Cross-Species Transferability of SSR Markers for Analyzing Genetic Diversity of Different Vicia species Collections
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Genomic DNA Extraction
2.2. SSR Genotyping by PCR Multiplex Amplification and Data Analysis
2.3. Population Structure and Genetic Diversity Analysis
2.4. Field Characterization of Agromorphological Traits
2.5. Statistical Analyses
3. Results
3.1. Cross-Species Transferability of SSR Markers to Species of the Genus Vicia
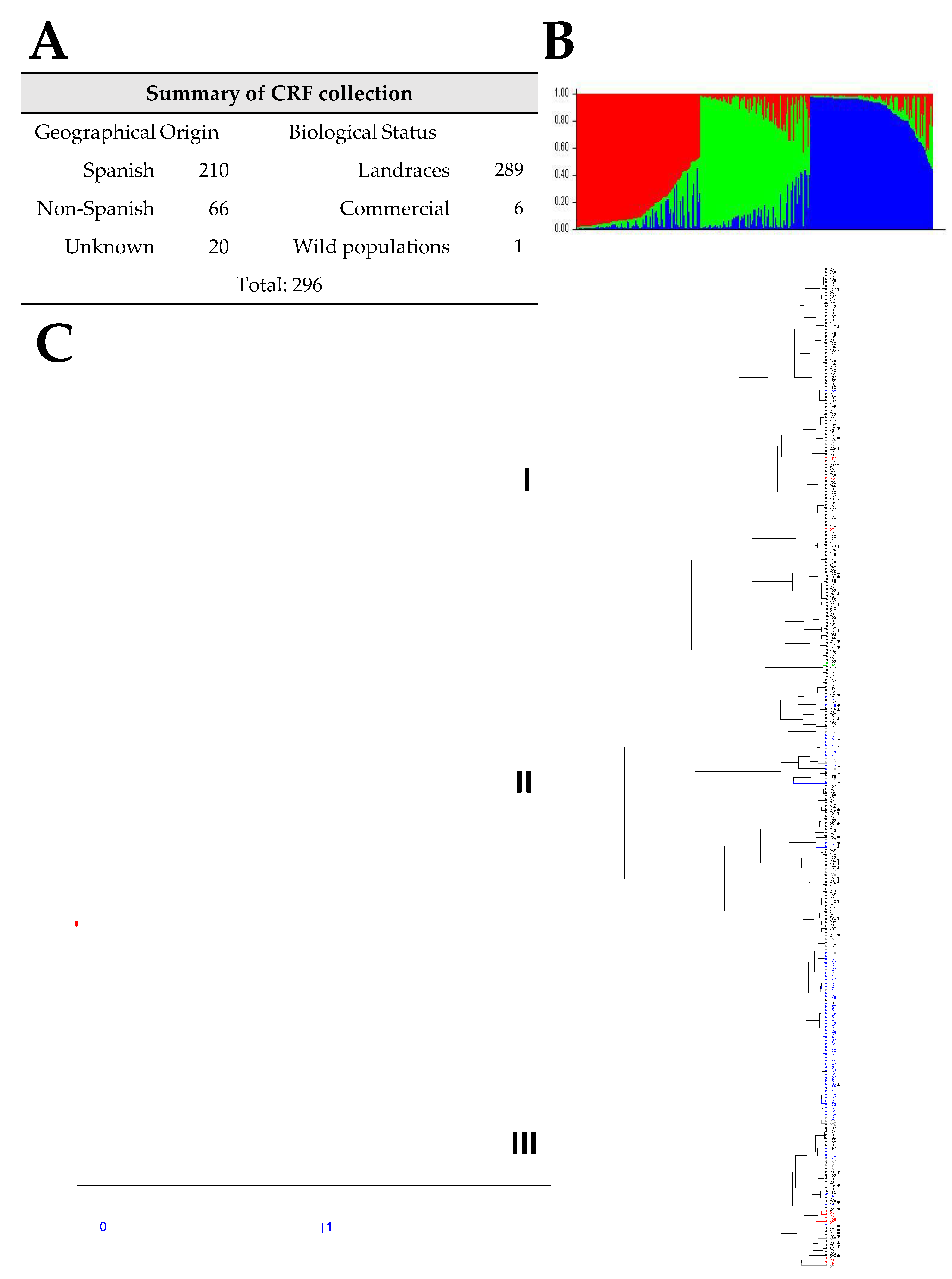
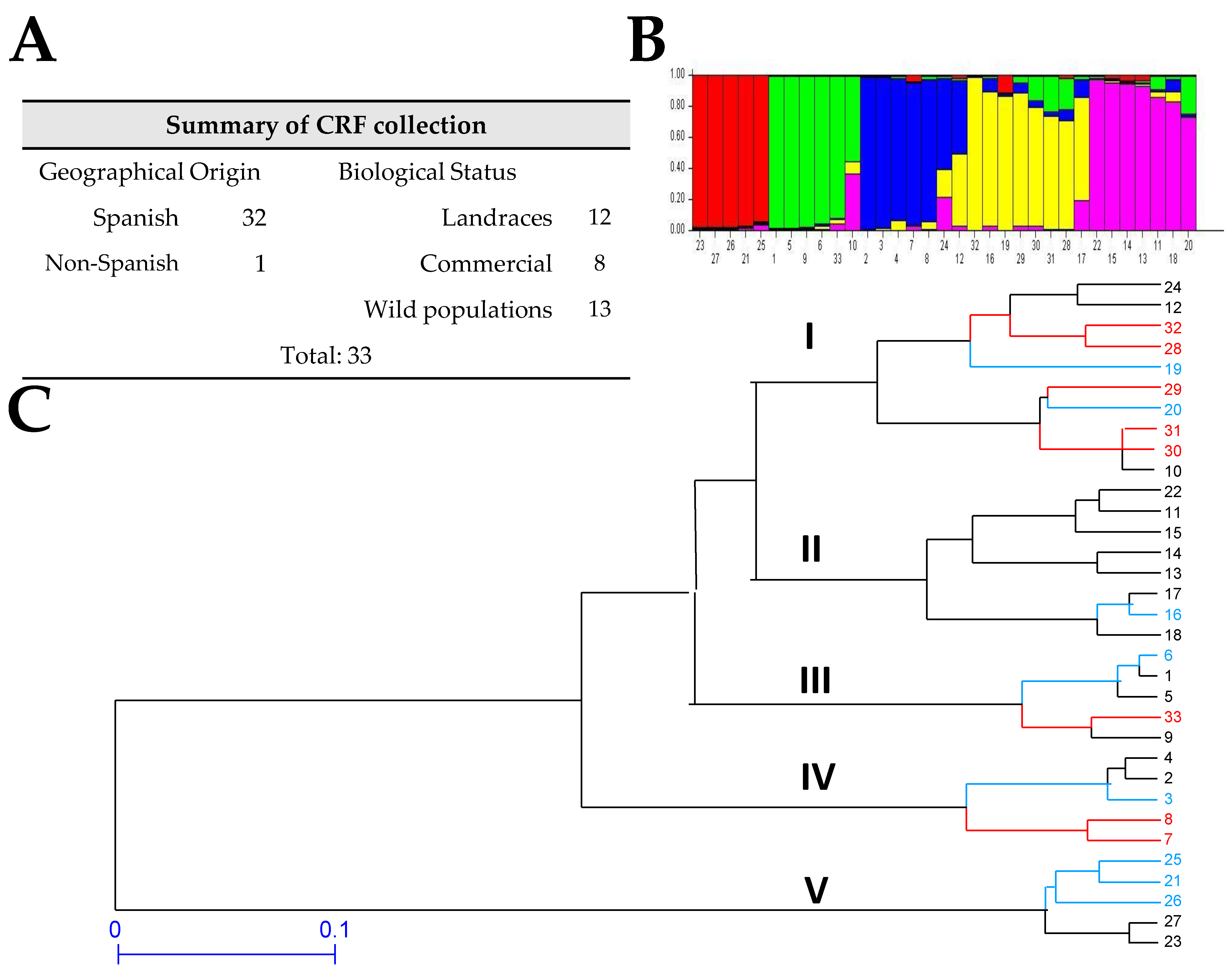
3.2. Genetic Diversity and Cluster Analysis of the V. articulata Collection
3.3. Genetic Diversity and Cluster Analysis of the V. ervilia Collection
3.4. Genetic Diversity and Cluster Analysis of the V. narbonensis Collection
3.5. Field Evaluation of Vicia Collections Phenotypic Traits
3.6. Building Core Collections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smýkal, P.; Coyne, C.J.; Ambrose, M.J.; Maxted, N.; Schaefer, H.; Blair, M.W.; Berger, J.; Greene, S.L.; Nelson, M.N.; Besharat, N.; et al. Legume Crops Phylogeny and Genetic Diversity for Science and Breeding. Crit. Rev. Plant Sci. 2015, 34, 43–104. [Google Scholar] [CrossRef]
- Maxted, N. An ecogeographical study of Vicia subgenus Vicia. In Systematic and Ecogeographic Studies on Crop Genepools; International Board for Plant Genetic Resources: Rome, Italy, 1995; Volume 8, p. 184. [Google Scholar]
- Maxted, N. A phenetic investigation of Vicia L. subgenus Vicia (Leguminosae, Vicieae). Bot. J. Linn. Soc. 1993, 111, 155–182. [Google Scholar] [CrossRef]
- Leht, M. Phylogenetics of Vicia (Fabaceae) based on morphological data. Feddes Repert. 2009, 120, 379–393. [Google Scholar] [CrossRef]
- Mateo-Box, J.M. Leguminosas de Grano; Ed. Salvat: Barcelona, Spain, 1961; p. 550. [Google Scholar]
