The PAT Gene Family in Citrus: Genome-Wide Identification and Its Potential Implications for Organic Acid Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification of PAT Family in Citrus Genome
2.3. Phylogenetic Analysis and Chromosomal Location of PAT Family
2.4. Gene Structure, Conserved Motif and Domain Analysis of CitPATs
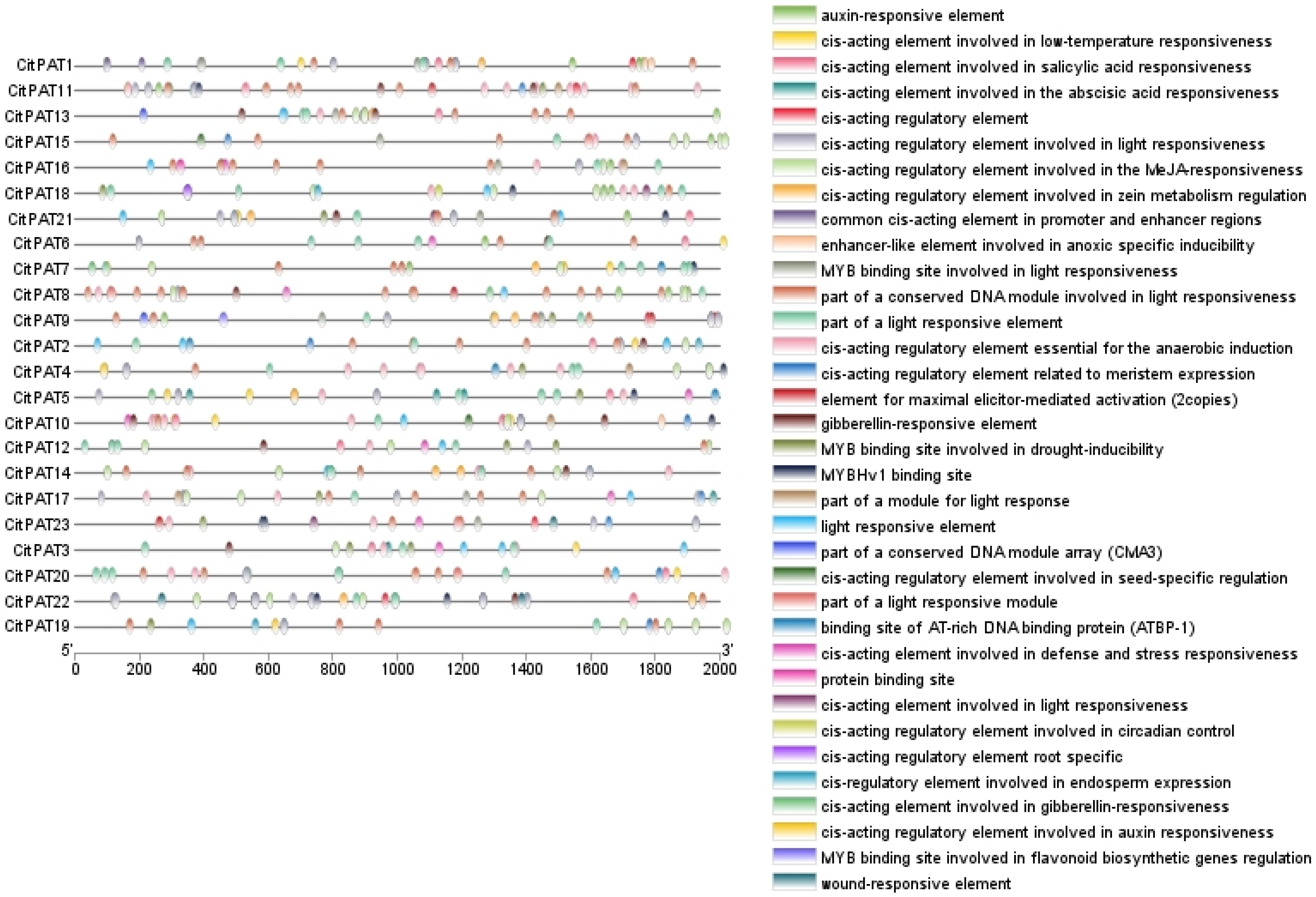
2.5. Promoter Cis-Regulatory Elements Analysis of CitPATs
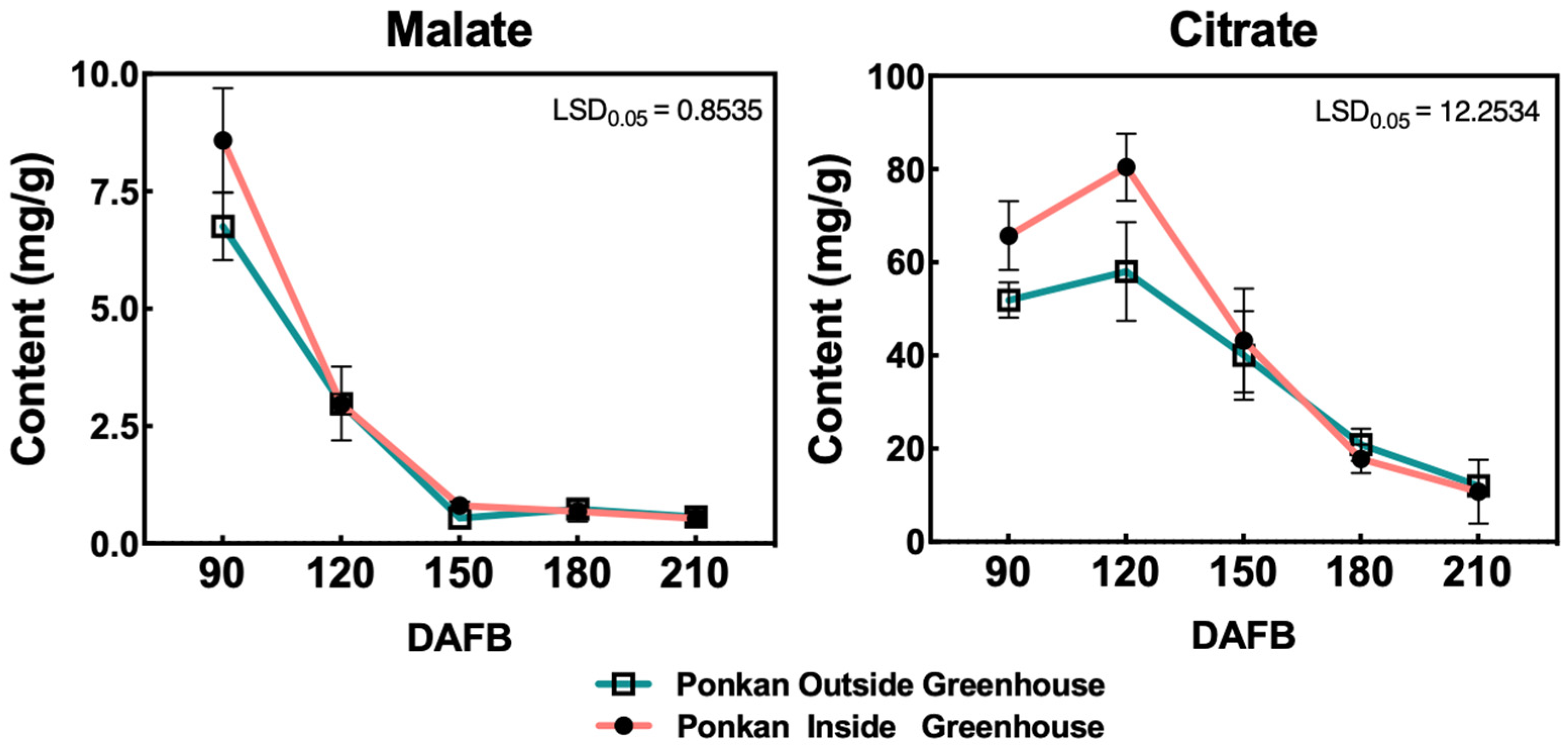
2.6. Organic Acid Measurement
2.7. RNA Extraction and cDNA Synthesis
2.8. RNA-seq
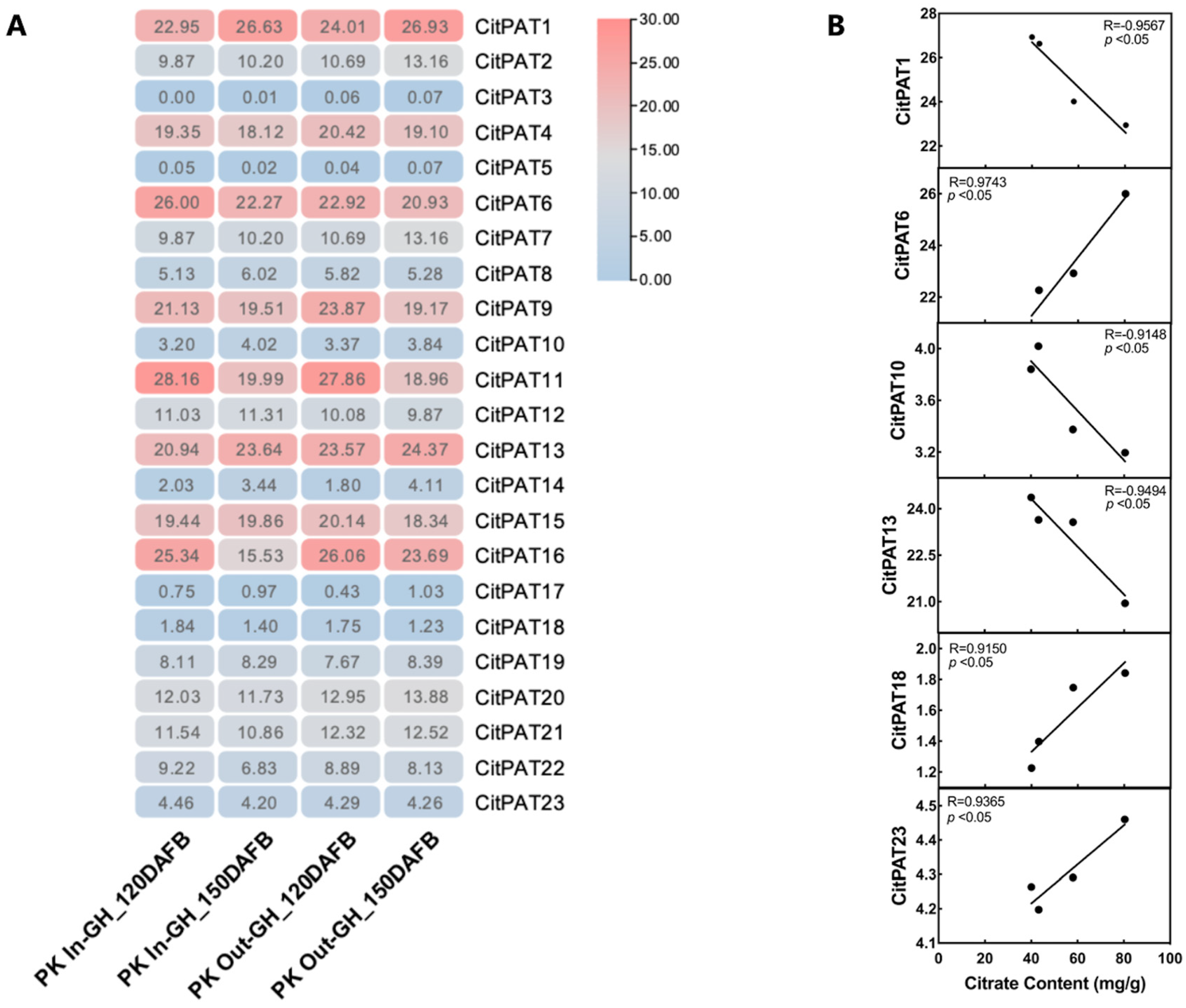
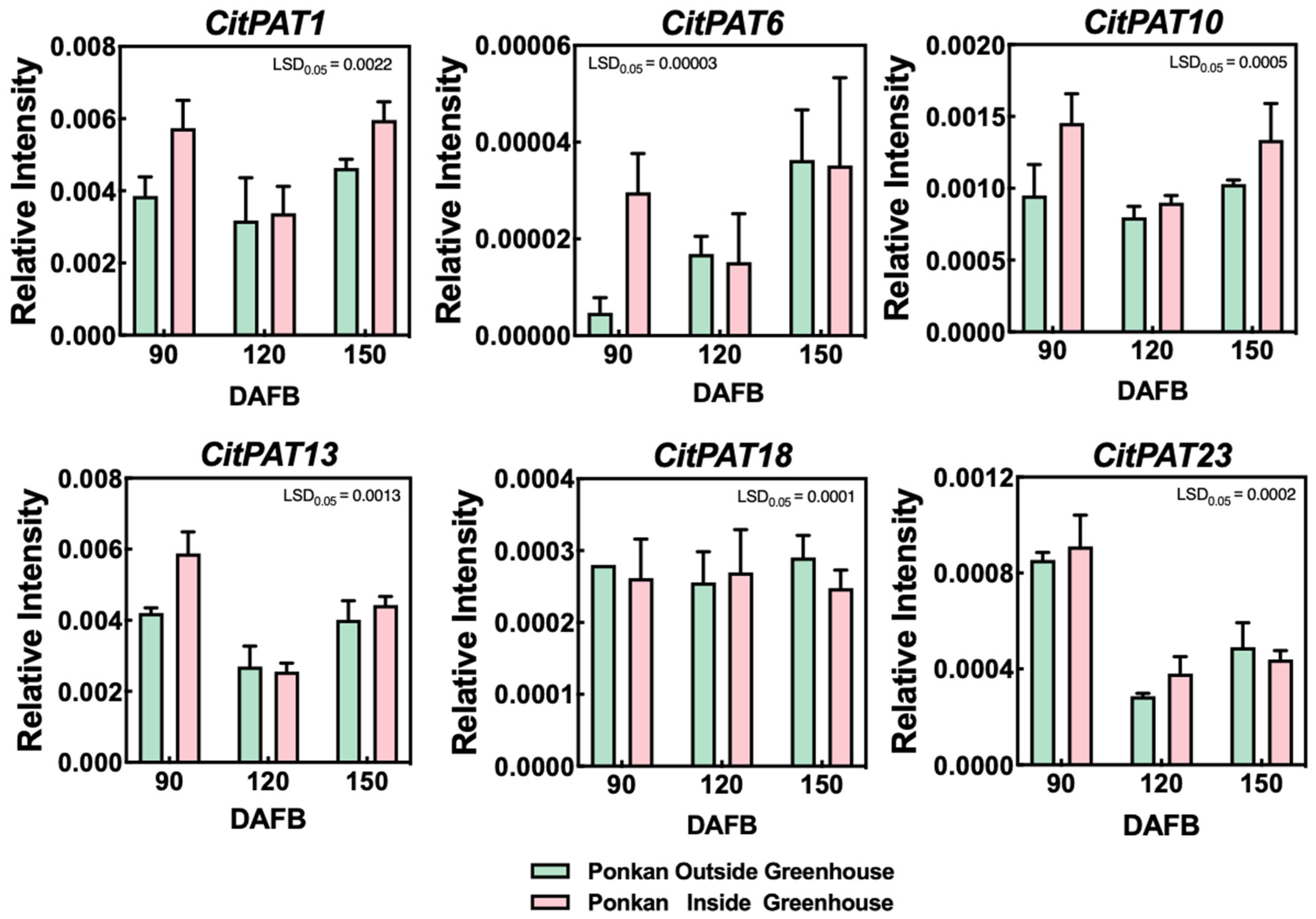
2.9. Expression Analysis of CitPATs
2.10. Statistical Analysis
3. Results
3.1. Identification of CitPATs Family in C. clementina Genome
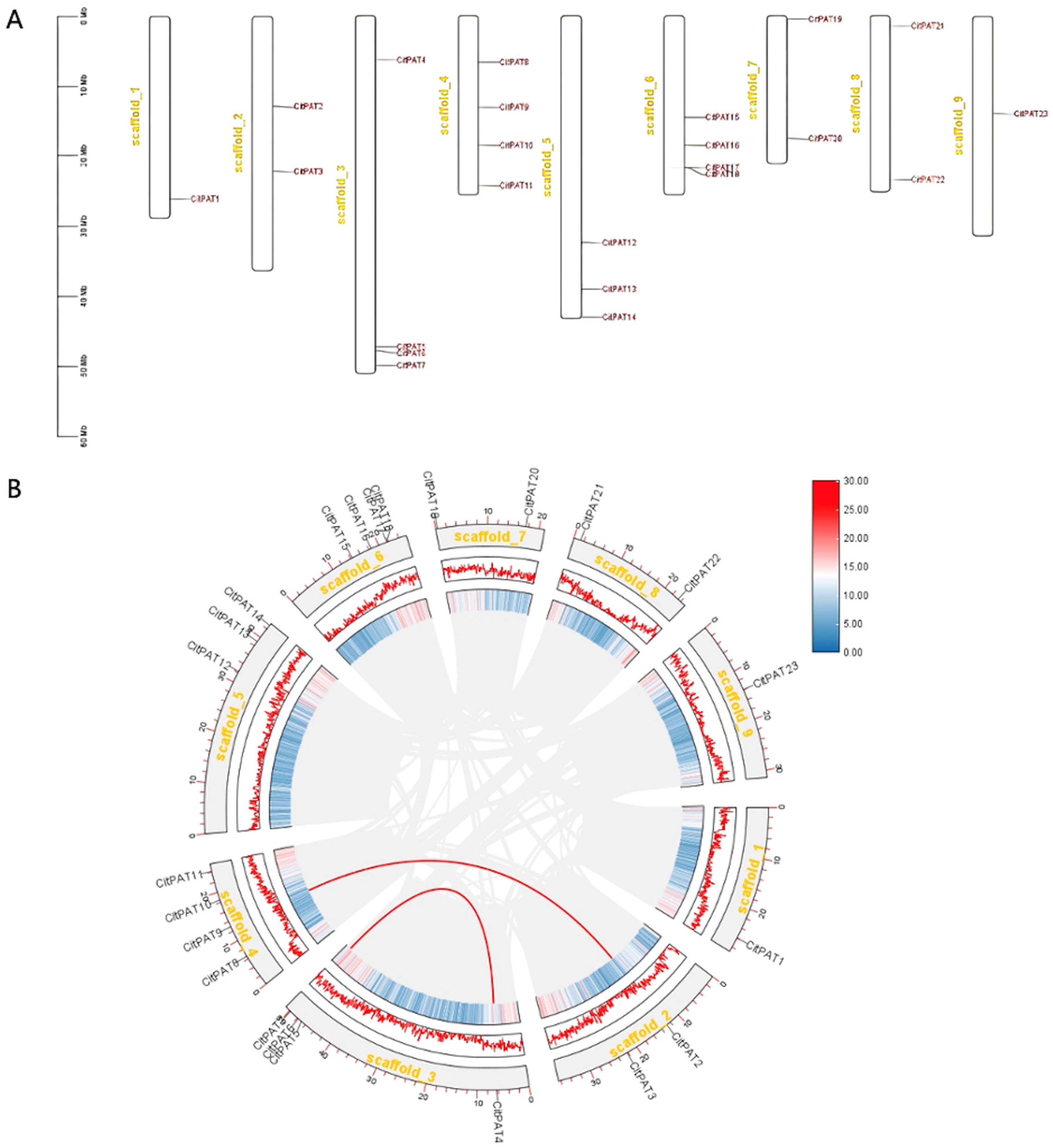
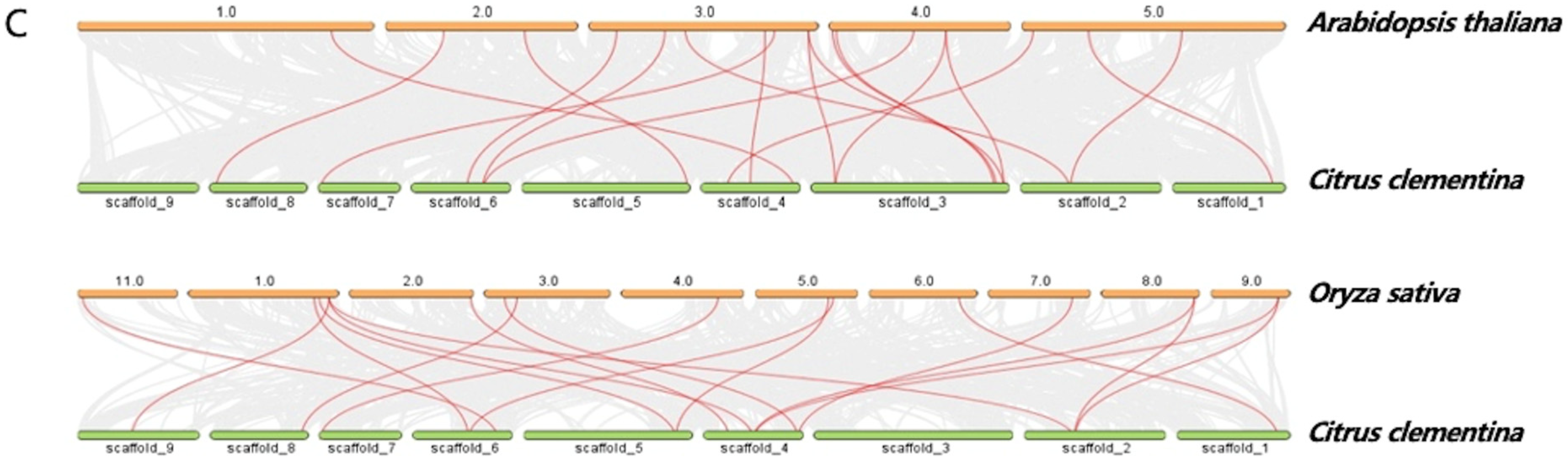
3.2. Chromosomal Location and Syntenic Relationships of CitPATs
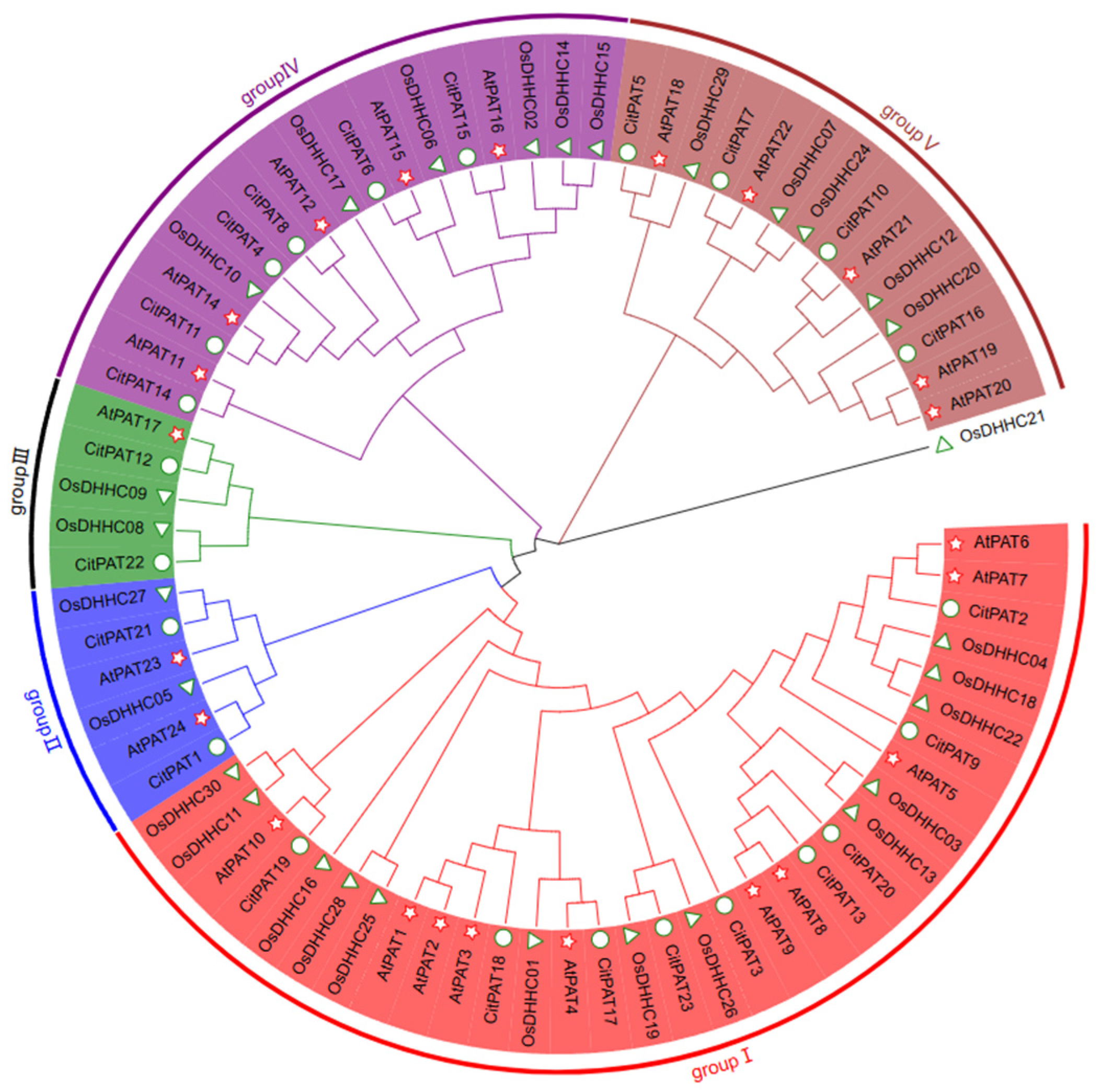
3.3. Phylogenetic Analysis of PAT Family
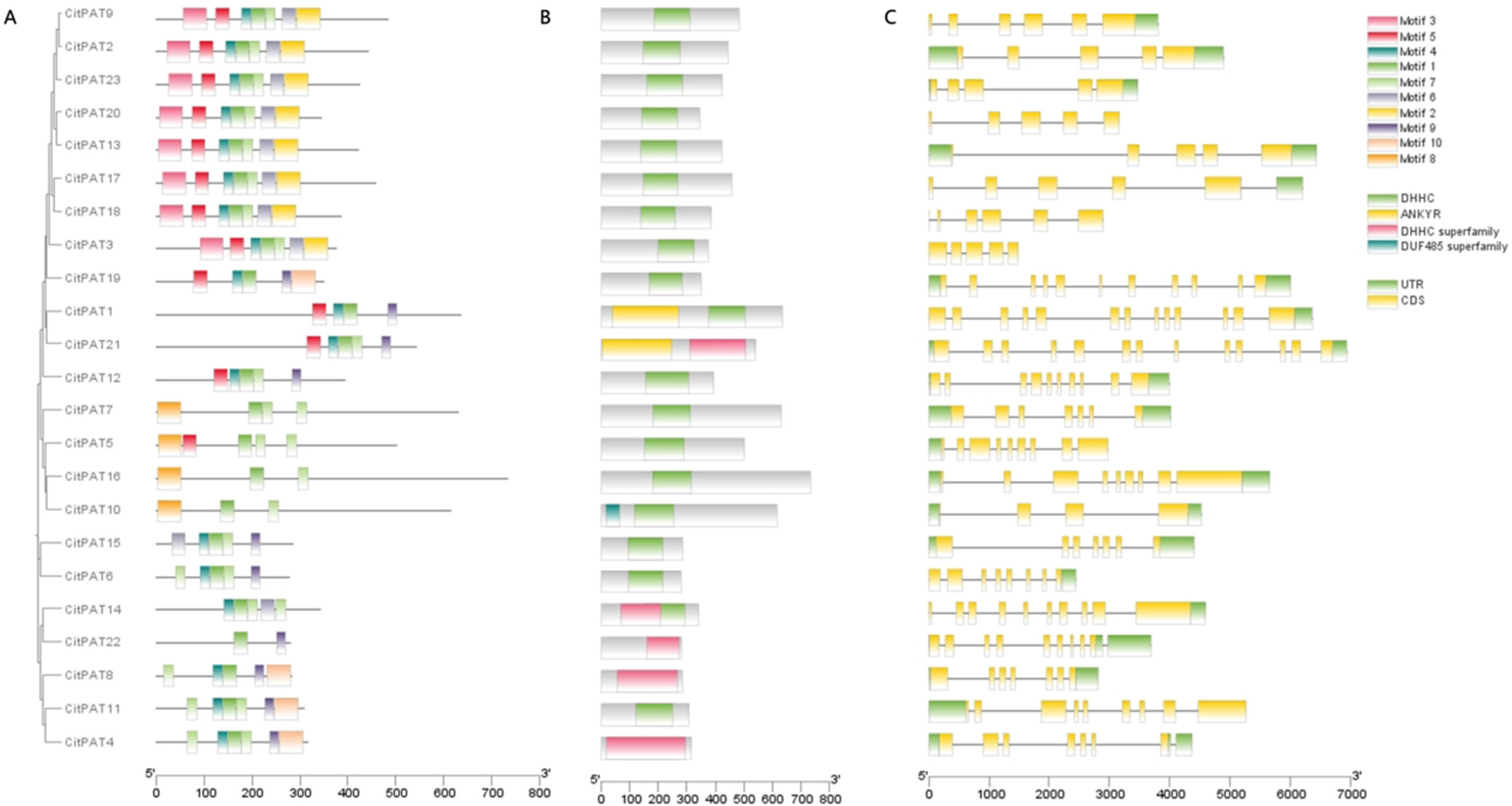
3.4. Gene Structure, Conserved Motif and Domain Analysis of CitPATs
3.5. Promoter Cis-Regulatory Elements Analysis of CitPATs
3.6. Changes in Organic Acid During ‘Ponkan’ Fruit Development
