Genome-Wide Association Study and Transcriptome Analysis Identify QTL and Candidate Genes Involved in Nitrogen Response Mechanisms in Sorghum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Trait Assessment and Derivation of Indices
2.3. GWAS Analysis
2.4. RNA-seq Analysis
2.5. Haplotype Analysis
3. Results
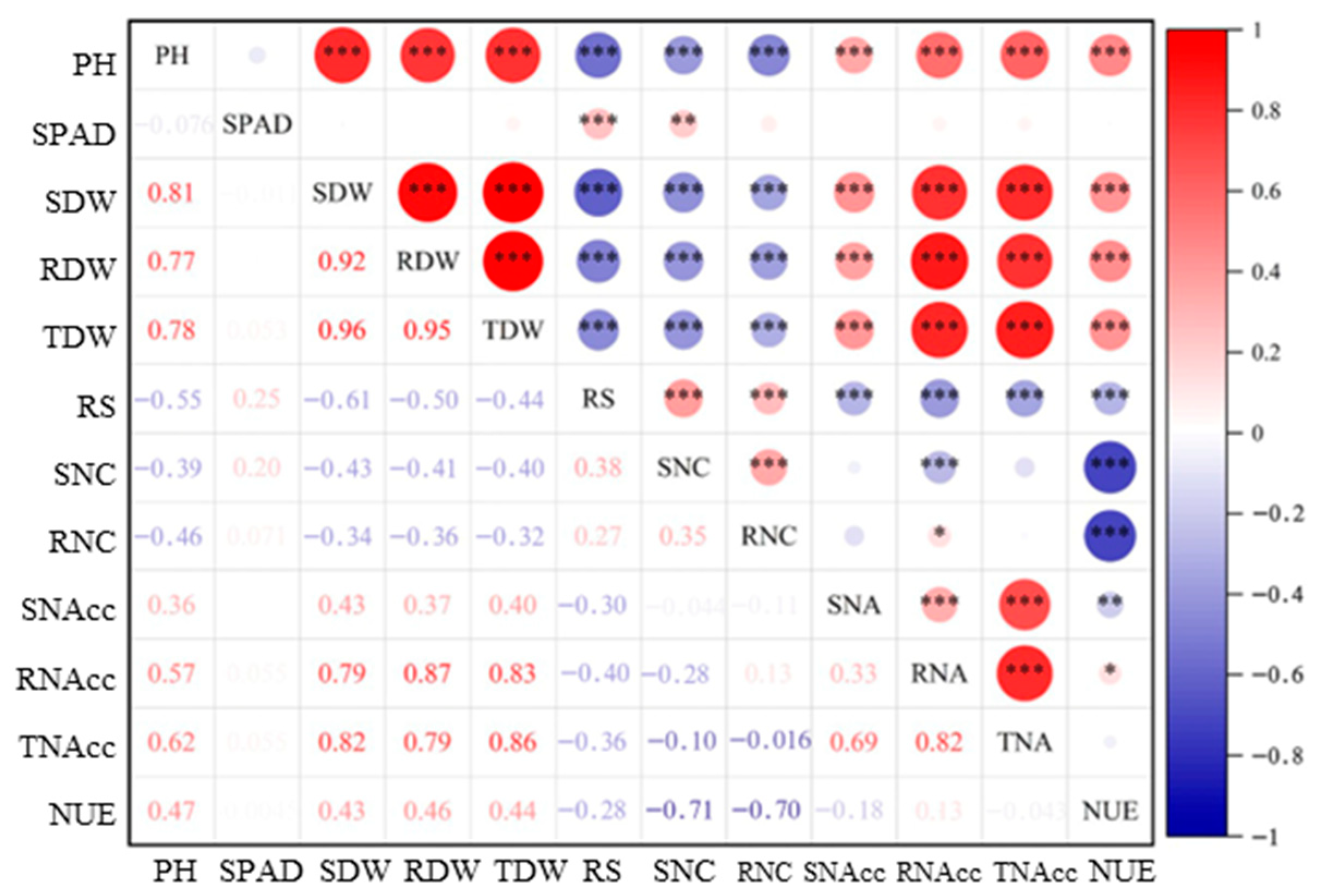
3.1. Phenotypic Variation of 232 Sorghum Accessions in Response to LN
3.2. Identification of QTL Regions and Significant Loci Involved in LN Response at the Seedling Stage via GWAS
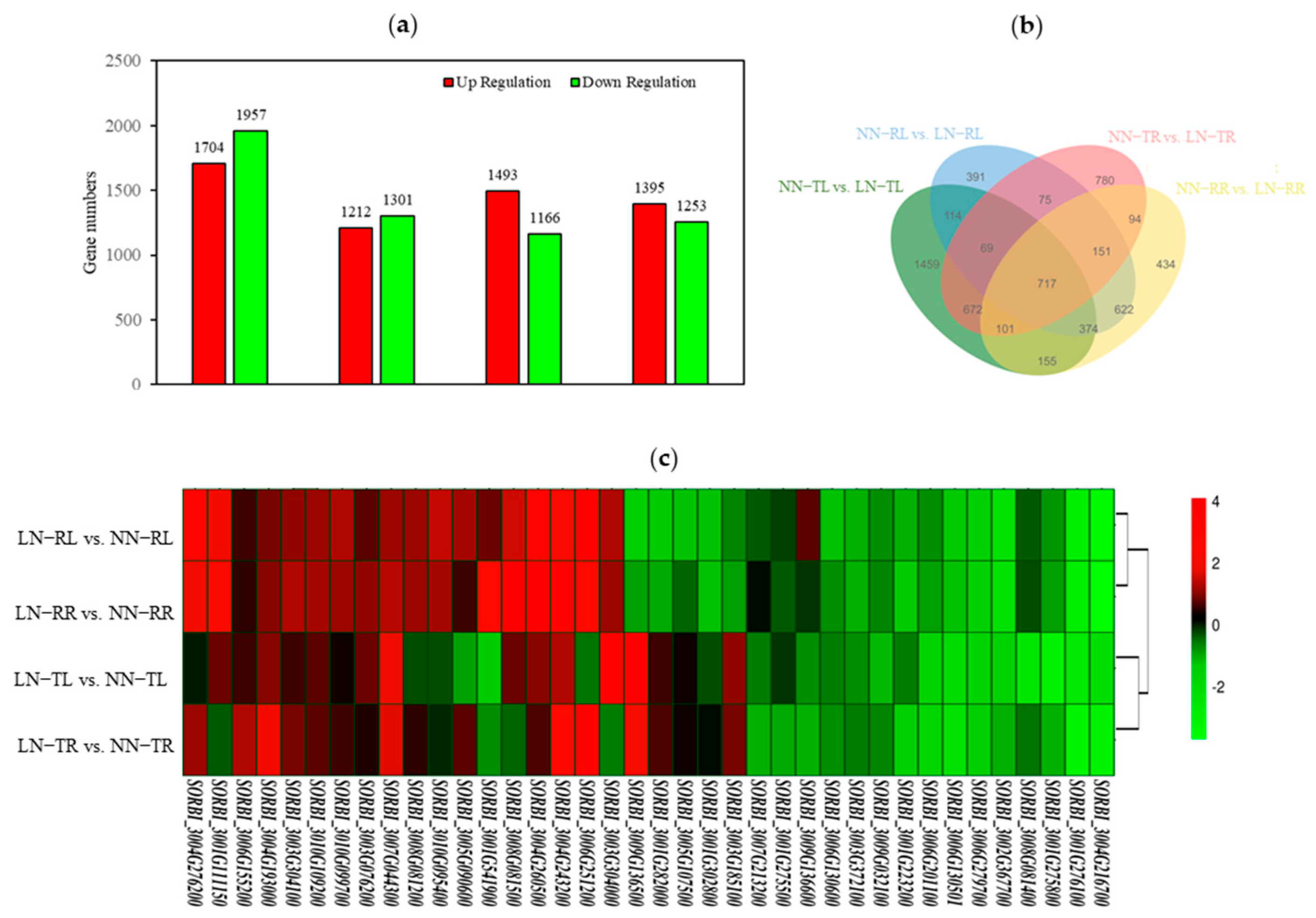
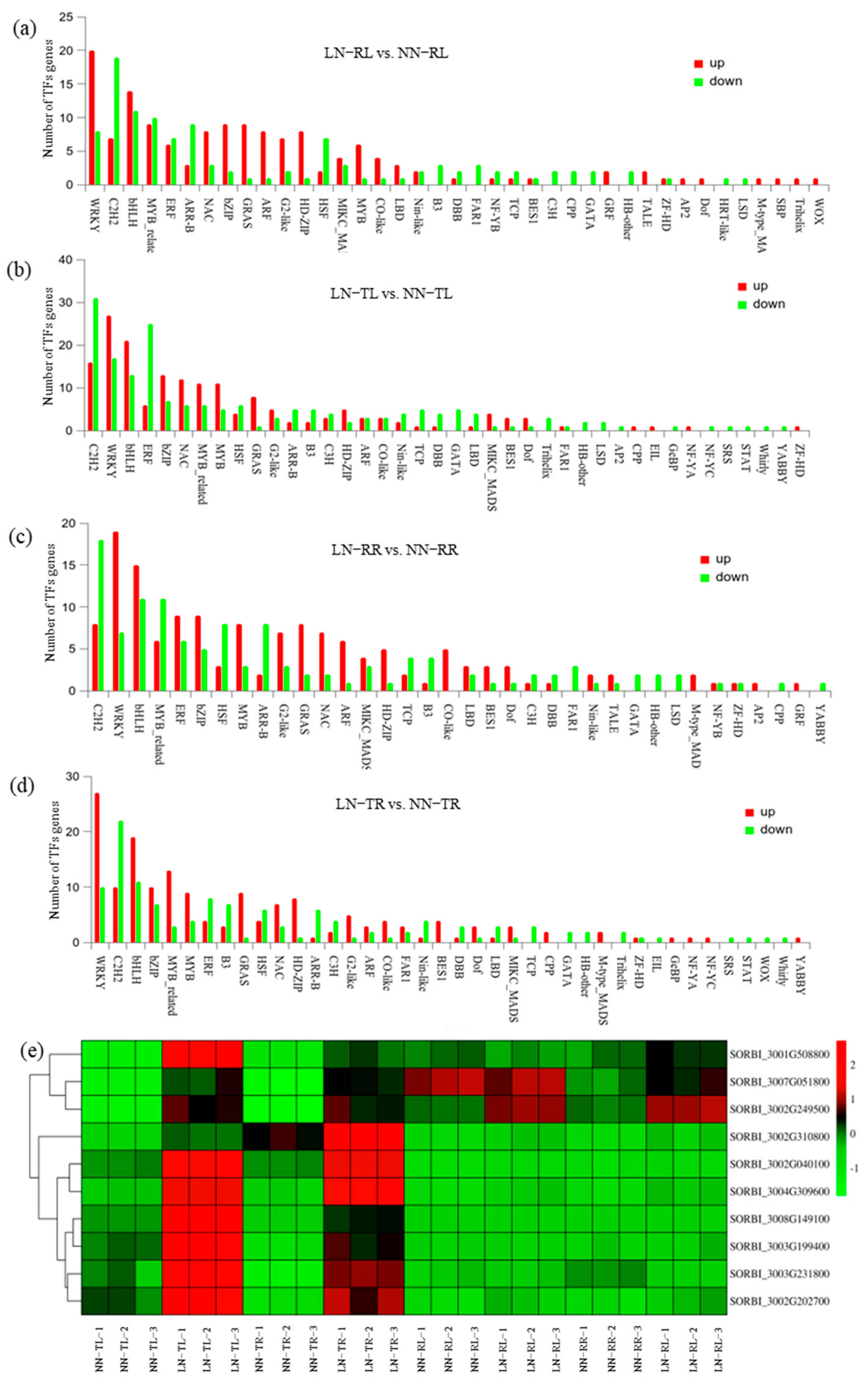
3.3. Differentially Expressed Genes Between LN-Responsive and LN-Tolerant Sorghum Accessions
3.4. Integrating GWAS and RNA-seq Data to Prioritize Candidate Genes
4. Discussion
4.1. Effects of LN Stress Conditions on Sorghum Phenotype
4.2. Candidate Genes Identified in the Detected QTL Regions Under LN Conditions
4.3. Nitrogen Transporter Family Members and Differential TFs Regulating LN Stress Responses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Zhuang, M.; Liang, X.; Lam, S.K.; Chen, D.; Malik, A.; Li, M.; Lenzen, M.; Zhang, L.; Zhang, R.; et al. Localized nitrogen management strategies can halve fertilizer use in Chinese staple crop production. Nat. Food. 2024, 5, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Bin, S.; Iqbal, A.; He, L.; Wei, S.; Zheng, H.; Yuan, P.; Liang, H.; Ali, I.; Xie, D.; et al. High Sink Capacity Improves Rice Grain Yield by Promoting Nitrogen and Dry Matter Accumulation. Agronomy 2022, 12, 1688. [Google Scholar] [CrossRef]
- Ma, R.; Jiang, C.; Shou, N.; Gao, W.; Yang, X. An Optimized Nitrogen Application Rate and Basal Topdressing Ratio Improves Yield, Quality, and Water- and N-use Efficiencies for Forage Maize (Zea mays L.). Agronomy 2023, 13, 181. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Jiang, Z.; Wang, W.; Xu, R.; Wang, Q.; Zhang, Z.; Li, A.; Liang, Y.; Ou, S.; et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef]