- Pellegrini, P.A.; Balatti, G.E. Noah’s arks in the XXI century. A typology of seed banks. Biodivers. Conserv. 2016, 25, 2753–2769. [Google Scholar] [CrossRef]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and future use of wild relatives in crop breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Ramirez-Villegas, J.; Jarvis, A.; Fujisaka, S.; Hanson, J.; Leibing, C. Crop and Forage Genetic Resources: International Interdependence in the Face of Climate Change; Routledge: Abingdon, UK, 2013; pp. 78–98. [Google Scholar]
- Van Hintum, T.; Brink, M. Technological and policy challenges to utilisation of plant genetic resources. In Sustaining Global Food Security: The Nexus of Science and Policy; Zeigler, R.S., Ed.; CSIRO Publishing: Clayton South, Australia, 2019; pp. 36–48. [Google Scholar]
- Hodgkin, T.; Rao, R.; Cibrian-Jaramillo, A.; Gaiji, S. The use of ex situ conserved plant genetic resources. Plant Genet. Resour. Charact. Util. 2003, 1, 19–29. [Google Scholar] [CrossRef]
- Westengen, O.T.; Skarbø, K.; Mulesa, T.H.; Berg, T. Access to genes: Linkages between genebanks and farmers’ seed systems. Food Secur. 2018, 10, 9–25. [Google Scholar] [CrossRef]
- GENESYS. Online Platform of Plant Genetic Resources for Food and Agriculture (PGRFA) Conserved in Genebanks Worldwide. Available online: https://www.genesys-pgr.org (accessed on 5 December 2023).
- Pathirana, R.; Carimi, F. Management and Utilization of Plant Genetic Resources for a Sustainable Agriculture. Plants 2022, 11, 2038. [Google Scholar] [CrossRef]
- Khazaei, H.; O’Sullivan, D.M.; Stoddard, F.L.; Adhikari, K.N. Recent advances in faba bean genetic and genomic tools for crop improvement. Legum. Sci. 2021, 3, e75. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, L.; López-Román, M.I.; González, J.M.; Zambrana, E.; Marcos-Prado, T.; Ramírez-Parra, E. Common vetch, valuable germplasm for resilient agriculture: Genetic characterization and Spanish core collection development. Front. Plant Sci. 2021, 12, 617873. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Parra, E.; De la Rosa, L. Designing Novel Strategies for Improving Old Legumes: An Overview from Common Vetch. Plants 2023, 12, 1275. [Google Scholar] [CrossRef] [PubMed]