3.7. Expression Pattern of CitPATs During ‘Ponkan’ Fruit Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, F.; Qu, P.-Y.; Li, J.-P.; Yang, L.-N.; Geng, Y.-J.; Lu, J.-Y.; Zhang, Y.; Li, S. Arabidopsis protein S-acyl transferases positively mediate BR signaling through S-acylation of BSK1. Proc. Natl. Acad. Sci. USA 2024, 121, e2322375121. [Google Scholar] [CrossRef] [PubMed]
- Hemsley, P.A. Progress in understanding the mechanisms and functional importance of protein-membrane interactions in plants. New Phytol. 2014, 204, 741–753. [Google Scholar] [CrossRef]
- Das, T.; Yount, J.S.; Hang, H.C. Protein S-palmitoylation in immunity. Open Biol. 2021, 11, 200411. [Google Scholar] [CrossRef]
- Ding, Y.J.; Han, Y.Y.; Zhou, J.W. Research progress of palmitoylated proteins and palmitoylation of proteins. Subtrop. Plant Sci. 2018, 47, 395–403. [Google Scholar]
- Thinon, E.; Fernandez, J.P.; Molina, H.; Hang, H.C. Selective enrichment and direct analysis of protein S-palmitoylation sites. J. Proteome Res. 2018, 17, 1907–1922. [Google Scholar] [CrossRef] [PubMed]
- Hemsley, P.A.; Kemp, A.C.; Grierson, C.S. The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 2005, 17, 2554–2563. