1. Introduction
Plant diseases are a major threat to worldwide agriculture, causing substantial yield losses and impacting food security and quality [
1]. Timely and accurate disease diagnosis is crucial for implementing effective management strategies in sustainable agriculture. These practices aim to contribute to more effective and precise plant protection measures due to more customized phytosanitary treatments regarding time, location, product used, and dose. However, traditional diagnostic methods often fail to detect diseases before visible symptoms emerge, limiting their effectiveness in proactive disease management [
2,
3]. Innovative plant disease monitoring and diagnosis methods involving different state-of-the-art sensing approaches have recently been explored for precise and in vivo and in situ disease assessment. Recent strides in innovative sensing techniques, particularly hyperspectral spectroscopy (HS), offer promising avenues for precise disease diagnosis [
2,
4].
Changes in the host plant’s physiological, biochemical, and metabolic properties caused by pathogens result in altered optical and metabolic features. Proximal optical sensors, including HS devices, can detect these changes, along with the monitorization of the spatiotemporal pattern of disease development [
5], which allows the development of several methods of diagnosis.
Plant pigments are one of the first host compounds to be affected and degraded by pathogens, resulting in changes in plant’s optical behavior. Chlorophylls (Chl) a and b are the major pigments of plants (accounting for almost 65% of the total pigment content), and their spectral absorption range is mostly concentrated in the 410–430 and (Chl a), 450–470 nm (Chl b), and 600–690 nm (Chl a) bands, located in the blue and red regions, respectively. Green radiation, on the other hand, is less strongly absorbed. In healthy plants, chlorophyll concentration is approximately ten times higher than that of other pigments (e.g., carotenoids and flavonoids, among others), thus masking out the specific absorption features of these compounds [
6].
With the disease development and onset, other photosynthetic pigment levels are increasingly more affected, namely carotenoids and polyphenols. The first type of pigment absorbs most effectively between 440 and 480 nm and extends its absorption action into the blue-green region. They include compounds such as yellow lutein pigments, β-carotenes, and xanthophylls (e.g., violaxanthin and zeaxanthin). In turn, polyphenols (e.g., brown pigments) start to appear only when the plant tissues begin to necrose [
7,
8,
9,
10]. They include compounds such as flavonoids and anthocyanins, which absorb radiation from blue to red spectral ranges with higher intensity in the shorter wavelengths [
7,
8,
9,
10].
Moreover, the optical spectral properties of host plants are also affected in the Near-Infrared (NIR) region (700–1300 nm) and short-wave infrared (SWIR, 1000–2500 nm) when plant leaves structure (e.g., cell layers, cell size, structural components—lignin, and proteins, among others), air spaces, and water content is affected [
9,
11].
In this regard, it is possible to see those changes in plant leaves’ biochemistry and cellular composition result in changes in a plant’s spectral characteristics. Nevertheless, it is important to mention that a leaf’s spectral properties are not a static phenomenon over time. Indeed, they continuously change during growth, maturity, senescence, decay, or stress (e.g., plant disease development).
Hence, despite the potential of HS in plant disease diagnosis, challenges persist in harnessing its full potential due to the complexity of hyperspectral data and the need for efficient processing methodologies to extract relevant information [
12,
13]. Addressing these challenges is crucial to unlocking the full potential of HS in improved disease diagnosis and management strategies.
HS is known for acquiring data in narrow wavebands (<10 nm), with high precision and resolution, and being able to capture detailed information from the electromagnetic spectrum [
12]. Nevertheless, despite this evident benefit, the measurement of this large number of variables (i.e., features, wavelengths) results in the data’s high dimensionality, which increases the complexity of its processing to produce relevant information. Furthermore, the spectral data assessed in near-contiguous variables likely present similar or overlapping information. This potential data redundancy also increases the complexity of its analysis interpretation and the chance of overfitting occurrence [
14]. Dimensionality reduction methods were developed to mitigate the effects of high dimensionality and collinearity, mostly based on identifying and extracting the most relevant and distinctive spectral features (without losing relevant information) [
13].
The computation of spectral Vegetation Indices (VIs) is one of the most widespread Feature Selection (FS) approaches for retrieving crop biophysical information, especially due to their intrinsic simplicity. It consists of a user-defined mathematical combination of two or more wavelengths that improves crop biophysical information extraction from data, i.e., identifying spectral relationships that unravel specific plant properties. Hence, VIs are considered as parametric, physiological-driven methods. Nonetheless, it is important to note that when narrowband hyperspectral data are used, Vis can be a restrictive formulation, since they only use some of the available wavelengths, failing to leverage the complete wealth of information in the continuous spectral data [
15]. Besides that, some of the VIs that have already been developed were designed to estimate specific vegetation traits (e.g., plant biomass and photosynthetic pigments research), which might not entirely suit the assessment of plant disease. The ones developed for studying specific plant–pathogen interactions (e.g., [
16]) are usually only applicable in analyzing that specific pathosystem (usually in similar environmental conditions), mostly in symptomatic conditions, and are unsuitable for generalized disease assessment. Disease studies are usually modeled as a classification approach, which adds difficulty to the application of the index.