- Román, B.; Satovic, Z.; Pozarkova, D.; Macas, J.; Dolezel, J.; Cubero, J.; Torres, A. Development of a composite map in Vicia faba, breeding applications and future prospects. Theor. Appl. Genet. 2004, 108, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Link, W.; Dixkens, C.; Singh, M.; Schwall, M.; Melchinger, A.E. Genetic diversity in European and Mediterranean faba bean germ plasm revealed by RAPD markers. Theor. Appl. Genet. 1995, 90, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Zeid, M.; Schön, C.-C.; Link, W. Genetic diversity in recent elite faba bean lines using AFLP markers. Theor. Appl. Genet. 2003, 107, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-F.; Zong, X.-X.; Guan, J.-P.; Yang, T.; Sun, X.-L.; Ma, Y.; Redden, R. Genetic diversity and relationship of global faba bean (Vicia faba L.) germplasm revealed by ISSR markers. Theor. Appl. Genet. 2012, 124, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.; Batley, J. Plant genome sequencing: Applications for crop improvement. Plant Biotechnol. J. 2010, 8, 2–9. [Google Scholar] [CrossRef]
- Kumar, J.; Gupta, D.S.; Gupta, S.; Dubey, S.; Gupta, P.; Kumar, S. Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell Rep. 2017, 36, 1187–1213. [Google Scholar] [CrossRef]
- Raveendar, S.; Lee, G.A.; Jeon, Y.A.; Lee, Y.J.; Lee, J.R.; Cho, G.T.; Cho, J.H.; Park, J.H.; Ma, K.H.; Chung, J.W. Cross-amplification of Vicia sativa subsp. sativa microsatellites across 22 other Vicia species. Molecules 2015, 20, 1543–1550. [Google Scholar] [CrossRef]
- El Fatehi, S.; Béna, G.; Filali-Maltouf, A.; Ater, M. Genetic diversity of moroccan bitter vetch Vicia ervilia (L.) Willd. landraces revealed by morphological and SSR markers. Aust. J. Crop Sci. 2016, 10, 717–725. [Google Scholar] [CrossRef]
- Osman, S.A.; Ali, H.B.; El-Ashry, Z.M.; El-Khodary, S.E. Karyotype variation and biochemical analysis of five Vicia species. Bull. Natl. Res. Cent. 2020, 44, 91. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr (accessed on 5 December 2023).