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Roux, M.E.; Sæmundsson, H.P.; Müller, M.; Bosch, S.M.; Petersen, M. The Arabidopsis thaliana mRNA decay factor PAT1 functions in osmotic stress responses and decaps ABA-responsive genes. FEBS Lett. 2021, 595, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Qi, B.; Doughty, J.; Hooley, R. A Golgi and tonoplast localized S-acyl transferase is involved in cell expansion, cell division, vascular patterning and fertility in Arabidopsis. New Phytol. 2013, 200, 444–456. [Google Scholar] [CrossRef]
- Zhou, L.Z.; Li, S.; Feng, Q.N.; Zhang, Y.L.; Zhao, X.; Zeng, Y.L.; Wang, H.; Jiang, L.; Zhang, Y. Protein S-ACYL Transferase10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 2013, 25, 1093–1107. [Google Scholar] [CrossRef]
- Rao, M.J.; Zuo, H.; Xu, Q. Genomic insights into citrus domestication and its important agronomic traits. Plant Commun. 2021, 2, 100138. [Google Scholar] [CrossRef]
- Albertini, M.V.; Carcouet, E.; Pailly, O.; Gambotti, C.; Luro, F.; Berti, L. Changes in organic acids and sugars during early stages of development of acidic and acidless citrus fruit. J. Agric. Food Chem. 2006, 54, 8335–8339. [Google Scholar] [CrossRef]
- Etienne, A.; Génard, M.; Lobit, P.; Mbeguié-A-Mbéguié, D.; Bugaud, C. What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 2013, 64, 1451–1469. [Google Scholar] [CrossRef]