Another emerging strategy, recently employed for exploring hyperspectral data, is applying different advanced techniques (e.g., machine learning algorithms) that search for relationships between spectral data and biophysical variables (also known as nonparametric, data-driven methods). They mostly consider all the spectral features measured by the hyperspectral sensors, which constitutes an important benefit compared to the VIs [
17]. These methods can be based on linear or nonlinear predictive methods.
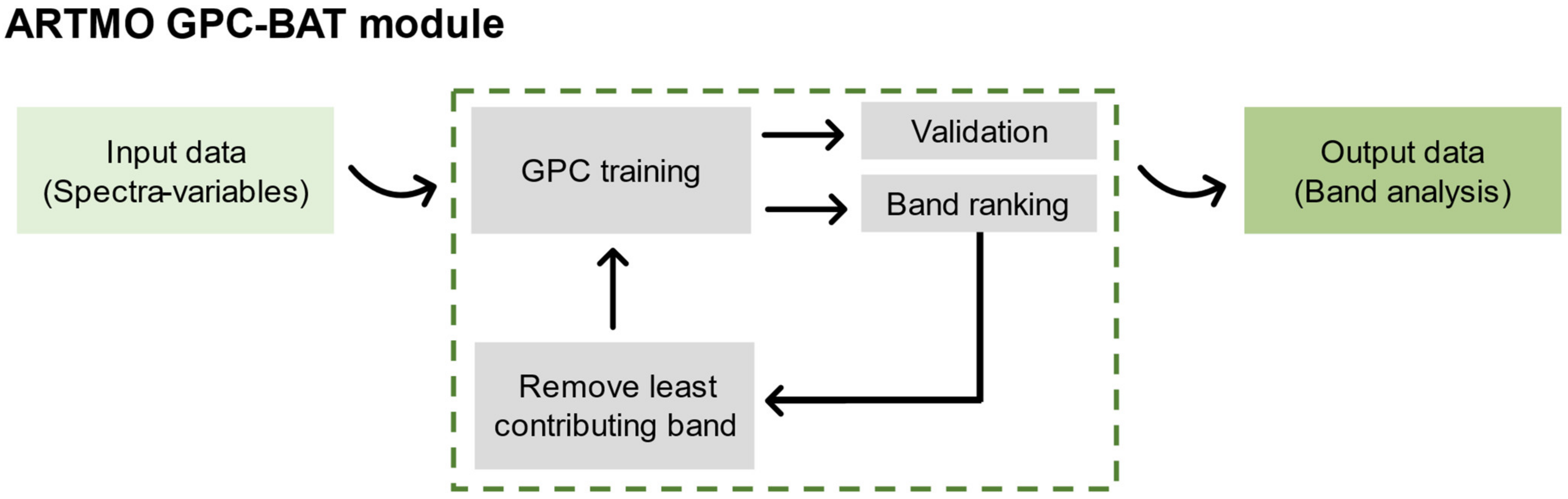
Furthermore, automated band analysis tools have been developed in the domain of machine learning classification algorithms (MLCAs). Following a band selection method earlier introduced in regression [
18], this paper introduces an automated spectral band analysis tool (BAT) based on Gaussian process classification (GPC) for the spectral analysis of bacterial plant diseases. Briefly, starting from using all bands, GPC-BAT sequentially removes the least informative band in GPC until one band is left. By tracking the accuracy of statistics, GPC-BAT allows (1) to identify the most informative bands relating spectral data to a classification problem, and (2) to find the least number of bands that preserve optimized accurate classification tasks.
