De Novo Synthesis of Anticholinergic Hyoscyamine and Scopolamine in Nicotiana benthamiana Based on Elucidating Tropane Alkaloid Biosynthetic Pathway of Anisodus luridus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Sequencing and Expression Profiling Analyses
2.3. Real-Time PCR Analysis
2.4. Homologous Gene Retrieval
2.5. Bioinformatics Analysis
2.6. Functional Verification of Candidate Genes in N. benthamiana
2.7. Quantification of TAs by LC-MS
3. Results
3.1. Transcriptome Sequencing and Quality Assessment
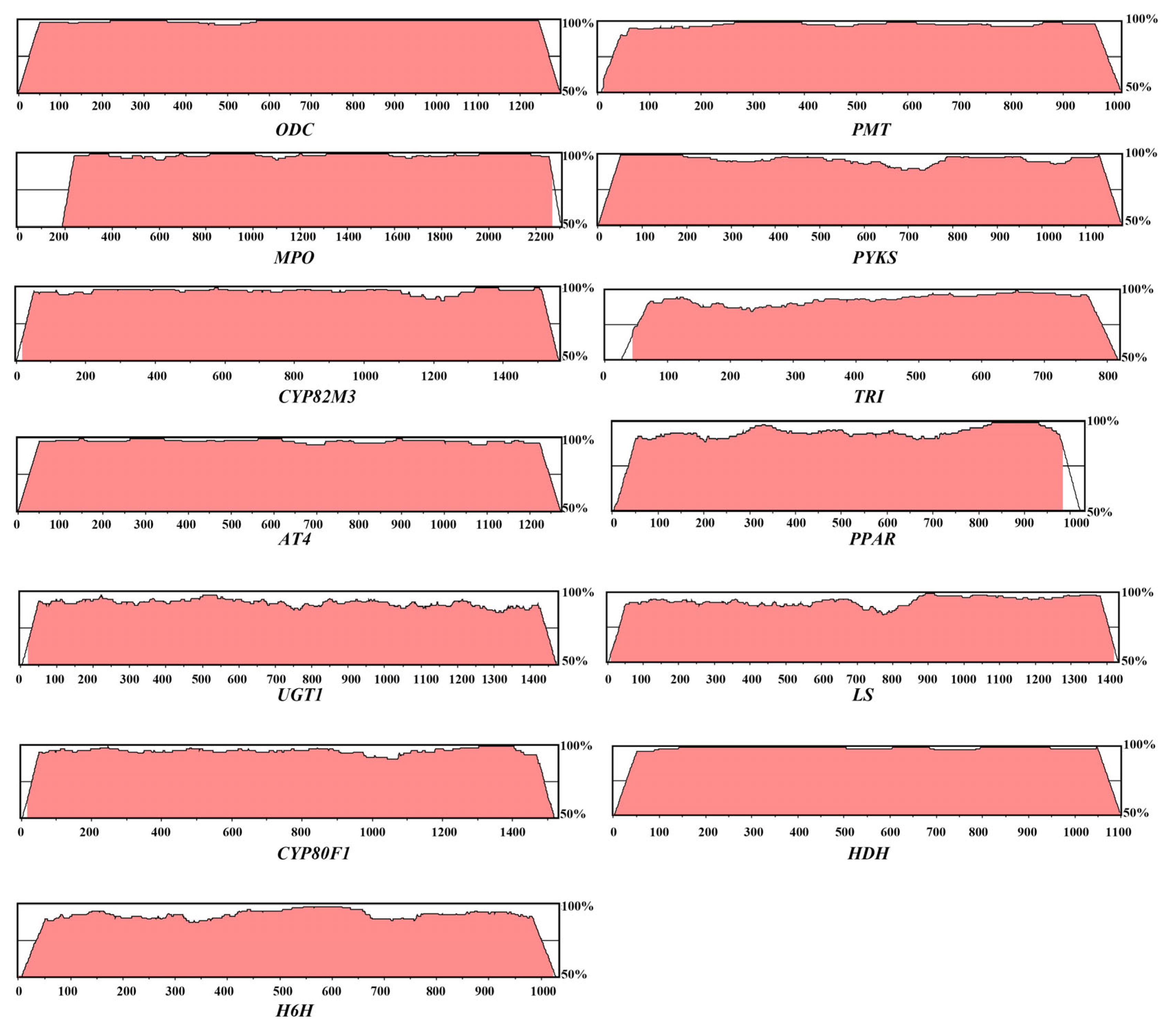
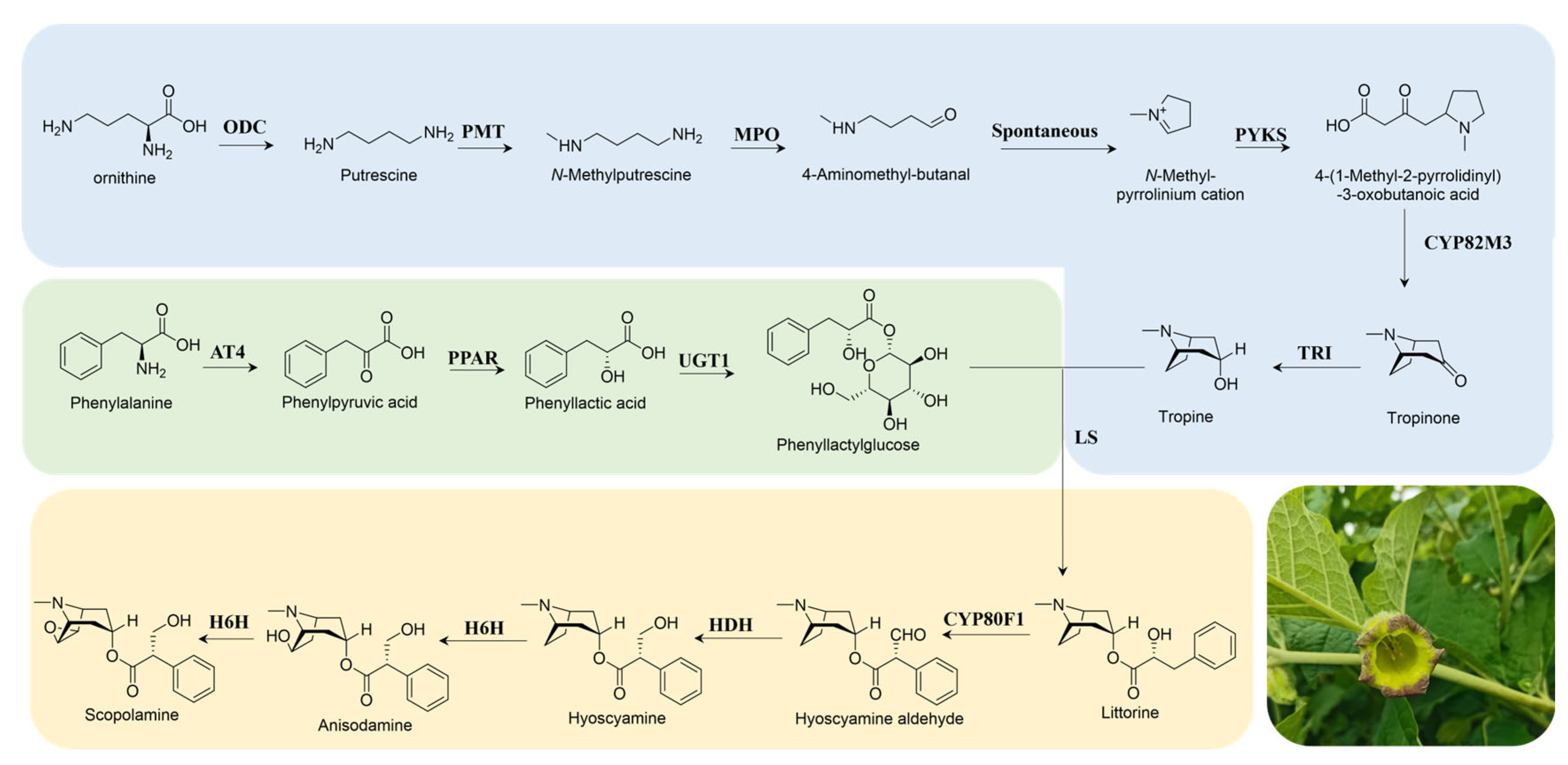
3.2. The Identification of Genes Involved in the Biosynthesis of TAs
3.3. Analysis of Physical and Chemical Properties and Subcellular Locations
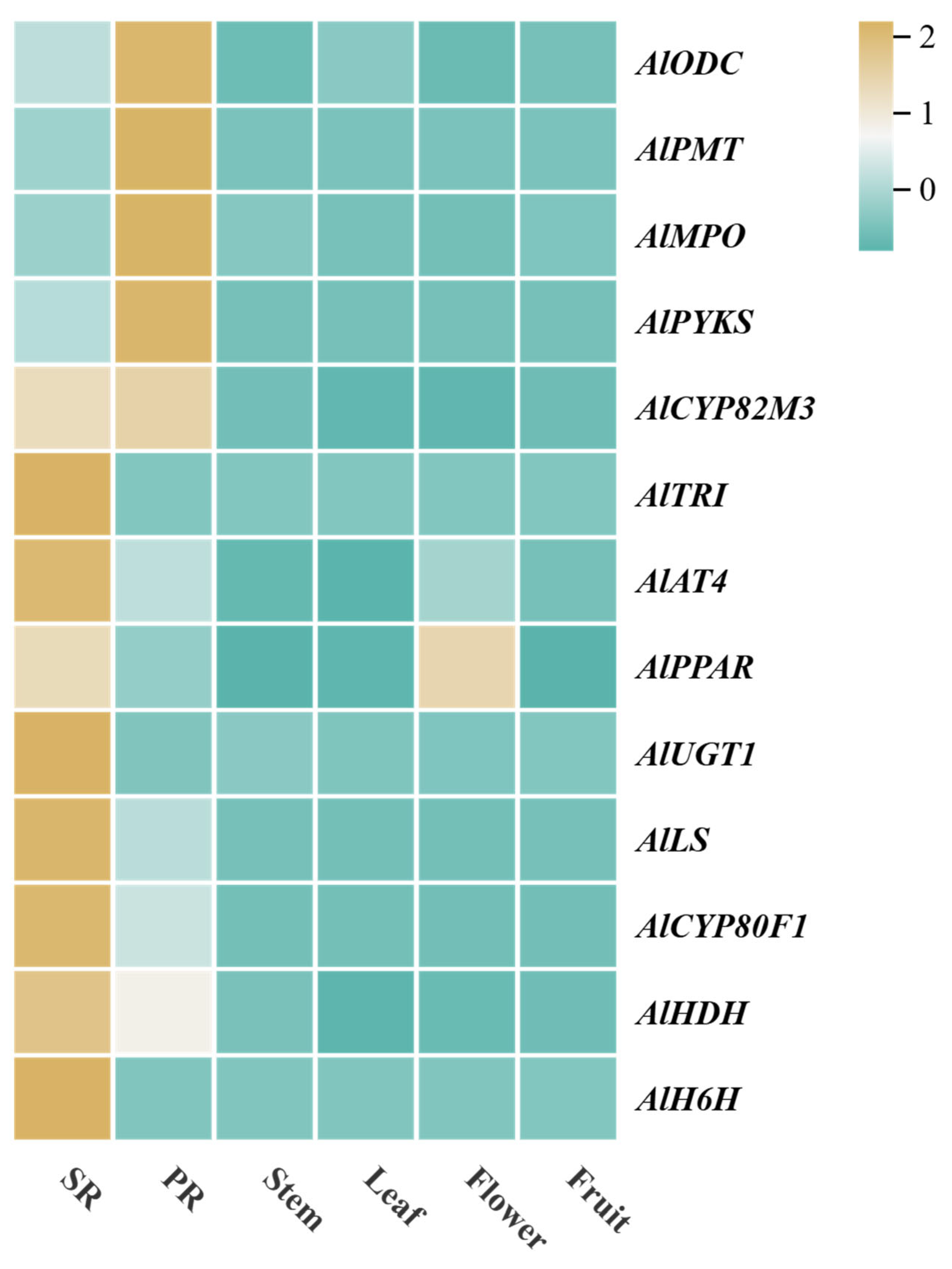
3.4. Expression Analysis Based on RNA-Seq
3.5. RT-PCR Validation
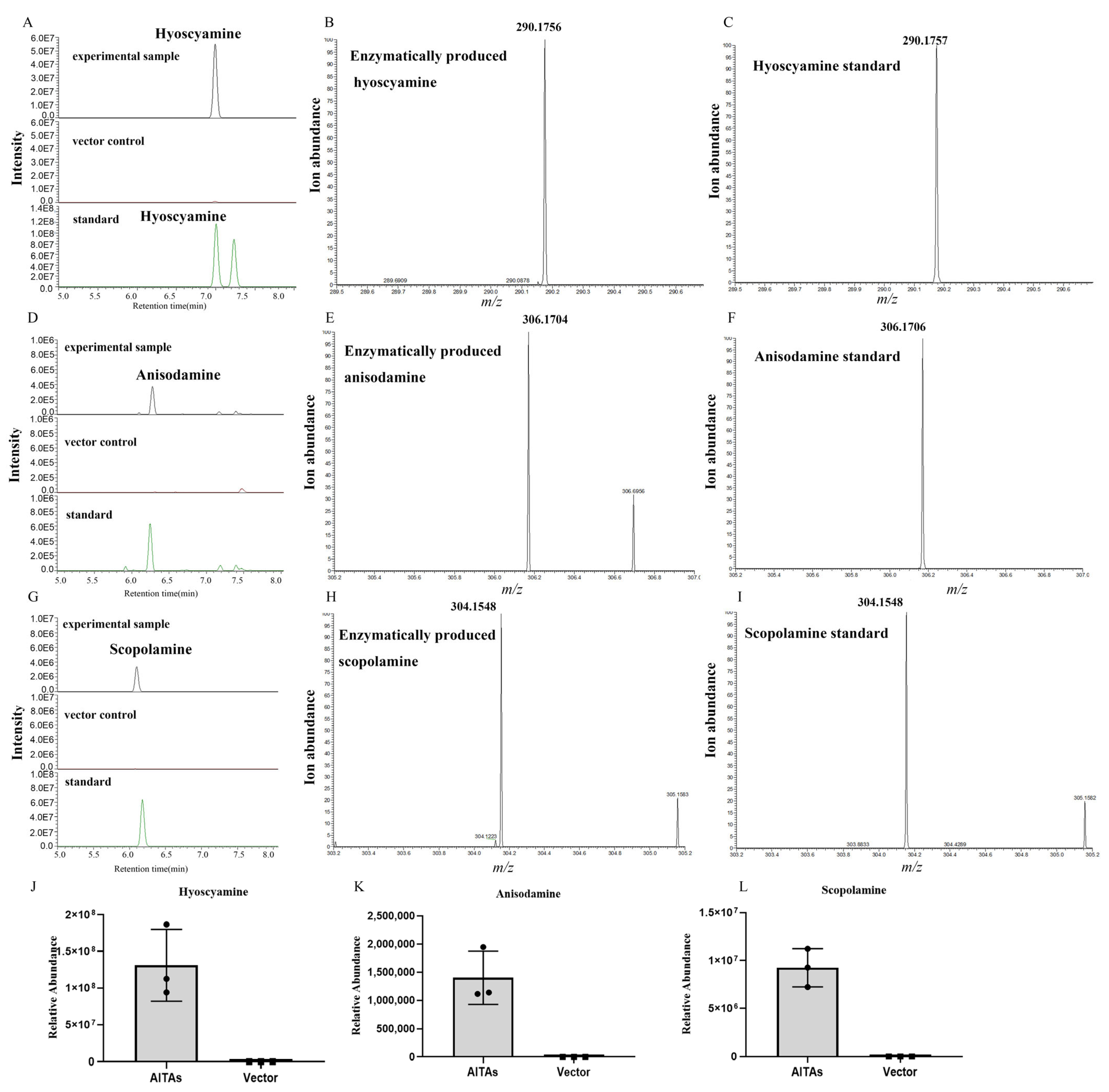
3.6. De Novo Synthesis of TAs in N. benthamiana
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lan, X.; Zeng, J.; Liu, K.; Zhang, F.; Bai, G.; Chen, M.; Liao, Z.; Huang, L. Comparison of two hyoscyamine 6β-hydroxylases in engineering scopolamine biosynthesis in root cultures of Scopolia lurida. Biochem. Biophys. Res. Commun. 2018, 497, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Mehrotra, S.; Mishra, D.S. Tropane Alkaloids Pathways, Potential and Biotechnological Applications: Pathways, Potential and Biotechnological Applications; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar] [CrossRef]
- Ullrich, S.F.; Hagels, H.; Kayser, O. Scopolamine: A journey from the field to clinics. Phytochem. Rev. 2017, 16, 333–353. [Google Scholar] [CrossRef]
- Kohnen-Johannsen, K.L.; Kayser, O. Tropane Alkaloids: Chemistry, Pharmacology, Biosynthesis and Production. Molecules 2019, 24, 796. [Google Scholar] [CrossRef]