- Yadav, M.R.; Kumar, S.; Lal, M.K.; Kumar, D.; Kumar, R.; Yadav, R.K.; Kumar, S.; Nanda, G.; Singh, J.; Udawat, P.; et al. Mechanistic Understanding of Leakage and Consequences and Recent Technological Advances in Improving Nitrogen Use Efficiency in Cereals. Agronomy 2023, 13, 527. [Google Scholar] [CrossRef]
- Moore, J.W.; Ditmore, M.; Tebeest, D.O. The effects of cropping history on grain sorghum yields and anthracnose severity in Arkansa. Crop Prot. 2009, 28, 737–743. [Google Scholar] [CrossRef]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Analysis and Interpretation of Factors Which Contribute to Efficiency of Nitrogen Utilization. Agron. J. 1962, 74, 562–564. [Google Scholar] [CrossRef]
- Chen, Z.; Li, L.; Halford, N.G.; Xu, H.; Huang, L.; Gao, R.; Lu, R.; Liu, C. Advances in Barley Breeding for Improving Nitrogen Use Efficiency. Agronomy 2022, 12, 1682. [Google Scholar] [CrossRef]
- Saini, D.K.; Chopra, Y.; Singh, J.; Sandhu, K.S.; Kumar, A.; Bazzer, S.; Srivastava, P. Comprehensive evaluation of mapping complex traits in wheat using genome-wide association studies. Mol Breed. 2022, 42, 1. [Google Scholar] [CrossRef]
- Yang, X.; Xia, X.; Zhang, Z.; Nong, B.; Zeng, Y.; Xiong, F.; Wu, Y.; Gao, J.; Deng, G.; Li, D. QTL Mapping by Whole Genome Re-sequencing and Analysis of Candidate Genes for Nitrogen Use Efficiency in Rice. Front. Plant Sci. 2017, 8, 1634. [Google Scholar] [CrossRef]
- Wei, D.; Cui, K.; Ye, G.; Pan, J.; Xiang, J.; Huang, J.; Nie, L. QTL mapping for nitrogen-use efficiency and nitrogen-deficiency tolerance traits in rice. Plant Soil 2012, 359, 281–295. [Google Scholar] [CrossRef]
- Li, P.; Chen, F.; Cai, H.; Liu, J.; Pan, Q.; Liu, Z.; Gu, R.; Mi, G.; Zhang, F.; Yuan, L. A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J. Exp. Bot. 2015, 66, 3175–3188. [Google Scholar] [CrossRef]
- Han, M.; Wong, J.; Su, T.; Beatty, P.H.; Good, A.G. Identification of Nitrogen Use Efficiency Genes in Barley: Searching for QTLs Controlling Complex Physiological Traits. Front. Plant Sci. 2016, 7, 1587. [Google Scholar] [CrossRef]