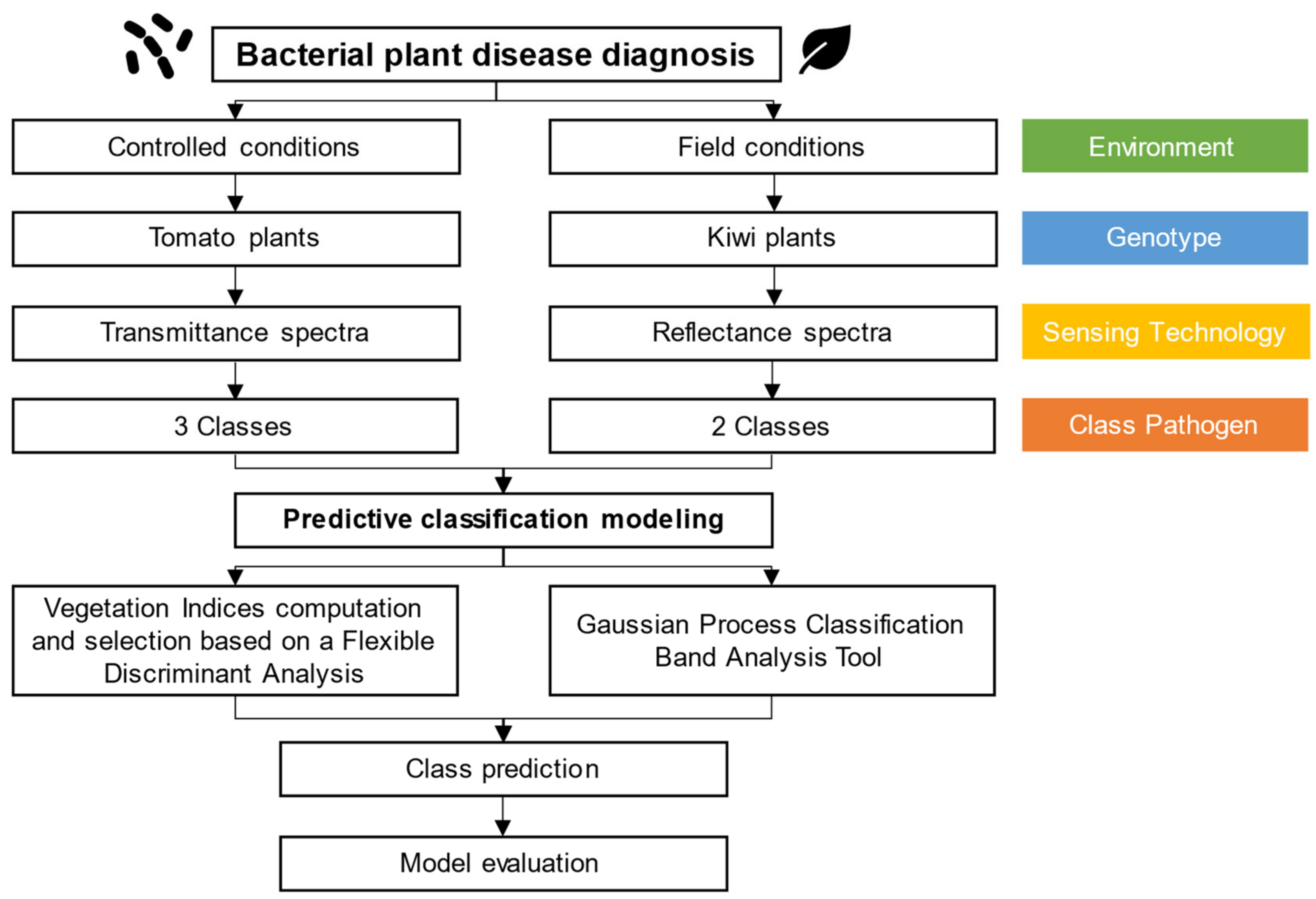
Hence, despite the development and availability of diverse methods for extracting meaningful spectral information in the context of plant bacterial disease diagnosis, it is necessary to address their suitability and performance when leading with different pathosystems. Therefore, the main objective of the present work aimed to explore, test, and validate the application of proximal optical sensed data for the early assessment and diagnosis of bacterial plant diseases. In this regard, the specific goals of this study were: (i) explore the suitability of different VIs for extracting relevant spectral features for performing plant bacterial disease diagnosis, using both reflectance and transmittance hyperspectral data (physiological driven approach); (ii) investigate the potential of a GPC-BAT for performing plant bacterial disease diagnosis using reflectance and transmission hyperspectral data (data-driven approach); (iii) compare and contrast the performance of VIs and GPC-BAT in discerning spectral features crucial for differentiating between healthy and diseased plant tissues; and (iv) uncover the biological significance of the identified spectral features concerning specific plant–pathogen interactions and their implications for early disease diagnosis. To achieve this, two case studies were conducted on tomato (controlled conditions) and kiwi (field conditions) plants, aiming to explore the capabilities of the developed approaches for performing bacterial disease diagnosis in distinct species in different environmental conditions.