- Hussain, S.B.; Shi, C.Y.; Guo, L.X.; Kamran, H.M.; Sadka, A.; Liu, Y.Z. Recent Advances in the Regulation of Citric Acid Metabolism in Citrus Fruit. Crit. Rev. Plant Sci. 2017, 36, 241–256. [Google Scholar] [CrossRef]
- Lin, Q.; Qian, J.; Zhao, C.; Wang, D.; Liu, C.; Wang, Z.; Sun, C.; Chen, K. Low temperature induced changes in citrate metabolism in ‘Ponkan’ (Citrus reticulata Blanco cv. ‘Ponkan’) fruit during maturation. PLoS ONE 2016, 11, e0156703. [Google Scholar] [CrossRef]
- Li, S.; Yin, X.; Wang, W.; Liu, X.; Zhang, B.; Chen, K. Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J. Exp. Bot. 2017, 68, 3419–3426. [Google Scholar] [CrossRef]
- Shimada, T.; Nakano, R.; Shulaev, V.; Sadka, A.; Blumwald, E. Vacuolar citrate/H+ symporter of citrus juice cells. Planta 2006, 224, 472–480. [Google Scholar] [CrossRef]
- Strazzer, P.; Spelt, C.E.; Li, S.; Bliek, M.; Federici, C.T.; Roose, M.L.; Koes, R.; Quattrocchio, F.M. Hyperacidification of Citrus fruits by a vacuolar proton-pumping P-ATPase complex. Nat. Commun. 2019, 10, 744. [Google Scholar] [CrossRef]
- Huang, Y.; He, J.; Xu, Y.; Zheng, W.; Wang, S.; Chen, P.; Zeng, B.; Yang, S.; Jiang, X.; Liu, Z.; et al. Pangenome analysis provides insight into the evolution of the orange subfamily and a key gene for citric acid accumulation in citrus fruits. Nat. Genet. 2023, 55, 1964–1975. [Google Scholar] [CrossRef]