- Huang, J.P.; Wang, Y.J.; Tian, T.; Wang, L.; Yan, Y.; Huang, S.X. Tropane alkaloid biosynthesis: A centennial review. Nat. Prod. Rep. 2021, 38, 1634–1658. [Google Scholar] [CrossRef]
- Zhang, F.; Qiu, F.; Zeng, J.; Xu, Z.; Tang, Y.; Zhao, T.; Gou, Y.; Su, F.; Wang, S.; Sun, X.; et al. Revealing evolution of tropane alkaloid biosynthesis by analyzing two genomes in the Solanaceae family. Nat. Commun. 2023, 14, 1446. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, Y.; Zhang, P.; Ma, J.; Yao, Y.J.; Ma, Y.L.; Zhang, L.; Yang, Y.; Zhao, C.; Wu, J.; et al. Multiple independent losses of the biosynthetic pathway for two tropane alkaloids in the Solanaceae family. Nat. Commun. 2023, 14, 8457. [Google Scholar] [CrossRef]
- Michael, A.J.; Furze, J.M.; Rhodes, M.J.; Burtin, D. Molecular cloning and functional identification of a plant ornithine decarboxylase cDNA. Biochem. J. 1996, 314 Pt 1, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Yamada, Y.; Hashimoto, T. Expression of Atropa belladonna putrescine N-methyltransferase gene in root pericycle. Plant Cell Physiol. 1999, 40, 289–297. [Google Scholar] [CrossRef]
- Huang, J.P.; Fang, C.; Ma, X.; Wang, L.; Yang, J.; Luo, J.; Yan, Y.; Zhang, Y.; Huang, S.X. Tropane alkaloids biosynthesis involves an unusual type III polyketide synthase and non-enzymatic condensation. Nat. Commun. 2019, 10, 4036. [Google Scholar] [CrossRef]
- Bedewitz, M.A.; Jones, A.D.; D’Auria, J.C.; Barry, C.S. Tropinone synthesis via an atypical polyketide synthase and P450-mediated cyclization. Nat. Commun. 2018, 9, 5281. [Google Scholar] [CrossRef]
- Nakajima, K.; Hashimoto, T.; Yamada, Y. Two tropinone reductases with different stereospecificities are short-chain dehydrogenases evolved from a common ancestor. Proc. Natl. Acad. Sci. USA 1993, 90, 9591–9595. [Google Scholar] [CrossRef] [PubMed]