- Hu, B.; Wang, W.; Chen, J.; Liu, Y.; Chu, C. Genetic improvement toward nitrogen-use efficiency in rice: Lessons and perspectives. Mol. Plant 2023, 16, 64–74. [Google Scholar] [CrossRef]
- Poudel, A.; Phogat, S.; Roy, J.; Saini, M.R.; Shivaprasad, K.M.; Madhavan, J.; Chinnusamy, V.; Vinod, K.K.; Sevanthi, A.M.; Mandal, P.K. Unlocking genetic hotspots: GWAS reveals key nitrogen responsive genomic regions and key genes for root and yield traits in indica rice. Mol. Genet. Genom. 2025, 300, 53. [Google Scholar] [CrossRef]
- Wang, M.H.; Cordell, H.J.; Van Steen, K. Statistical methods for genome-wide association studies. Semin. Cancer Biol. 2019, 55, 53–60. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Li, Q.; Lu, X.; Wang, C.; Shen, L.; Dai, L.; He, J.; Yang, L.; Li, P.; Hong, Y.; Zhang, Q.; et al. Genome-wide association study and transcriptome analysis reveal new QTL and candidate genes for nitrogen-deficiency tolerance in rice. Crop J. 2022, 10, 942–951. [Google Scholar] [CrossRef]
- Zhao, L.; Cai, B.; Zhang, X.; Zhang, B.; Feng, J.; Zhou, D.; Chen, Y.; Zhang, M.; Qi, D.; Wang, W.; et al. Physiological and Transcriptional Characteristics of Banana Seedlings in Response to Nitrogen Deficiency Stress. Horticulturae 2024, 10, 290. [Google Scholar] [CrossRef]
- Li, G.; Yang, D.; Hu, Y.; Xu, J.; Lu, Z. Genome-Wide Identification and Expression Analysis of Nitrate Transporter (NRT) Gene Family in Eucalyptus grandis. Genes 2024, 15, 930. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.; Liu, R.; Cheng, X.; Wang, Y.; Lv, X.; Chu, J.; Niu, H.; Yan, H.; Wang, Y.; Fan, F.; et al. Integrated transcriptome, GWAS, and metabolome revealed the mechanism of seed germination in sorghum. Front. Plant Sci. 2025, 16, 1601899. [Google Scholar] [CrossRef]