- Liu, S.C.; Li, Y.C.; Fan, Y.J.; Xu, M.J.; Huang, Z.Y.; Wang, D.L.; Sun, C.D.; Li, S.J. CitSAR-mediated coordination of sucrose and citrate metabolism in citrus fruits. Plant Physiol. 2025, 198, 2499–2518. [Google Scholar] [CrossRef]
- Lin, X.; Li, S.; Shi, Y.; Ma, Y.; Li, Y.; Tan, H.; Zhang, B.; Xu, C.; Chen, K. CitGATA7 interact with histone acetyltransferase CitHAG28 to promote citric acid degradation by regulating the glutamine synthetase pathway in citrus. Mol. Hortic. 2025, 5, 8. [Google Scholar] [CrossRef]
- Xin, W.; Zheng, H.; Yang, L.; Xie, S.; Xia, S.; Wang, J.; Wang, M.; Liu, Y.; Zou, D.; Liu, H.; et al. Genome-Wide Association Studies Identify OsNLP6 as a Key Regulator of Nitrogen Use Efficiency in Rice. Plant Biotechnol. J. 2025, 1–16. [Google Scholar] [CrossRef]
- Jaina, M.; Sara, C.; Lowri, W.; Matloob, Q.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef]
- Bailey, L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef]
- Batistic, O. Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiol. 2012, 160, 1597–1612. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.; Zhou, L. Protein S-acyltransferases and acyl protein thioesterases, regulation executors of protein S-acylation in plants. Front. Plant Sci. 2022, 13, 956231. [Google Scholar] [CrossRef]