4. Discussion
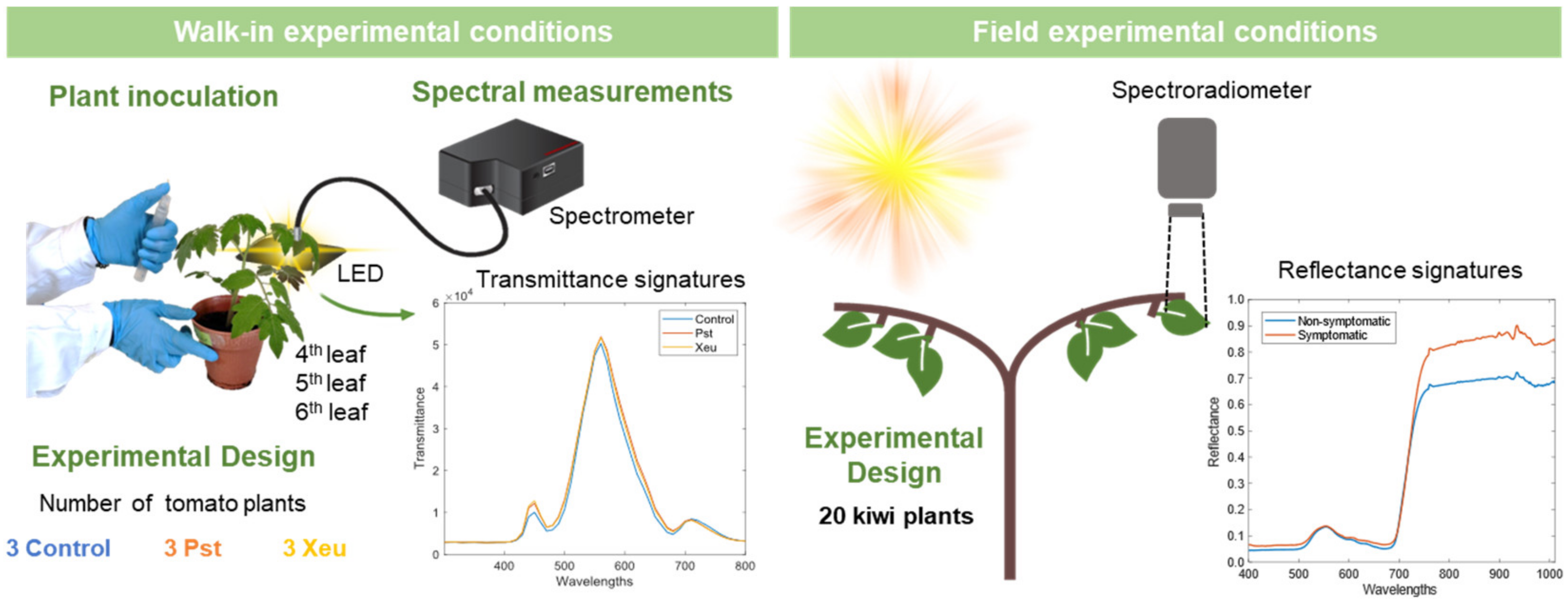
This work analyzed two methodologies for performing bacterial disease classification in tomato (assay performed in controlled environmental conditions, using a transmittance-based sensor) and kiwi (assay made in the field, using a reflectance-based sensor) plants. One approach combines the calculation of different VIs described in the literature and a machine learning algorithm with a built-in FS method (FDA). Another approach uses the two distinct hyperspectral datasets combined through the ARTMO GPC-BAT. In both approaches, the most relevant spectral wavelengths for class detection were identified and linked to their biological significance. The first approach uses VIs developed according to the physiological information of plants. In contrast, the second approach constitutes a data-driven approach. The tomato experiment (three classes: Control vs. Pst and Xeu), compared to the kiwi experiment (binary), represents a more complex classification model. The sensor used in the tomato experiment allows obtaining spectral information from the visible spectrum up to 800 nm, with wavelength resampling to 10 nm intervals (approximately), whereas the sensor used in the field experiment with kiwi allows data from the visible spectrum up to 1000 nm, with a more precise step of 1 nm wavelength.
It is possible to observe that both approaches allowed the identification of the different classes in the study, using the tomato and kiwi datasets. Nevertheless, the strategy involving the application of VIs as an FS technique showed lower classification metrics than the methodology that used the GPC-BAT, which may not be high enough to justify their future application. This may be related to the fact that the VIs used, despite being well established in the literature, were developed for specific plant traits and situations differing from the bacterial plant disease diagnosing problem in the study. Furthermore, since they are calculated using only available spectral features, they may not use all the information in spectral narrowband, high-dimensional hyperspectral data [
5]. Hence, this modeling approach must be enhanced aiming at a more effective class discrimination. Exploring distinct wavelength combinations in the VIs, different FS models (e.g., Random Forest), and performing hyperparameter tunning are some of the strategies that may be performance.
In contrast, GPC-BAT considered all the available spectral features and performed a selection according to their relevance for identifying the class in the study.