- Gautam, V.P.; Mishra, S.; Ahmed, H. Comparison of Total Nitrogen estimation by Kjeldahl Method and CHNS Analyzer in Dry Tropical Grassland. Int. J. Plant Environ. 2023, 9, 180–182. [Google Scholar] [CrossRef]
- Yun, P.; Kaya, C.; Shabala, S. Hormonal and epigenetic regulation of root responses to salinity stress. Crop J. 2024, 12, 1309–1320. [Google Scholar] [CrossRef]
- de Almeida, C.P.; Barbosa, R.R.; Ferraz, C.G.; de Castro, R.D.; Ribeiro, P.R. Genome-wide identification of the GDSL-type esterase/lipase protein (GELP) gene family in Ricinus communis and its transcriptional regulation during germination and seedling establishment under different abiotic stresses. Plant Physiol. Biochem. 2024, 214, 108939. [Google Scholar] [CrossRef]
- Qiao, X.; Yu, T.; Ruan, M.; Cui, C.; Chen, C.; Zhu, Y.; Li, F.; Wang, S.; Na, X.; Wang, X.; et al. Mitochondrial uncoupling protein contributes to the regulation of carbon and nitrogen metabolism and seed yield under low nitrogen stress in Arabidopsis. Cell. Mol. Life Sci. 2021, preprint. [Google Scholar] [CrossRef]
- Krapp, A.; David, L.C.; Chardin, C.; Girin, T.; Marmagne, A.; Leprince, A.-S.; Chaillou, S.; Ferrario-Méry, S.; Meyer, C.; Daniel-Vedele, F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.-X.; Cheng, H.; Ma, Z.; Xu, Q.; Wang, H.; Yan, Y.; Sun, M.; Wang, R.; Liu, W.; Ullah, S.; et al. Rapeseed transcription factor BnaC9bHLH35 promotes root growth and improves nitrogen use efficiency under low nitrate conditions. Theor. Appl. Genet. 2025, 138, 162. [Google Scholar] [CrossRef]
- Shi, H.; Wang, W.; Gao, L.; Wu, J.; Hu, C.; Yan, H.; Shi, Y.; Li, N.; Ma, Y.; Zhou, Y.; et al. Genome-wide association study of seedling nitrogen-use efficiency-associated traits in common wheat (Triticum aestivum L.). Crop J. 2024, 12, 222–231. [Google Scholar] [CrossRef]
- Boopathi, N.M. Genetic Mapping and Marker Assisted Selection; Springer: Singapore, 2020; pp. 253–326. [Google Scholar] [CrossRef]