- Bedewitz, M.A.; Góngora-Castillo, E.; Uebler, J.B.; Gonzales-Vigil, E.; Wiegert-Rininger, K.E.; Childs, K.L.; Hamilton, J.P.; Vaillancourt, B.; Yeo, Y.S.; Chappell, J.; et al. A root-expressed L-phenylalanine:4-hydroxyphenylpyruvate aminotransferase is required for tropane alkaloid biosynthesis in Atropa belladonna. Plant Cell 2014, 26, 3745–3762. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Yang, C.; Yuan, L.; Xiang, D.; Lan, X.; Chen, M.; Liao, Z. A Phenylpyruvic Acid Reductase Is Required for Biosynthesis of Tropane Alkaloids. Org. Lett. 2018, 20, 7807–7810. [Google Scholar] [CrossRef]
- Qiu, F.; Zeng, J.; Wang, J.; Huang, J.P.; Zhou, W.; Yang, C.; Lan, X.; Chen, M.; Huang, S.X.; Kai, G.; et al. Functional genomics analysis reveals two novel genes required for littorine biosynthesis. New Phytol. 2020, 225, 1906–1914. [Google Scholar] [CrossRef]
- Li, R.; Reed, D.W.; Liu, E.; Nowak, J.; Pelcher, L.E.; Page, J.E.; Covello, P.S. Functional genomic analysis of alkaloid biosynthesis in Hyoscyamus niger reveals a cytochrome P450 involved in littorine rearrangement. Chem. Biol. 2006, 13, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, P.; Smolke, C.D. Biosynthesis of medicinal tropane alkaloids in yeast. Nature 2020, 585, 614–619. [Google Scholar] [CrossRef]
- Hashimoto, T.; Matsuda, J.; Yamada, Y. Two-step epoxidation of hyoscyamine to scopolamine is catalyzed by bifunctional hyoscyamine 6 beta-hydroxylase. FEBS Lett. 1993, 329, 35–39. [Google Scholar] [CrossRef]
- Wang, Y.J.; Tain, T.; Yu, J.Y.; Li, J.; Xu, B.; Chen, J.; D’Auria, J.C.; Huang, J.P.; Huang, S.X. Genomic and structural basis for evolution of tropane alkaloid biosynthesis. Proc. Natl. Acad. Sci. USA 2023, 120, e2302448120. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, C.; Hao, X.; Chen, F.; Huang, Q.; Liu, T.; Xu, J.; Guo, S.; Liao, B.; Liu, Z.; et al. A chromosome-level genome assembly of anesthetic drug-producing Anisodus acutangulus provides insights into its evolution and the biosynthesis of tropane alkaloids. Plant Commun. 2024, 5, 100680. [Google Scholar] [CrossRef]