- Erayman, M.; İlhan, E.; Güzel, Y.; Eren, A.H. Transferability of SSR markers from distantly related legumes to Glycyrrhiza species. Turk. J. Agric. For. 2014, 38, 32–38. [Google Scholar] [CrossRef]
- Gutierrez, M.; Vaz Patto, M.; Huguet, T.; Cubero, J.; Moreno, M.; Torres, A. Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops. Theor. Appl. Genet. 2005, 110, 1210–1217. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Woullard, F.; Marong, I.; Guo, B. Transferability of soybean SSR markers in peanut (Arachis hypogaea L.). Peanut Sci. 2006, 33, 22–28. [Google Scholar] [CrossRef]
- Choudhary, S.; Sethy, N.K.; Shokeen, B.; Bhatia, S. Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor. Appl. Genet. 2009, 118, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.; Rathour, R.; Kumar, N.; Katoch, P.; Sharma, T. Cross-genera legume SSR markers for analysis of genetic diversity in Lens species. Plant Breed. 2010, 129, 514–518. [Google Scholar] [CrossRef]
- Gupta, M.; Verma, B.; Kumar, N.; Chahota, R.K.; Rathour, R.; Sharma, S.K.; Bhatia, S.; Sharma, T.R. Construction of intersubspecific molecular genetic map of lentil based on ISSR, RAPD and SSR markers. J. Genet. 2012, 91, 279–287. [Google Scholar] [CrossRef]
- Datta, J.; Lal, N. Application of molecular markers for genetic discrimination of Fusarium wilt pathogen races affecting chickpea and pigeonpea in major regions of India. Cell. Mol. Biol. 2012, 58, 55–65. [Google Scholar]
- El Fatehi, S.; Béna, G.; Sbabou, L.; Filali-Maltouf, A.; Ater, M. Preliminary Results For Use SSR Markers in Bitter Vetch “Vicia ervilia (L.) Willd”. Int. J. Res. 2013, 1, 2311–2476. [Google Scholar]
- Russi, L.; Acuti, G.; Trabalza-Marinucci, M.; Porta, R.; Rubini, A.; Damiani, F.; Cristiani, S.; Dal Bosco, A.; Martuscelli, G.; Bellucci, M. Genetic characterisation and agronomic and nutritional value of bitter vetch (Vicia ervilia), an under-utilised species suitable for low-input farming systems. Crop Pasture Sci. 2019, 70, 606–614. [Google Scholar] [CrossRef]
- Röder, M.S.; Wendehake, K.; Korzun, V.; Bredemeijer, G.; Laborie, D.; Bertrand, L.; Isaac, P.; Rendell, S.; Jackson, J.; Cooke, R.J.; et al. Construction and analysis of a microsatellite-based database of European wheat varieties. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2002, 106, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Bredemeijer, M.; Cooke, J.; Ganal, W.; Peeters, R.; Isaac, P.; Noordijk, Y.; Rendell, S.; Jackson, J.; Röder, S.; Wendehake, K.; et al. Construction and testing of a microsatellite database containing more than 500 tomato varieties. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2002, 105, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Botta, R.; Costantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreira-Monteiro, F.; et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- GRIN. Germplasm Resource Information Network (GRIN) The GRIN-Global Database Platform. Available online: https://npgsweb.ars-grin.gov/gringlobal/taxon/taxonomysearchwep (accessed on 5 December 2023).