- Kadirvel, P.; Senthilvel, S.; Geethanjali, S.; Sujatha, M.; Varaprasad, K.S. Genetic Markers, Trait Mapping and Marker-Assisted Selection in Plant Breeding. In Plant Biology and Biotechnology; Bahadur, B., Venkat Rajam, M., Sahijram, L., Krishnamurthy, K., Eds.; Springer: New Delhi, India, 2015; pp. 65–88. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef]
- Gelli, M.; Mitchell, S.E.; Liu, K.; Clemente, T.E.; Weeks, D.P.; Zhang, C.; Holding, D.R.; Dweikat, I.M. Mapping QTLs and association of differentially expressed gene transcripts for multiple agronomic traits under different nitrogen levels in sorghum. BMC Plant Biol. 2016, 16, 16. [Google Scholar] [CrossRef]
- Liu, C.; Gu, W.; Li, B.; Feng, Y.; Liu, C.; Shi, X.; Zhou, Y. Screening key sorghum germplasms for low-nitrogen tolerance at the seedling stage and identifying from the carbon and nitrogen metabolism. Front. Plant Sci. 2024, 15, 1340509. [Google Scholar] [CrossRef]
- He, F.; Wei, C.; Zhang, Y.; Long, R.; Li, M.; Wang, Z.; Yang, Q.; Kang, J.; Chen, L. Genome-Wide Association Analysis Coupled with Transcriptome Analysis Reveals Candidate Genes Related to Salt Stress in Alfalfa. Front. Plant Sci. 2022, 12, 826584. [Google Scholar] [CrossRef]
- Ying, W.; Wang, Y.; Wei, H.; Luo, Y.; Ma, Q.; Zhu, H.; Janssens, H.; Vukašinović, N.; Kvasnica, M.; Winne, J.M.; et al. Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export. Science 2024, 383, eadj4591. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Ye, R.; Liu, X.; He, Q.; Qiao, J.; Charrier, L.; Geisler, M.; Gao, Z.; Qian, Q.; Qi, Y. The OsbHLH166-OsABCB4 module regulates grain length and weight via altering auxin efflux. Sci. Bull. 2025, 70, 2125–2136. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Hasegawa, Y.; Suzuki, Y. Identification of a GDSL-motif carboxylester hydrolase from rice bran (Oryza sativa L.). J. Cereal Sci. 2012, 55, 100–105. [Google Scholar] [CrossRef]
- Zhao, H.; Ma, B.; Duan, K.-X.; Li, X.-K.; Lu, X.; Yin, C.-C.; Tao, J.-J.; Wei, W.; Zhang, W.-K.; Xin, P.-Y.; et al. The GDSL Lipase MHZ11 Modulates Ethylene Signaling in Rice Roots. Plant Cell 2020, 32, 1626–1643. [Google Scholar] [CrossRef]