- Golubova, D.; Tansley, C.; Su, H.; Patron, N.J. Engineering Nicotiana benthamiana as a platform for natural product biosynthesis. Curr. Opin. Plant Biol. 2024, 81, 102611. [Google Scholar] [CrossRef]
- Zhang, Y.; Wiese, L.; Fang, H.; Alseekh, S.; Perez de Souza, L.; Scossa, F.; Molloy, J.; Christmann, M.; Fernie, A.R. Synthetic biology identifies the minimal gene set required for paclitaxel biosynthesis in a plant chassis. Mol. Plant 2023, 16, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.; Orme, A.; El-Demerdash, A.; Owen, C.; Martin, L.B.B.; Misra, R.C.; Kikuchi, S.; Rejzek, M.; Martin, A.C.; Harkess, A.; et al. Elucidation of the pathway for biosynthesis of saponin adjuvants from the soapbark tree. Science 2023, 379, 1252–1264. [Google Scholar] [CrossRef]
- Martin, L.B.B.; Kikuchi, S.; Rejzek, M.; Owen, C.; Reed, J.; Orme, A.; Misra, R.C.; El-Demerdash, A.; Hill, L.; Hodgson, H.; et al. Complete biosynthesis of the potent vaccine adjuvant QS-21. Nat. Chem. Biol. 2024, 20, 493–502. [Google Scholar] [CrossRef]
- Berman, P.; de Haro, L.A.; Jozwiak, A.; Panda, S.; Pinkas, Z.; Dong, Y.; Cveticanin, J.; Barbole, R.; Livne, R.; Scherf, T.; et al. Parallel evolution of cannabinoid biosynthesis. Nat. Plants 2023, 9, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Bajsa-Hirschel, J.; Vaughn, J.N.; Rimando, A.M.; Baerson, S.R.; Duke, S.O. In vivo assembly of the sorgoleone biosynthetic pathway and its impact on agroinfiltrated leaves of Nicotiana benthamiana. New Phytol. 2021, 230, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Kruse, L.H.; Sunstrum, F.G.; Garcia, D.; López Pérez, G.; Jancsik, S.; Bohlmann, J.; Irmisch, S. Improved production of the antidiabetic metabolite montbretin A in Nicotiana benthamiana: Discovery, characterization, and use of Crocosmia shikimate shunt genes. Plant J. 2024, 117, 766–785. [Google Scholar] [CrossRef]