- Mitchell, D.A.; Vasudevan, A.; Linder, M.E.; Deschenes, R.J. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 2006, 47, 1118–1127. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, X.; Xu, N.; Luo, X.; Zhang, Y.; Xu, R. Genomics and expression analysis of DHHC-cysteine-rich domain S-acyltransferase protein family in apple. Genes Genom. 2016, 38, 671–684. [Google Scholar] [CrossRef]
- Tian, Y.; Zeng, H.; Wu, J.; Huang, J.; Gao, Q.; Tang, D.; Cai, L.; Liao, Z.; Wang, Y.; Liu, X.; et al. Screening DHHCs of S-acylated proteins using an OsDHHC cDNA library and bimolecular fluorescence complementation in rice. Plant J. 2022, 110, 1763–1780. [Google Scholar] [CrossRef]
- Silaiyiman, S.; Zheng, Q.; Wang, Y.; Ouyang, L.; Guo, Z.; Yu, J.; Chen, R.; Peng, R.; Shen, C. Comprehensive Genome-Wide Investigation and Transcriptional Regulation of the DHHC Gene Family in Cotton Seed and Fiber Development. Agronomy 2024, 14, 1214. [Google Scholar] [CrossRef]
- Pang, H.; Yan, Q.; Zhao, S.; He, F.; Xu, J.; Qi, B.; Zhang, Y. Knockout of the S-acyltransferase Gene, PbPAT14, Confers the Dwarf Yellowing Phenotype in First Generation Pear by ABA Accumulation. Int. J. Mol. Sci. 2019, 20, 6347. [Google Scholar] [CrossRef]