- Luo, J.; Hang, J.; Wu, B.; Wei, X.; Zhao, Q.; Fang, Z. Co-overexpression of genes for nitrogen transport, assimilation, and utilization boosts rice grain yield and nitrogen use efficiency. Crop J. 2023, 11, 785–799. [Google Scholar] [CrossRef]
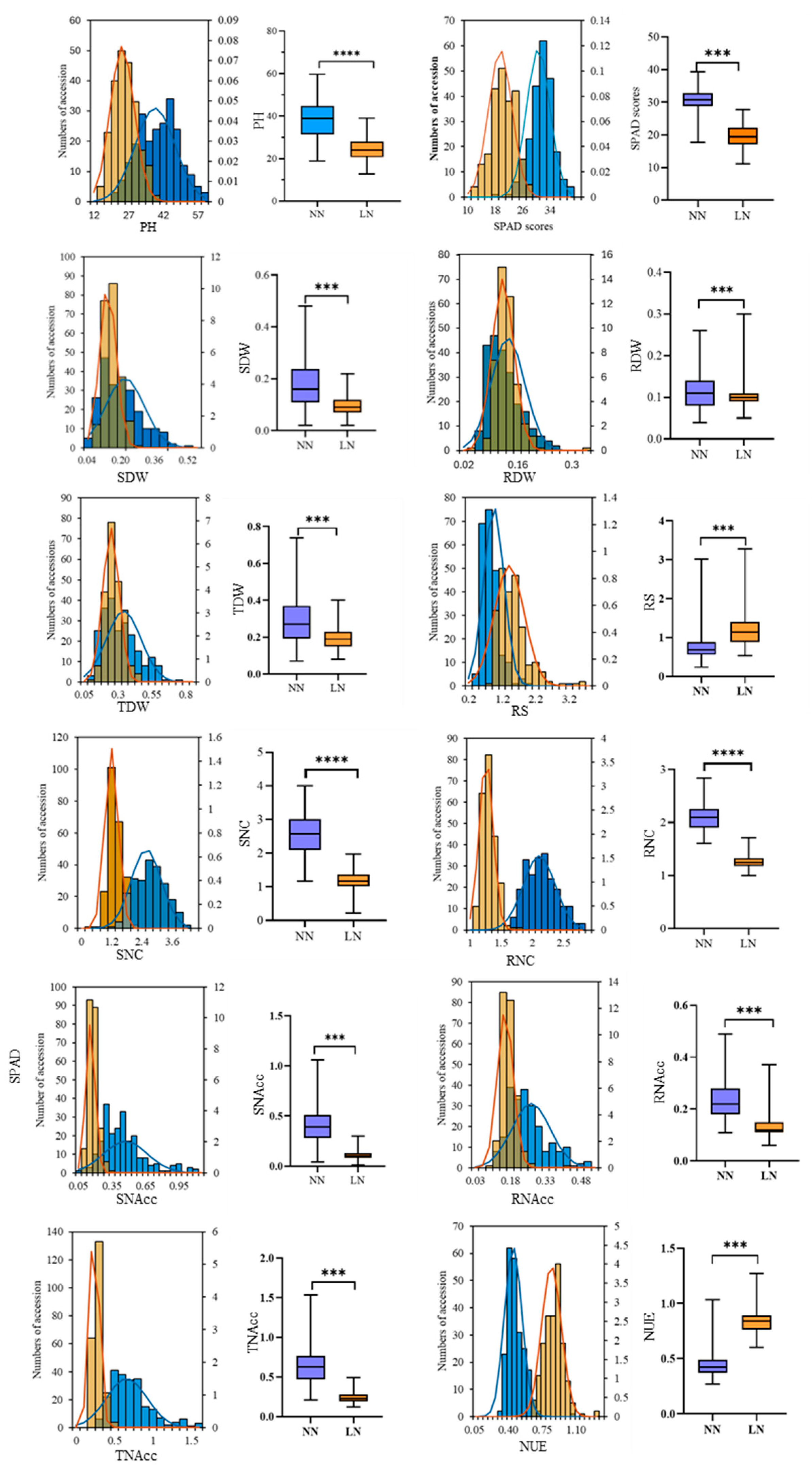

| Trait | NN | LN | NRC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Range | Mean | CV% | Range | Mean | CV% | Range | Mean | CV% | |
| PH | 18.75–59.60 | 38.47 | 22.23 | 12.13–39.00 | 24.29 | 23.61 | 0.38–1.02 | 0.64 | 15.11 |
| SPAD | 17.68–39.35 | 30.57 | 11.06 | 11.05–27.78 | 19.47 | 17.51 | 0.38–0.89 | 0.64 | 14.79 |
| SDW | 0.02–0.48 | 0.18 | 51.80 | 0.02–0.22 | 0.09 | 40.93 | 0.18–1.71 | 0.61 | 41.42 |
| RDW | 0.04–0.26 | 0.12 | 37.66 | 0.05–0.30 | 0.10 | 27.79 | 0.45–1.81 | 0.95 | 26.14 |
| TDW | 0.07–0.74 | 0.29 | 44.42 | 0.08–0.40 | 0.20 | 30.33 | 0.28–1.72 | 0.75 | 32.53 |
| RS | 0.24–3.02 | 0.75 | 39.84 | 0.53–3.28 | 1.21 | 36.82 | 0.90–5.00 | 1.68 | 32.38 |
| SNC | 1.17–4.00 | 2.58 | 23.40 | 0.21–1.97 | 1.20 | 22.14 | 0.16–0.88 | 0.47 | 18.88 |
| RNC | 1.60–2.84 | 2.11 | 12.27 | 1.00–1.71 | 1.26 | 8.76 | 0.00–0.84 | 0.60 | 14.40 |
| SNAcc | 0.04–1.06 | 0.42 | 46.76 | 0.01–0.26 | 0.11 | 38.18 | 0.02–3.18 | 0.29 | 74.93 |
| RNAcc | 0.11–0.49 | 0.24 | 34.62 | 0.06–0.37 | 0.13 | 26.41 | 0.27–1.14 | 0.57 | 26.65 |
| TNAcc | 0.21–1.53 | 0.66 | 40.03 | 0.12–0.49 | 0.24 | 26.88 | 0.17–0.95 | 0.39 | 29.58 |
| NUE | 0.27–1.03 | 0.44 | 20.45 | 0.61–1.27 | 0.83 | 12.10 | 0.83–2.80 | 1.94 | 14.85 |
| QTL | Chr. | Significant SNP Position (nt) | Physical Region | No. of SNPs Within the Genes | Detected Trait | −log10(P) |
|---|---|---|---|---|---|---|
| q1 | 3 | 8637974 | 8,593,974–8,681,978 | 42 | SDW, TDW, TNAcc, SNAcc | 6.81, 7.29, 6.68, 6.78 |
| q1 | 3 | 8637978 | 8,593,974–8,681,978 | SDW, TDW, TNAcc, RDW, SNAcc | 7.07, 7.50, 6.81, 6.58, 6.94 | |
| q1 | 3 | 8637994 | 8,593,974–8,681,996 | TDW | 6.78 | |