The GPC-BAT, when applied to the analysis encompassing all wavelengths captured by the hyperspectral sensors, exhibited lower classification metrics than the VIs approach in both the tomato and kiwi case studies. For instance, in the kiwi study, where the hyperspectral data indicated a narrowband field of 1 nm, the model’s performance using all available features was as poor as when only three wavelengths were utilized (data not displayed). This may be related to hyperspectral data being super-imposed in the recorded spectra at different interference scales [
25], (i.e., the data collected corresponds to several structural and metabolic plant compounds present in the area measured) and to the significant amount of redundant information embedded in contiguous wavelengths. As a result, only a few specific spectral variables are relevant to identify diseased plant tissues [
90,
91].
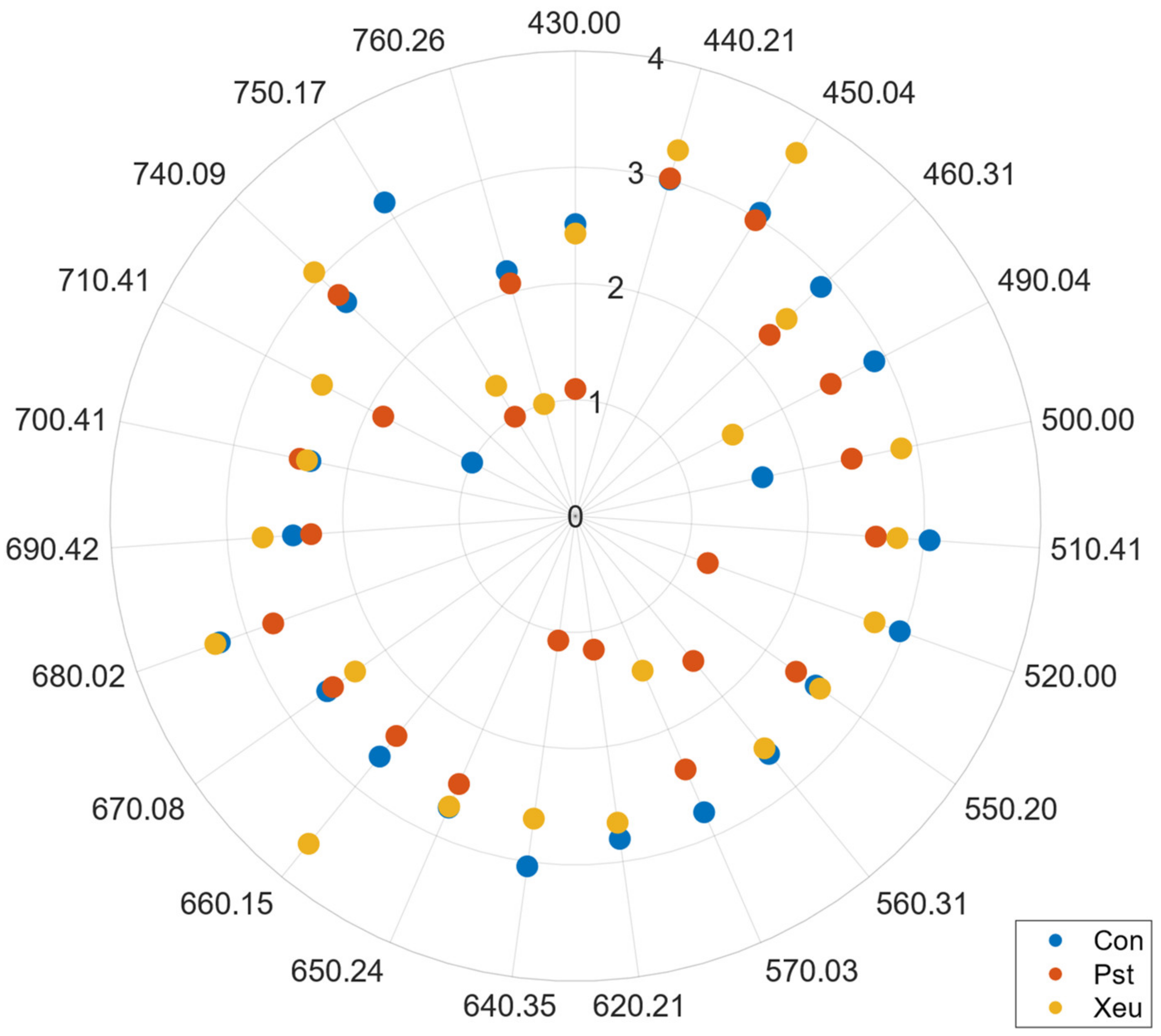
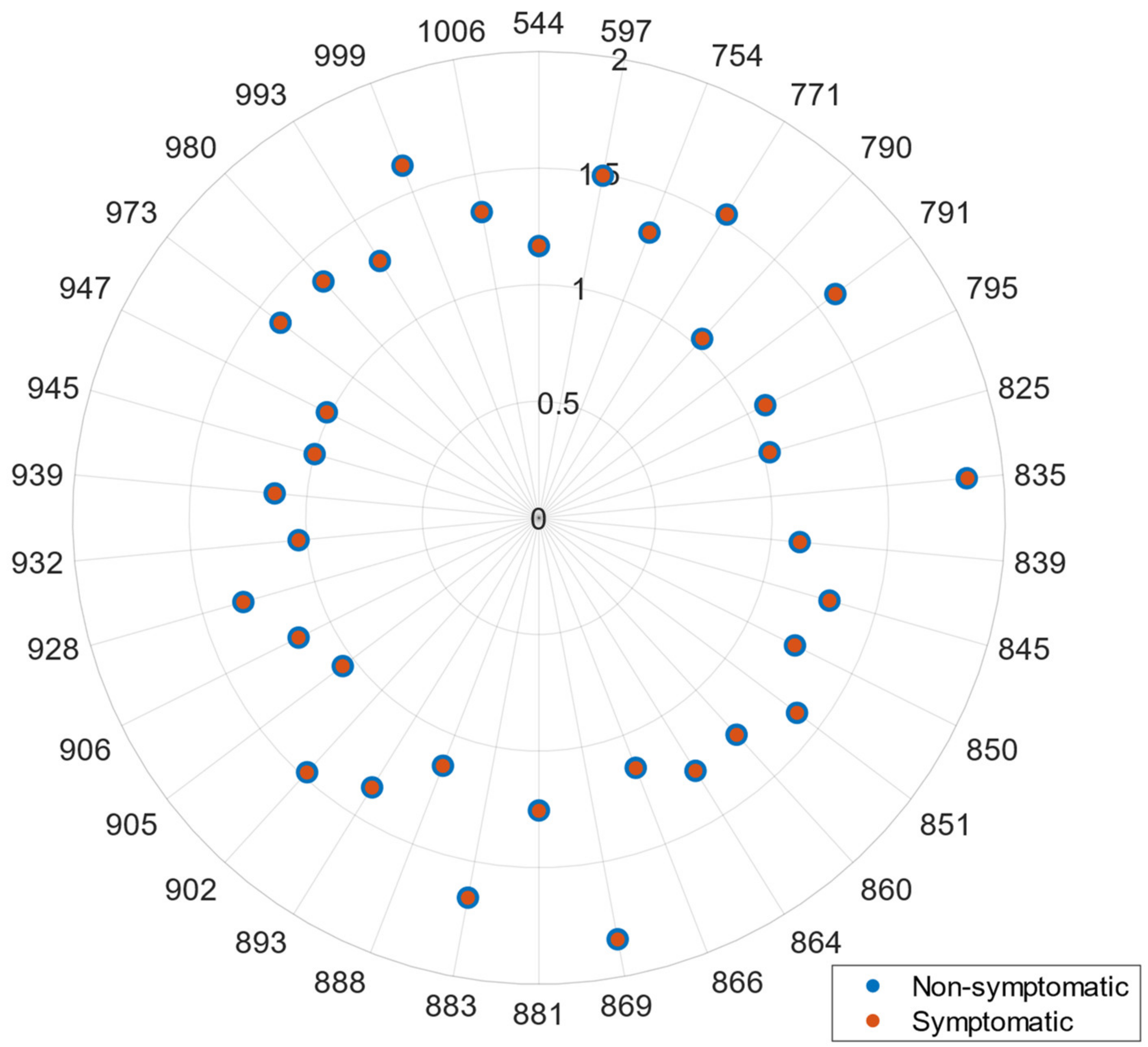
In this regard, Feature Selection or spectral reduction techniques are, thus, recommended to overcome this hurdle. In this work, two approaches were analyzed namely an FDA algorithm and the BAT of ARMO. Given the selected wavelengths, both studied strategies (VI-based vs. GPC-BAT) presented comparable results for both case studies, notably when