- Van de Wouw, M.; Enneking, D.; Robertson, L.; Maxted, N. Vetches (Vicia L.). In Plant Genetic Resources of Legumes in the Mediterranean; Maxted, N., Bennett, S.J., Eds.; Current Plant Science and Biotechnology in Agriculture; Springer: Dordrecht, The Netherlands, 2001; Volume 39. [Google Scholar] [CrossRef]
- Enneking, D.; Lahlou, A.; Noutfia, A.; Bounejmate, M. A note on Vicia ervilia cultivation, utilisation and toxicity in Morocco. Al Awamia 1995, 89, 141–148. [Google Scholar]
- Renzi, J.; Cantamutto, M. Vicias: Bases Agronómicas Para el Manejo en la Región Pampeana; Ediciones INTA, Ed.; Ediciones INTA: Buenos Aires, Argentina, 2013; ISBN 978-987-521-470-5. [Google Scholar]
- Vincent, H.; Wiersema, J.; Kell, S.; Fielder, H.; Dobbie, S.; Castañeda-Álvarez, N.P.; Maxted, N. A prioritized crop wild relative inventory to help underpin global food security. Biol. Conserv. 2013, 167, 265–275. [Google Scholar] [CrossRef]
- Raina, S.N.; Ogihara, Y. Chloroplast DNA diversity in Vicia faba and its close wild relatives: Implications for reassessment. Theor. Appl. Genet. 1994, 88, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Birch, A.N.E.; Tithecott, M.T.; Bisby, F.A. Vicia johannis and wild relatives of the faba bean: A taxonomic study. Econ. Bot. 1985, 39, 177–190. [Google Scholar] [CrossRef]
- Sillero, J.C.; Moreno, M.T.; Rubiales, D. Sources of resistance to crenate broomrape among species of Vicia. Plant Dis. 2005, 89, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Inci, N.E.; Toker, C. Screening and selection of faba beans (Vicia faba L.) for cold tolerance and comparison to wild relatives. Genet. Resour. Crop Evol. 2011, 58, 1169–1175. [Google Scholar] [CrossRef]
- Rubiales, D.; Fondevilla, S.; Chen, W.; Gentzbittel, L.; Higgins, T.J.V.; Sánchez, M.Á.C.; Rispail, N. Achievements and Challenges in Legume Breeding for Pest and Disease Resistance. Crit. Rev. Plant Sci. 2014, 34, 195–236. [Google Scholar] [CrossRef]
- Siddique, K.; Loss, S.; Enneking, D. Narbon bean (Vicia narbonensis L.): A promising grain legume for low rainfall areas of south-western Australia. Aust. J. Exp. Agric. 1996, 36, 53–62. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia, Europe and the Nile Valley, 3rd ed.; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Maxted, N.; Bennett, S. Plant Genetic Resources of Legumes in the Mediterranean; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2001; Volume 39. [Google Scholar]
- Gu, R.; Fan, S.; Wei, S.; Li, J.; Zheng, S.; Liu, G. Developments on Core Collections of Plant Genetic Resources: Do We Know Enough? Forests 2023, 14, 926. [Google Scholar] [CrossRef]

| SSR Marker |
Donor Species | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VsSSR310 | VsSSR102 | VsSSR179 | VsSSR185 | VsSSR073 | VsSSR140 | VsSSR217 | VsSSR129 | VsSSR138 | VsSSR115 | VsSSRO | VsSSRP | VsSSRS | VsSSRN | VeSSR02 | VeSSR05 | VeSSR07 | VeSSR09 | V. sativa (VsSSRs) | V. ervilia (VsSSRs) | ||
| Target Species | V. sativa | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 100% (14/14) | 100% (4/4) |