- Wang, Y.J.; Huang, J.P.; Tian, T.; Yan, Y.; Chen, Y.; Yang, J.; Chen, J.; Gu, Y.C.; Huang, S.X. Discovery and Engineering of the Cocaine Biosynthetic Pathway. J. Am. Chem. Soc. 2022, 144, 22000–22007. [Google Scholar] [CrossRef]
- Hong, B.; Grzech, D.; Caputi, L.; Sonawane, P.; López, C.E.R.; Kamileen, M.O.; Hernández Lozada, N.J.; Grabe, V.; O’Connor, S.E. Biosynthesis of strychnine. Nature 2022, 607, 617–622. [Google Scholar] [CrossRef]
- Florean, M.; Luck, K.; Hong, B.; Nakamura, Y.; O’Connor, S.E.; Köllner, T.G. Reinventing metabolic pathways: Independent evolution of benzoxazinoids in flowering plants. Proc. Natl. Acad. Sci. USA 2023, 120, e2307981120. [Google Scholar] [CrossRef]
- Nett, R.S.; Lau, W.; Sattely, E.S. Discovery and engineering of colchicine alkaloid biosynthesis. Nature 2020, 584, 148–153. [Google Scholar] [CrossRef]
- Yin, X.; Liu, J.; Kou, C.; Lu, J.; Zhang, H.; Song, W.; Li, Y.; Xue, Z.; Hua, X. Deciphering the network of cholesterol biosynthesis in Paris polyphylla laid a base for efficient diosgenin production in plant chassis. Metab. Eng. 2023, 76, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Zhu, J.; Huang, S.; Xing, X.; Zhou, S.; Yao, H.; Yang, Z.; Liu, L.; Huang, S.; Miao, Y.; et al. Metabolome profiling and transcriptome analysis filling the early crucial missing steps of piperine biosynthesis in Piper nigrum L. Plant J. 2024, 117, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Leete, E.; Marion, L.; Spenser, I.D. Biogenesis of hyoscyamine. Nature 1954, 174, 650–651. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Sainsbury, F.; Thuenemann, E.C.; Lomonossoff, G.P. pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 2009, 7, 682–693. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Facchini, P.J. Alkaloid Biosynthesis in Plants: Biochemistry, Cell Biology, Molecular Regulation, and Metabolic Engineering Applications. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 29–66. [Google Scholar] [CrossRef]
