum (updated) | Tetraploid | 2019 | ~2.3 | >10 Mb | PacBio, BioNano, Hi-C | Chromosome-scale assembly; improved annotation | |
| G. barbadense (updated) | Tetraploid | 2019 | ~2.3 | >10 Mb | PacBio, BioNano, Hi-C | Repeat resolution; functional gene discovery | |
| Pangenome resources | Multi-accession panels | Mixed | 2022–2024 | 1.5–2.3 | Near-chromosome scale | Hybrid assembly + Hi-C + ONT | Structural variation, PAVs, CNVs, haplotype diversity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghorbanzadeh, Z.; Panahi, B.; Purhang, L.; Hossein Panahi, Z.; Zeinalabedini, M.; Mardi, M.; Hamid, R.; Ghaffari, M.R. Integrative Genomics and Precision Breeding for Stress-Resilient Cotton: Recent Advances and Prospects. Agronomy 2025, 15, 2393. https://doi.org/10.3390/agronomy15102393
Ghorbanzadeh Z, Panahi B, Purhang L, Hossein Panahi Z, Zeinalabedini M, Mardi M, Hamid R, Ghaffari MR. Integrative Genomics and Precision Breeding for Stress-Resilient Cotton: Recent Advances and Prospects. Agronomy. 2025; 15(10):2393. https://doi.org/10.3390/agronomy15102393
Chicago/Turabian StyleGhorbanzadeh, Zahra, Bahman Panahi, Leila Purhang, Zhila Hossein Panahi, Mehrshad Zeinalabedini, Mohsen Mardi, Rasmieh Hamid, and Mohammad Reza Ghaffari. 2025. "Integrative Genomics and Precision Breeding for Stress-Resilient Cotton: Recent Advances and Prospects" Agronomy 15, no. 10: 2393. https://doi.org/10.3390/agronomy15102393
APA StyleGhorbanzadeh, Z., Panahi, B., Purhang, L., Hossein Panahi, Z., Zeinalabedini, M., Mardi, M., Hamid, R., & Ghaffari, M. R. (2025). Integrative Genomics and Precision Breeding for Stress-Resilient Cotton: Recent Advances and Prospects. Agronomy, 15(10), 2393. https://doi.org/10.3390/agronomy15102393