dealing with the more complex tomato dataset especially with the tomato dataset. Equivalent wavelengths were found in the blue (450 nm), green (550 nm), and red-edge regions (680, 690, 700, and 750 nm). VIs further highlighted the 800 nm wavelength in the NIR region. In the kiwi case study, the features selected by the two algorithms were similar but not entirely coincident, namely in the green region (where VIs selected the 530, 553, and 554 nm, and the GPC-BAT chosen the 544, 597 nm), red-edge region (Vis identified the 670, 677, 700, 705, 730, and 750 nm as relevant, and GPC-BAT only considered the 754 nm), and NIR region (Vis picked a lower amount of wavelengths, namely 780, 800, 994, and 1000 nm, when compared to the GPC-BAT which took into account the following wavelengths 771, 790, 791, 795, 825, 835, 839, 845, 850, 851, 860, 864, 866, 869, 881, 883, 888, 893, 902, 905, 906, 928, 932, 939, 945, 947, 973, 980, 993, 999, and 1006 nm). Only the Vis presented features in the blue region 400 and 450 nm. Thus, it is important to address that despite the two modeling strategies applied to work with different types of spectra (reflectance vs. transmittance), having different spectral resolution (~10–10 nm vs. 1–1 nm) and presenting different pre-processing methods, they selected similar wavelengths.