- Zeng, J.; Liu, X.; Dong, Z.; Zhang, F.; Qiu, F.; Zhong, M.; Zhao, T.; Yang, C.; Zeng, L.; Lan, X.; et al. Discovering a mitochondrion-localized BAHD acyltransferase involved in calystegine biosynthesis and engineering the production of 3β-tigloyloxytropane. Nat. Commun. 2024, 15, 3623. [Google Scholar] [CrossRef]
- Moyano, E.; Jouhikainen, K.; Tammela, P.; Palazón, J.; Cusidó, R.M.; Piñol, M.T.; Teeri, T.H.; Oksman-Caldentey, K.M. Effect of pmt gene overexpression on tropane alkaloid production in transformed root cultures of Datura metel and Hyoscyamus muticus. J. Exp. Bot. 2003, 54, 203–211. [Google Scholar] [CrossRef]
- Lee, O.-S.; Kang, Y.-M.; Jung, H.-Y.; Min, J.-Y.; Kang, S.-M.; Karigar, C.S.; Prasad, D.T.; Bahk, J.-D.; Choi, M.-S. Enhanced production of tropane alkaloids in Scopolia parviflora by introducing the PMT (putrescine N-methyltransferase) gene. In Vitro Cell. Dev.-Plant 2005, 41, 167–172. [Google Scholar] [CrossRef]
- Richter, U.; Rothe, G.; Fabian, A.K.; Rahfeld, B.; Dräger, B. Overexpression of tropinone reductases alters alkaloid composition in Atropa belladonna root cultures. J. Exp. Bot. 2005, 56, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Yun, D.J.; Hashimoto, T.; Yamada, Y. Metabolic engineering of medicinal plants: Transgenic Atropa belladonna with an improved alkaloid composition. Proc. Natl. Acad. Sci. USA 1992, 89, 11799–11803. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, P.; Smolke, C.D. Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids. Nat. Commun. 2019, 10, 3634. [Google Scholar] [CrossRef] [PubMed]