| q1 | 3 | 8637996 | 8,593,974–8,681,996 | TDW | 6.78 | |
| q2 | 3 | 60352480 | 60,308,480–60,396,480 | 76 | SDW, TDW, SNAcc, TNAcc | 6.98, 7.35, 6.59, 6.84 |
| q3 | 4 | 62886807 | 62,842,807–62,930,912 | 35 | SDW, TDW | 6.74, 6.58 |
| q3 | 4 | 62886832 | 62,842,807–62,930,912 | SDW, RDW, TDW | 7.46, 7.23, 7.42 | |
| q3 | 4 | 62886912 | 62,842,807–62,930,912 | SDW, RDW, TDW | 7.03, 6.63, 6.88 | |
| q3 | 4 | 62888412 | 62,844,412–62,932,412 | SNAcc | 6.58 | |
| q4 | 4 | 62949098 | 62,905,098–62,993,098 | 112 | PH | 7.12 |
| q5 | 4 | 63207939 | 63,163,939–63,251,939 | 16 | SDW, TDW, SNAcc | 6.95, 7.17, 6.56 |
| q6 | 6 | 42546320 | 42,502,320–42,590,320 | 48 | SNAcc, TNAcc | 6.62, 6.58 |
| q7 | 6 | 5956208 | 5,912,208–6,000,214 | 12 | NUE | 6.56 |
| q7 | 6 | 5956214 | 5,912,208–6,000,214 | NUE | 6.91 | |
| q8 | 8 | 15182381 | 15,138,381–15,226,381 | 2 | RNC | 6.60 |
| q9 | 10 | 34207226 | 34,163,226–34,251,226 | 7 | RNAcc, TDW | 6.92, 6.78 |
| q10 | 10 | 2040239 | 1,996,239–2,084,239 | 52 | RS | 6.85 |
| QTL | Locus ID | Log2(Fold Change) for DEGs | NsSNP No. | Haplotype No. | Functional Annotation | |||
|---|---|---|---|---|---|---|---|---|
| LN-RL vs. NN-RL | LN-TL vs. NN-TL | LN-RR vs. NN-RR | LN-TR vs. NN-TR | |||||
| q1 | SORBI_3004G286700 | Ns | 1.08 | Ns | Ns | 2 | 2 | GDSL esterase/lipase At5g55050-like |
| q1 | SORBI_3004G287100 | Ns | −1.37 | Ns | Ns | 1 | 2 | tRNA pseudouridine synthase A, mitochondrial isoform X1 |
| q1 | SORBI_3004G287400 | 1.19 | Ns | 1.15 | Ns | 3 | 4 | mechanosensitive ion channel protein 5 |
| q2 | SORBI_3003G097900 | 5.25 | Ns | 4.63 | Ns | 1 | 2 | putative E3 ubiquitin-protein ligase SINA-like 6 |
| q2 | SORBI_3003G098100 | Ns | Ns | 2.97 | 2.69 | 3 | 4 | aquaporin NIP4-1 |
| q2 | SORBI_3003G098200 | Ns | −1.63 | Ns | −1.19 | 3 | 3 | uncharacterized protein LOC8079071 |
| q3 | SORBI_3003G266600 | Ns | 1.06 | Ns | 1.07 | 1 | 2 | Phosphomethy lethanolamine N-methyltransferase |
| q3 | SORBI_3003G267300 | 1.38 | 1.28 | 1.44 | 1.29 | 2 | 3 | ABC transporter B family member 11 |