- Gu, S.; Nie, X.; George, A.; Tyler, K.; Xing, Y.; Qin, L.; Qi, B. Bioinformatics and Expression rofiling of the DHHC-CRD S-Acyltransferases Reveal Their Roles in Growth and Stress Response in Woodland Strawberry (Fragaria vesca). Plants 2025, 14, 127. [Google Scholar] [CrossRef]
- Jiang, H.; Ma, Q.-J.; Zhong, M.-S.; Gao, H.-N.; Li, Y.-Y.; Hao, Y.-J. The apple palmitoyltransferase MdPAT16 influences sugar content and salt tolerance via an MdCBL1–MdCIPK13–MdSUT2.2 pathway. Plant J. 2021, 106, 689–705. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, X.; Dai, M.; Fan, Y.; Yang, Z.; Sun, Y.; Yu, X.; Song, R.; Zhang, M.; Lan, H.; et al. Citrate accumulation mediated through GhCS6 enhances antioxidant modulation under Cd2+ stress in cotton. Plant Physiol. Biochem. 2025, 228, 110226. [Google Scholar] [CrossRef]
- Liu, G.; Chen, Q.; Li, D.; Mai, H.; Zhou, Y.; Lin, M.; Feng, X.; Lin, X.; Lu, X.; Chen, K.; et al. GmSTOP1-3 Increases Soybean Manganese Accumulation Under Phosphorus Deficiency by Regulating GmMATE2/13 and GmZIP6/GmIREG3. Plant Cell Environ. 2025, 48, 1812–1828. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.-J.; Morgan, C.; Bomblies, K.; Yang, W.; Qi, B. Both male and female gametogenesis require a fully functional protein S-acyl transferase 21 in Arabidopsis thaliana. Plant J. 2019, 100, 754–767. [Google Scholar] [CrossRef] [PubMed]