| V. ervilia | + | − | + | − | + | − | − | + | − | − | − | + | + | + | + | + | + | + | 50% (7/14) | 100% (4/4) | |
| V. narbonensis | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | 93% (13/14) | 100% (4/4) | |
| V. articulata | + | + | + | − | + | + | − | + | − | − | − | + | + | + | − | − | − | − | 64% (9/14) | 0% (0/4) | |
| Locus | Na | Ne | F | I | PIC | Ho | He | uHe |
|---|---|---|---|---|---|---|---|---|
| VsSSR073 | 7 | 1.538 | 0.503 | 0.804 | 0.549 | 0.174 | 0.350 | 0.351 |
| VsSSR102 | 7 | 3.370 | 0.555 | 1.373 | 0.337 | 0.313 | 0.703 | 0.706 |
| VsSSR129 | 9 | 1.794 | 0.941 | 1.076 | 0.648 | 0.026 | 0.443 | 0.445 |
| VsSSR140 | 4 | 1.073 | 0.232 | 0.187 | 0.643 | 0.052 | 0.068 | 0.068 |
| VsSSR179 | 5 | 2.067 | −0.162 | 0.894 | 0.067 | 0.600 | 0.516 | 0.519 |
| VsSSR310 | 7 | 2.353 | 0.773 | 1.242 | 0.435 | 0.130 | 0.575 | 0.578 |
| VsSSRN | 10 | 1.316 | 0.421 | 0.628 | 0.431 | 0.139 | 0.240 | 0.241 |
| VsSSRP | 7 | 3.298 | 0.014 | 1.355 | 0.768 | 0.687 | 0.697 | 0.700 |
| VsSSRS | 9 | 4.895 | 0.268 | 1.781 | 0.235 | 0.583 | 0.796 | 0.799 |
| Mean | 7.222 | 2.412 | 0.394 | 1.038 | 0.457 | 0.300 | 0.488 | 0.49 |
| SE | 0.641 | 0.410 | 0.117 | 0.157 | 0.209 | 0.086 | 0.079 | 0.08 |
| Population | N | Na | Ne | Ho | He | uHe | F | PIC | H | N. Private Alleles |
|---|---|---|---|---|---|---|---|---|---|---|
| Landraces | 111 | 7.11 ± 0.66 | 2.40 ± 0.41 | 0.30 ± 0.09 | 0.49 ± 0.08 | 0.49 ± 0.08 | 0.37 ± 0.12 | 0.45 ± 0.23 | 1.03 ± 0.16 | 43 |
| Wild Relatives | 2 | 1.14 ± 0.24 | 1.40 ± 0.21 | 0.28 ± 0.15 | 0.18 ± 0.09 | 0.24 ± 0.12 | 0.60 ± 0.23 | 0.15 ± 0.22 | 10.27 ± 0.14 | 0 |
| Commercial Cultivars | 2 | 1.78 ± 0.28 | 1.66 ± 024 | 0.22 ± 0.12 | 0.29 ± 0.10 | 0.39 ± 0.13 | 0.25 ± 0.25 | 0.24 ± 0.24 | 0.45 ± 0.15 | 1 |
| Locus | Na | Ne | F | I | PIC | Ho | He | uHe |
|---|---|---|---|---|---|---|---|---|
| VeSSR02 | 8 | 2.585 | 0.392 | 1.171 | 0.556 | 0.095 | 0.613 | 0.614 |
| VeSSR05 | 8 | 3.187 | 0.791 | 1.329 | 0.632 | 0.338 | 0.686 | 0.687 |
| VeSSR07 | 10 | 2.586 | 0.603 | 1.306 | 0.587 | 0.135 | 0.613 | 0.614 |
| VeSSR09 | 10 | 4.810 | 0.850 | 1.750 | 0.762 | 0.216 | 0.792 | 0.793 |
| VsSSR073 | 5 | 1.221 | 0.839 | 0.142 | 0.172 | 0.027 | 0.181 | 0.181 |
| VsSSR129 | 28 | 17.114 | 0.888 | 2.974 | 0.939 | 0.152 | 0.942 | 0.943 |
| VsSSR179 | 10 | 4.055 | 0.625 | 1.579 | 0.719 | 0.084 | 0.753 | 0.755 |
| VsSSR310 | 4 | 2.314 | 0.846 | 0.963 | 0.516 | 0.213 | 0.568 | 0.569 |
| VsSSRN | 6 | 1.304 | 0.508 | 0.470 | 0.227 | 0.142 | 0.233 | 0.234 |
| VsSSRP | 11 | 4.434 | 0.780 | 1.770 | 0.750 | 0.162 | 0.774 | 0.776 |
| VsSSRS | 9 | 1.557 | 0.727 | 0.863 | 0.343 | 0.142 | 0.358 | 0.358 |
| Mean | 9.909 | 4.106 | 0.713 | 1.302 | 0.564 | 0.155 | 0.592 | 0.593 |
| SE | 1.933 | 1.352 | 0.048 | 0.216 | 0.237 | 0.025 | 0.073 | 0.073 |
| Group | N | Na | Ne | Ho | F | I | PIC | He | uHe | Priv. A. |
|---|---|---|---|---|---|---|---|---|---|---|
| LR | 285 | 9.91 ± 1.93 | 4.12 ± 1.37 | 0.16 ± 0.02 | 0.71 ± 0.05 | 1.36 ± 0.22 | 0.56 ± 0.23 | 0.59 ± 0.07 | 0.59 ± 0.07 | 68 |