These findings present biological significance, since the relationship between the plant host and the pathogen causes changes in photosynthetic pigment content, water levels, and structural composition (e.g., cellulose and lignin levels) [
92]. This ultimately leads to modifications in the tissues’ spectral behavior. In particular, the variance in spectral characteristics among diseased leaves infected by distinct bacteria could be linked to the generation of unique molecules by each pathogen, which may influence the spectral signature of the host. For instance, Pst bacteria produce a specific phytotoxin named coronatine that induces changes in chlorophyll fluorescence (by altering photosystem II—PSII), impacting tomato plant tissues’ absorption and scattering of light [
93]. Moreover, the host tomato plant can activate diverse defense responses upon encountering a pathogen, initiating a cascade of biochemical and molecular reactions that further contribute to spectral modifications in the visible wavelength ranges. Phytoalexins (e.g., flavonoids) serve as an example, with their production hypothesized to be linked to an increase in the spectral reflectance of plants in the VIS range [
94].
Previous studies performed by our team also reported similar outcomes. In particular, that study developed a methodology for early diagnosing two bacterial diseases of tomato, caused by Pst and Xeu bacteria, using hyperspectral transmittance data and an applied predictive modeling approach [
19]. A total of 3478 spectral measurements were normalized and subjected to a Linear Discriminant Analysis (LDA) aiming to reduce data dimensionality. This algorithm highlighted similar relevant wavelengths in the blue, green, and red spectral regions. Furthermore, a modeling approach using a Support Vector Machine was applied for spectral classification. It achieved an accuracy of 100% for samples measured on tissues inoculated with Pst and 74% for tissues inoculated with Xeu when samples collected before symptom appearance were used [
19]. Likewise, another study performed on a kiwi orchard allowed the identification of hyperspectral reflectance samples collected on nonsymptomatic and symptomatic Psa disease leaf tissues. Several methodologies involving different Feature Selection techniques combined with different Machine Learning algorithms were explored, and the one combining a stepwise forward various selection (SFVS) approach followed by the computation of an SVM algorithm was selected, achieving an overall accuracy of 85%. Similar to the other strategies explored, the SFVS elected the blue region, green region, and NIR region as the most relevant for sample classification.
Furthermore, other researchers reported similar classification findings to the ones found in the present work, namely when studying different tomato and kiwi diseases based on modeling hyperspectral spectroscopy data. The suitability of a portable hyperspectral spectrometer combined with various algorithms for FS and data modeling for early nondestructive diagnosis of tomato bacterial wilt disease (
Erwinia tracheiphila) in leaves was explored [
95]. The model presenting higher evaluation metrics (overall accuracy of 90.70%) applied Genetic Algorithms for FS and SVM to predict classification. The Simple Ratio Pigment Index (SRPI) was the VI and was found to have a higher contribution in the developed model. It considers 430 and 680 nm wavelengths and is sensitive to leaf nitrogen content and photosynthetic efficiency (and is similar to our findings) [
95].
Another study using tomato plants explored the usage of a portable high-resolution spectroradiometer combined with VIs, Principal Component Analysis (PCA), and a classification model K-nearest neighbor (KNN) for the diagnosis of late blight (
Phytophthora infestans), target (
Corynespora cassiicola), and bacterial spot (
Xanthomonas euvesicatoria) [
96]. They successfully identified the spectral samples collected on detached tomato leaflets with an accuracy reaching the 100% level even in nonsymptomatic stages (Error Rate of 9.50%), when the 15 VIs selected by PCA in the first principal component (PC) were considered. Interestingly, it is possible to observe that when 30 VIs selected by PCA and belonging to the first PC were used, the model showed a lower accuracy value (65.20%) and a higher error rate (28.6). In terms of VIs, the ones selected presented similar features to the ones found in our study (such as the 680 and 800 nm used in the Normalized difference index and the Simple Ratio, and structure-intensive pigment index, among others) [
96].