- Lodi, R.S.; Jia, X.; Yang, P.; Peng, C.; Dong, X.; Han, J.; Liu, X.; Wan, L.; Peng, L. Whole genome sequencing and annotations of Trametes sanguinea ZHSJ. Sci. Data 2025, 12, 1460. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Huang, Z.; Jiang, Z.; Fan, Y.; Sun, L.; Li, S. The PAT Gene Family in Citrus: Genome-Wide Identification and Its Potential Implications for Organic Acid Metabolism. Agronomy 2025, 15, 2350. https://doi.org/10.3390/agronomy15102350
Li Y, Huang Z, Jiang Z, Fan Y, Sun L, Li S. The PAT Gene Family in Citrus: Genome-Wide Identification and Its Potential Implications for Organic Acid Metabolism. Agronomy. 2025; 15(10):2350. https://doi.org/10.3390/agronomy15102350
Chicago/Turabian StyleLi, Yinchun, Ziyi Huang, Ziyan Jiang, Yijing Fan, Lifang Sun, and Shaojia Li. 2025. "The PAT Gene Family in Citrus: Genome-Wide Identification and Its Potential Implications for Organic Acid Metabolism" Agronomy 15, no. 10: 2350. https://doi.org/10.3390/agronomy15102350
APA StyleLi, Y., Huang, Z., Jiang, Z., Fan, Y., Sun, L., & Li, S. (2025). The PAT Gene Family in Citrus: Genome-Wide Identification and Its Potential Implications for Organic Acid Metabolism. Agronomy, 15(10), 2350. https://doi.org/10.3390/agronomy15102350