| WP | 1 | 1.18 ± 0.12 | 1.18 ± 0.12 | 0.18 ± 0.12 | 1.00 ± 0.00 | 0.13 ± 0.08 | 0.15 ± 0.28 | 0.09 ± 0.06 | 0.18 ± 0.12 | 0 |
| CC | 10 | 3.45 ± 0.39 | 2.41 ± 0.38 | 0.10 ± 0.03 | 0.73 ± 0.10 | 0.91 ± 0.14 | 0.45 ± 0.20 | 0.49 ± 0.07 | 0.52 ± 0.07 | 0 |
| Locus | Na | Ne | F | I | PIC | Ho | He | uHe |
|---|---|---|---|---|---|---|---|---|
| VsSSR310 | 4 | 3.346 | 0.477 | 1.273 | 0.644 | 0.367 | 0.701 | 0.713 |
| VsSSR102 | 5 | 3.430 | 0.872 | 1.342 | 0.655 | 0.091 | 0.708 | 0.719 |
| VsSSR179 | 7 | 3.184 | 0.116 | 1.430 | 0.646 | 0.606 | 0.686 | 0.697 |
| VsSSR185 | 8 | 5.158 | 1.000 | 1.866 | 0.786 | 0.000 | 0.806 | 0.836 |
| VsSSR073 | 6 | 4.055 | 0.834 | 1.542 | 0.714 | 0.125 | 0.753 | 0.765 |
| VsSSR140 | 3 | 1.575 | −0.198 | 0.675 | 0.335 | 0.438 | 0.365 | 0.371 |
| VsSSR129 | 10 | 5.461 | 0.617 | 1.965 | 0.796 | 0.313 | 0.817 | 0.830 |
| VsSSR138 | 4 | 1.761 | 0.649 | 0.801 | 0.390 | 0.152 | 0.432 | 0.439 |
| VsSSR115 | 2 | 1.031 | −0.015 | 0.079 | 0.029 | 0.030 | 0.030 | 0.030 |
| VsSSRO | 4 | 2.150 | 0.037 | 0.924 | 0.462 | 0.515 | 0.535 | 0.543 |
| VsSSRP | 7 | 4.755 | 0.655 | 1.671 | 0.758 | 0.273 | 0.790 | 0.802 |
| VsSSRS | 5 | 3.050 | 0.594 | 1.254 | 0.615 | 0.273 | 0.672 | 0.683 |
| VsSSRN | 3 | 1.603 | 0.275 | 0.689 | 0.344 | 0.273 | 0.376 | 0.382 |
| VeSSR02 | 8 | 3.692 | 0.668 | 1.615 | 0.700 | 0.242 | 0.729 | 0.740 |
| VeSSR05 | 5 | 3.755 | 0.050 | 1.446 | 0.691 | 0.697 | 0.734 | 0.745 |
| VeSSR07 | 3 | 1.598 | −0.296 | 0.611 | 0.315 | 0.485 | 0.374 | 0.380 |
| VeSSR09 | 6 | 2.373 | 0.633 | 1.105 | 0.518 | 0.212 | 0.579 | 0.587 |
| Mean | 5.294 | 3.057 | 0.410 | 1.193 | 0.553 | 0.299 | 0.593 | 0.604 |
| SE | 0.527 | 0.325 | 0.095 | 0.122 | 0.211 | 0.048 | 0.052 | 0.053 |
| Group | N | Na | Ne | F | I | PIC | Ho | He | uHe | Priv. A. |
|---|---|---|---|---|---|---|---|---|---|---|
| L | 17 | 4.29 ± 0.38 | 2.73 ± 0.24 | 0.38 ± 0.10 | 1.07 ± 0.10 | 0.52 ± 0.19 | 0.30 ± 0.05 | 0.57 ± 0.05 | 0.59 ± 0.05 | 15 |
| WP | 8 | 3.71 ± 0.35 | 2.87 ± 0.31 | 0.46 ± 0.09 | 1.05 ± 0.11 | 0.52 ± 0.21 | 0.28 ± 0.05 | 0.57 ± 0.05 | 0.61 ± 0.06 | 5 |
| CC | 8 | 3.41 ± 0.32 | 2.52 ± 0.33 | 0.35 ± 0.14 | 0.94 ± 0.11 | 0.47 ± 0.20 | 0.31 ± 0.07 | 0.51 ± 0.05 | 0.55 ± 0.06 | 6 |
| Quantitative Agromorphological Traits | ||||
|---|---|---|---|---|
| Average | SD | Max | Min | |
| Flowering | ||||
| Days to first flowering (d) | 159.4 | 6.0 | 175.0 | 139.0 |
| Days to 50% flowering (d) | 169.6 | 10.2 | 211.0 | 143.0 |
| Days to final flowering (d) | 208.6 | 11.4 | 228.0 | 175.0 |
| Days to maturity (d) | 221.5 | 6.4 | 232.0 | 196.0 |
| Plant | ||||
| Height (cm) | 70.1 | 19.7 | 121.1 | 35.8 |
| First pod height (cm) | 24.5 | 8.5 | 55.6 | 12.2 |
| Number of main branches | 7.24 | 2.75 | 15.60 | 2.90 |
| Pod/Seed | ||||
| Number of pods per plant | 74.74 | 36.99 | 195.70 | 21.10 |
| Number of seeds per pod | 2.68 | 0.27 | 3.40 | 2.00 |
| Seed length (mm) | 4.73 | 0.24 | 5.43 | 4.12 |
| Seed width (mm) | 4.62 | 0.21 | 5.14 | 4.05 |
| Seed thickness (mm) | 2.64 | 0.23 | 3.92 | 2.07 |