| Samples | Clean Reads | Clean Bases | GC Content | % ≥ Q30 | Total Reads |
|---|---|---|---|---|---|
| SR-1 | 21,416,963 | 6,412,990,514 | 42.12% | 96.15% | 42,833,926 |
| SR-2 | 21,508,499 | 6,439,821,856 | 43.19% | 95.27% | 43,016,998 |
| SR-3 | 21,400,303 | 6,406,596,650 | 42.29% | 94.68% | 42,800,606 |
| Flower-1 | 22,920,435 | 6,861,817,636 | 42.60% | 94.50% | 45,840,870 |
| Flower-2 | 24,844,633 | 7,438,750,734 | 42.58% | 94.81% | 49,689,266 |
| Flower-3 | 21,680,946 | 6,491,325,066 | 42.59% | 95.08% | 43,361,892 |
| Fruit-1 | 20,698,862 | 6,196,316,816 | 42.78% | 95.10% | 41,397,724 |
| Fruit-2 | 19,288,560 | 5,774,477,710 | 42.13% | 94.98% | 38,577,120 |
| Fruit-3 | 21,871,094 | 6,547,980,308 | 42.22% | 94.86% | 43,742,188 |
| Leaf-1 | 22,519,097 | 6,743,921,392 | 42.33% | 94.69% | 45,038,194 |
| Leaf-2 | 23,490,539 | 7,034,719,032 | 42.21% | 94.92% | 46,981,078 |
| Leaf-3 | 20,035,207 | 5,998,901,816 | 42.07% | 94.78% | 40,070,414 |
| PR-1 | 19,990,642 | 5,984,977,532 | 42.96% | 94.25% | 39,981,284 |
| PR-2 | 21,249,355 | 6,359,057,558 | 42.94% | 94.91% | 42,498,710 |
| PR-3 | 20,825,144 | 6,233,208,310 | 42.87% | 94.63% | 41,650,288 |
| Stem-1 | 21,476,136 | 6,423,857,126 | 42.19% | 95.02% | 42,952,272 |
| Stem-2 | 20,458,651 | 6,125,644,130 | 42.01% | 94.69% | 40,917,302 |
| Stem-3 | 21,037,941 | 6,296,989,810 | 42.03% | 94.74% | 42,075,882 |
| Name | Identity (%) A. luridus vs. A. belladonna |
|---|---|
| ODC | 97.67% |
| PMT | 97.34% |
| MPO | 90.35% |
| PYKS | 97.19% |
| CYP82M3 | 97.30% |
| TRI | 90.48% |
| AT4 | 97.16% |
| PPAR | 91.84% |
| UGT1 | 93.28% |
| LS | 92.65% |
| CYP80F1 | 95.47% |
| HDH | 98.63% |
| H6H | 90.41% |
| Gene Name | Length of CDS | Num. of AA | MW (Da) | pI | Subcellular Location |
|---|---|---|---|---|---|
| AlODC | 1293 | 430 | 46,526.93 | 5.61 | Plasma Membrane |
| AlPMT | 1017 | 338 | 37,176.54 | 5.73 | Cytoplasmic |