Hyperspectral VIS-NIR spectroscopy was, moreover, used for the nondestructive early diagnosis of tomato chlorosis virus (ToCV) [
97]. They used a Neighborhood component analysis (NCA) for performing FS and for selecting the most relevant VIs in the study, along with two ML models for data modeling (XY-fusion network—XY-F—and Multilayer Perceptron with Automated Relevance Determination—MLP-ARD). The best overall accuracy (92.1% before outlier removal and 100% after outlier removal) was obtained using MLP-ARD. In terms of relevant VIs, is possible to observe that wavelengths such as 550, 670, 700, 720, 740, and 800 nm, among others, were present in the most notable VIs formulae (such as Anthocyanin Reflectance Index—ARI, Pigment Specific Simple Ratio—PSSR, Red Edge Inflection Point—REIP, Simple Ratio—SR, and Vogelmann Index—VOG). In turn, from the 15 wavelengths selected by the NCA, these were mostly located in the blue (402.20 to 449.20 nm), green (556.40 to 566.40 nm), red-edge (676.40 to 726.30 nm), and NIR regions (862.10 nm). These outcomes coincided with our observations [
97].
The feasibility of multispectral data for predicting kiwifruit decline (probably caused by
Phytophtora spp. and
Phytopythium spp.) in diseased orchards was also tested [
98]. Multispectral data included the 550, 660, and 790 nm spectral features, and when combined with K-means clustering allowed the determination of kiwi plants’ vigor affected or not by the disease with 73% (or more) Accuracy and 82% Precision. These results are, thus, in line with ours also identifying the green, red, and NIR regions as relevant for estimating plant biophysical traits [
98].
The present outcomes demonstrate that hyperspectral transmittance and reflectance spectroscopy can identify healthy and diseased tissues, such as tomato (herbaceous) and kiwi (woody) crops, in laboratory or field conditions. Further research is advised to explore if specific host–pathogen interactions require customized modeling approaches to be predicted or if it is possible to elaborate a unified strategy that allows bacterial disease assessment. Nevertheless, it should be taken into consideration that model comparison may be challenging due to several factors: pathogen species in the study; the occurrence of specific host–pathogen interactions; the number of spectral points measured; the environmental conditions where the data are collected; and the stage of the disease cycle where the spectral assessments are made, among others. Furthermore, hyperspectral spectroscopy sensors present a relatively low Technology Readiness Level (TRL), indicating that these sensors have a large margin to be improved. In this regard, developing and enhancing effective FS strategies or Dimensionality Reduction approaches may be conducted to identify specific spectral regions valuable for performing plant disease diagnosis, which may be incorporated in multispectral sensors involving lower production and data processing costs. Moreover, the application of hyperspectral imaging sensors should also be explored in future studies, since it would offer a comprehensive spectral overview across spatial dimensions, allowing the capture of spectral signatures from several pixels in the image. This spatial information enables the identification of spatial variations in spectral features within the plant tissue sample, which can provide valuable insights into the spatial distribution of disease symptoms or other physiological changes. However, hyperspectral imaging typically requires more complex data processing and may involve higher equipment costs compared to point-measurement techniques, which must be taken into consideration in the technique development.
5. Conclusions
This study aimed to explore and compare two distinct modeling approaches, namely the parametric Spectral Vegetation Indices (VIs) and the Gaussian Process Classification based on an Automated Spectral Band Analysis Tool (GPC-BAT), for diagnosing bacterial diseases in plants using hyperspectral sensing. Our analysis covered two experimental conditions, namely controlled conditions where tomato plants were used and field conditions where kiwi plants were analyzed, revealing insights into the performances of each approach.
Both the VI- and GPC-BAT-based approaches demonstrated potential for class discrimination in both experiments. Nevertheless, the modeling strategy applying VIs showed moderate success in distinguishing healthy and Pst-inoculated tomato tissues, with accuracy values ranging from 63% to 71%. In turn, the GPC-BAT strategy exhibited enhanced accuracy in classifying healthy and diseased tissues, achieving overall accuracy values extending from 70% to 75% in the tomato and kiwi case studies. This approach also revealed greater potential for discriminating nonsymptomatic from symptomatic tissues in kiwi plants in field conditions.
The identified spectral bands, particularly in the blue, green, red-edge, and NIR regions, align with the absorption regions of several photosynthetic pigments and plant structural components. These spectral modifications correlate with bacterial infections, affecting chlorophylls, carotenoids, flavonoids, pheophytins, and water content in plant leaves.
Hence, while VIs offer a simplistic yet moderately effective means of disease diagnosis, GPC-BAT, with feature reduction, shows promise in improving accuracy. However, further refinements are necessary, especially for the early-stage diagnosis of specific bacterial infections.
The identified wavelengths hold biological significance, suggesting a correlation between bacterial infections and alterations in photosynthetic pigments and leaf structural components. Future research could focus on refining and integrating these approaches to develop more robust and accurate diagnostic tools for various plant–pathogen interactions, thereby aiding in early disease detection and management in diverse agricultural settings.