| 100-seed weight (g) | 4.19 | 0.43 | 5.40 | 3.36 |
| Protein content (mg/g seed) | 26.28 | 2.49 | 32.20 | 21.81 |
| Quantitative Agromorphological Traits | |||||
|---|---|---|---|---|---|
| Average | SD | Max | Min | ||
| Flowering | |||||
| Days to 50% flowering (d) | 154.4 | 8.2 | 189.0 | 133.0 | |
| Days to maturity (d) | 188.9 | 8.7 | 225.0 | 152.0 | |
| Plant | |||||
| Height (cm) | 42.5 | 6.7 | 64.9 | 23.2 | |
| First pod height (cm) | 24.7 | 5.7 | 41.3 | 7.2 | |
| Leaf | |||||
| Leaf length (mm) | 99.7 | 12.4 | 168.0 | 59.6 | |
| Length of basal leaflet (mm) | 14.6 | 2.0 | 20.1 | 10.0 | |
| Width of basal leaflet (mm) | 3.6 | 0.6 | 5.5 | 2.1 | |
| Flower | |||||
| Number of flowers by peduncle | 2.7 | 0.5 | 4.2 | 1.6 | |
| Pod/seed | |||||
| Racime number per plant | 25.4 | 23.3 | 123.2 | 3.6 | |
| Pod number per plant | 41.5 | 32.5 | 179.8 | 6.1 | |
| Seeds per pod | 2.9 | 0.4 | 3.9 | 1.1 | |
| 100-seed weight (g) | 3.8 | 0.6 | 5.9 | 2.6 | |
| Quantitative Agromorphological Traits | |||||
|---|---|---|---|---|---|
| Average | SD | Max | Min | ||
| Flowering | |||||
| Days to first flowering (d) | 127.6 | 7.6 | 151.0 | 116.0 | |
| Days to 50% flowering (d) | 134.4 | 11.3 | 179.0 | 121.0 | |
| Days to final flowering (d) | 183.4 | 8.5 | 208.0 | 156.0 | |
| Days to maturity (d) | 193.9 | 6.6 | 216.0 | 183.0 | |
| Plant | |||||
| Height (cm) | 61.4 | 13.8 | 89.5 | 37.8 | |
| First pod height (cm) | 25.2 | 7.3 | 39.5 | 12.7 | |
| Branches per plant | 3.6 | 1.6 | 7.6 | 2.1 | |
| Leaf | |||||
| Petiole length (mm) | 2.0 | 0.5 | 2.9 | 1.0 | |
| Leaflet area (mm2) | 863.0 | 224.3 | 1668.5 | 573.3 | |
| Leaflet length (mm) | 44.2 | 6.0 | 68.2 | 35.0 | |
| Leaflet width (mm) | 28.0 | 3.3 | 37.4 | 22.9 | |
| Pod/seed | |||||
| Pod number per plant | 15.2 | 10.8 | 56.7 | 1.5 | |
| Ovules per pod | 6.0 | 0.9 | 9.0 | 4.5 | |
| Harvest Index | 29.2 | 17.1 | 52.3 | 0.7 | |
| 100-seed weight (g) | 20.7 | 6.8 | 26.8 | 5.5 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Román, M.I.; De la Rosa, L.; Marcos-Prado, T.; Ramírez-Parra, E. Cross-Species Transferability of SSR Markers for Analyzing Genetic Diversity of Different Vicia species Collections. Agronomy 2024, 14, 326. https://doi.org/10.3390/agronomy14020326
López-Román MI, De la Rosa L, Marcos-Prado T, Ramírez-Parra E. Cross-Species Transferability of SSR Markers for Analyzing Genetic Diversity of Different Vicia species Collections. Agronomy. 2024; 14(2):326. https://doi.org/10.3390/agronomy14020326
Chicago/Turabian StyleLópez-Román, María Isabel, Lucía De la Rosa, Teresa Marcos-Prado, and Elena Ramírez-Parra. 2024. "Cross-Species Transferability of SSR Markers for Analyzing Genetic Diversity of Different Vicia species Collections" Agronomy 14, no. 2: 326. https://doi.org/10.3390/agronomy14020326
APA StyleLópez-Román, M. I., De la Rosa, L., Marcos-Prado, T., & Ramírez-Parra, E. (2024). Cross-Species Transferability of SSR Markers for Analyzing Genetic Diversity of Different Vicia species Collections. Agronomy, 14(2), 326. https://doi.org/10.3390/agronomy14020326