| AlMPO | 2118 | 705 | 79,090.01 | 6.38 | Cytoplasmic |
| AlPYKS | 1179 | 392 | 43,261.08 | 7.15 | Cytoplasmic |
| AlCYP82M3 | 1560 | 519 | 59,463.58 | 6.69 | Cytoplasmic |
| AlTRI | 801 | 266 | 28,652.85 | 6.45 | Cytoplasmic |
| AlAT4 | 1272 | 423 | 47,057.53 | 5.89 | Plasma Membrane |
| AlPPAR | 1029 | 342 | 38,254.44 | 5.92 | Cytoplasmic |
| AlUGT1 | 1476 | 491 | 54,927.14 | 5.54 | Cytoplasmic |
| AlLS | 1431 | 476 | 53,931.69 | 6.07 | Plasma Membrane |
| AlCYP80F1 | 1521 | 506 | 57,674.2 | 7.19 | Cytoplasmic |
| AlHDH | 1098 | 365 | 38,951.97 | 5.26 | Cytoplasmic |
| AlH6H | 1035 | 344 | 38,883.49 | 5.02 | Cytoplasmic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, M.; Zeng, J.; Qiu, F.; Zhang, F.; Liao, Z. De Novo Synthesis of Anticholinergic Hyoscyamine and Scopolamine in Nicotiana benthamiana Based on Elucidating Tropane Alkaloid Biosynthetic Pathway of Anisodus luridus. Agronomy 2024, 14, 2460. https://doi.org/10.3390/agronomy14112460
Wen M, Zeng J, Qiu F, Zhang F, Liao Z. De Novo Synthesis of Anticholinergic Hyoscyamine and Scopolamine in Nicotiana benthamiana Based on Elucidating Tropane Alkaloid Biosynthetic Pathway of Anisodus luridus. Agronomy. 2024; 14(11):2460. https://doi.org/10.3390/agronomy14112460
Chicago/Turabian StyleWen, Mengling, Junlan Zeng, Fei Qiu, Fangyuan Zhang, and Zhihua Liao. 2024. "De Novo Synthesis of Anticholinergic Hyoscyamine and Scopolamine in Nicotiana benthamiana Based on Elucidating Tropane Alkaloid Biosynthetic Pathway of Anisodus luridus" Agronomy 14, no. 11: 2460. https://doi.org/10.3390/agronomy14112460
APA StyleWen, M., Zeng, J., Qiu, F., Zhang, F., & Liao, Z. (2024). De Novo Synthesis of Anticholinergic Hyoscyamine and Scopolamine in Nicotiana benthamiana Based on Elucidating Tropane Alkaloid Biosynthetic Pathway of Anisodus luridus. Agronomy, 14(11), 2460. https://doi.org/10.3390/agronomy14112460