| q4 | SORBI_3004G286200 | 1.11 | Ns | 1.43 | Ns | 2 | 3 | uncharacterized protein LOC8073918 |
| q4 | SORBI_3004G286400 | 1.06 | Ns | 1.15 | Ns | 1 | 2 | uncharacterized protein LOC8076049 |
| q5 | SORBI_3004G291500 | Ns | −4.74 | Ns | −1.10 | 1 | 2 | 14 kDa proline-rich protein DC2.15 |
| q6 | SORBI_3006G065900 | Ns | Ns | Ns | 1.39 | 1 | 2 | uncharacterized protein At4g22758 |
| q8 | SORBI_3008G082200 | −4.02 | Ns | Ns | Ns | protein TWIN SISTER of FT | ||
| q10 | SORBI_3010G024500 | Ns | −1.05 | Ns | Ns | probable LRR receptor-like serine/threonine-protein kinase At1g34110 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, F.; Wang, Y.; Cheng, X.; Liu, R.; Wang, Y.; Ju, L.; Yan, H.; Niu, H.; Lv, X.; Chu, J.; et al. Genome-Wide Association Study and Transcriptome Analysis Identify QTL and Candidate Genes Involved in Nitrogen Response Mechanisms in Sorghum. Agronomy 2025, 15, 2250. https://doi.org/10.3390/agronomy15102250
Fan F, Wang Y, Cheng X, Liu R, Wang Y, Ju L, Yan H, Niu H, Lv X, Chu J, et al. Genome-Wide Association Study and Transcriptome Analysis Identify QTL and Candidate Genes Involved in Nitrogen Response Mechanisms in Sorghum. Agronomy. 2025; 15(10):2250. https://doi.org/10.3390/agronomy15102250
Chicago/Turabian StyleFan, Fangfang, Yao Wang, Xiaoqiang Cheng, Ruizhen Liu, Yubin Wang, Lan Ju, Haisheng Yan, Hao Niu, Xin Lv, Jianqiang Chu, and et al. 2025. "Genome-Wide Association Study and Transcriptome Analysis Identify QTL and Candidate Genes Involved in Nitrogen Response Mechanisms in Sorghum" Agronomy 15, no. 10: 2250. https://doi.org/10.3390/agronomy15102250
APA StyleFan, F., Wang, Y., Cheng, X., Liu, R., Wang, Y., Ju, L., Yan, H., Niu, H., Lv, X., Chu, J., Ping, J., & Jiao, X. (2025). Genome-Wide Association Study and Transcriptome Analysis Identify QTL and Candidate Genes Involved in Nitrogen Response Mechanisms in Sorghum. Agronomy, 15(10), 2250. https://doi.org/10.3390/agronomy15102250






