Comparative Transcriptome Analysis and Expression of Genes Associated with Polysaccharide Biosynthesis in Dendrobium officinale Diploid and Tetraploid Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. RNA Isolation, cDNA Library Preparation, Transcriptome Sequencing
2.3. De Novo Assembly and Functional Annotation
2.4. Identification of Differentially Expressed Genes (DEGs)
2.5. Quantitative Real-Time PCR (qRT-PCR) Validation
3. Results
3.1. Transcriptome Sequencing and De Novo Assembly
3.2. Functional Annotation of D. officinale Transcriptome
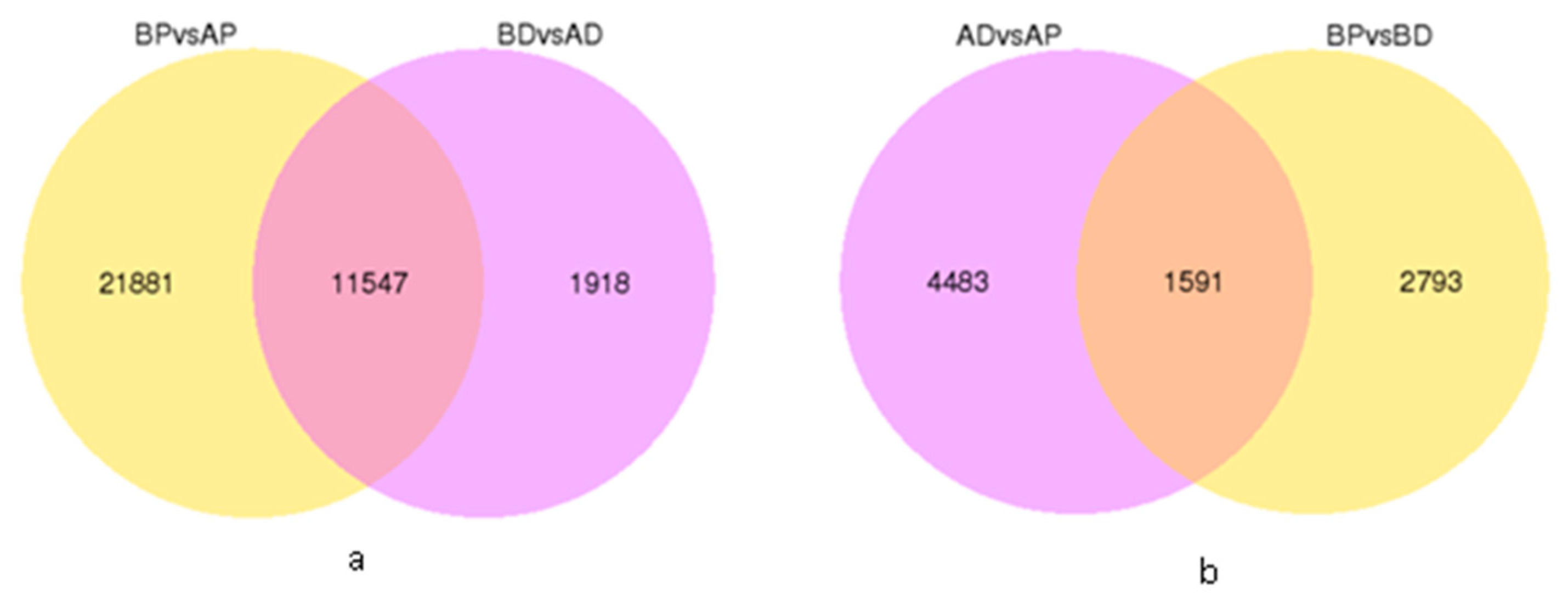
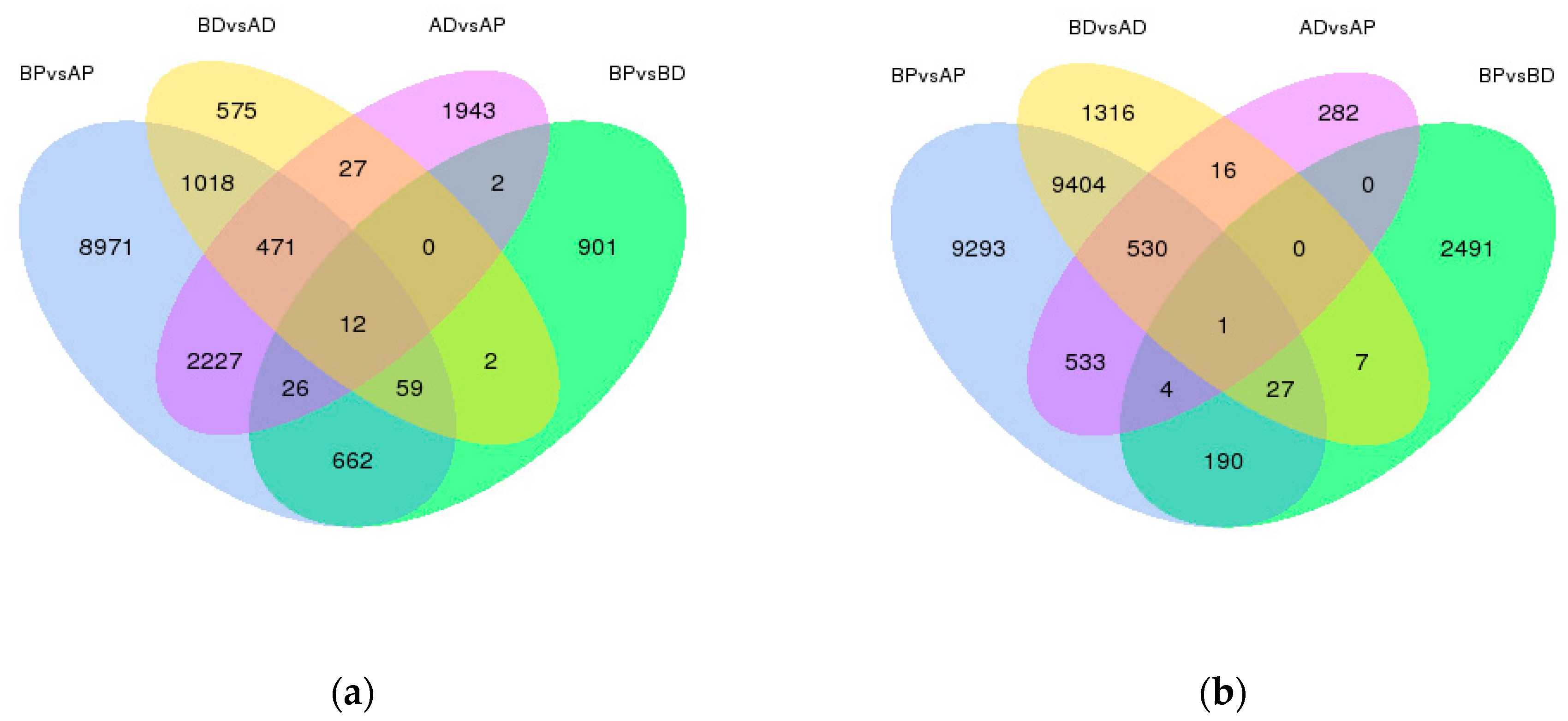
3.3. Screening and Identification of DEGs
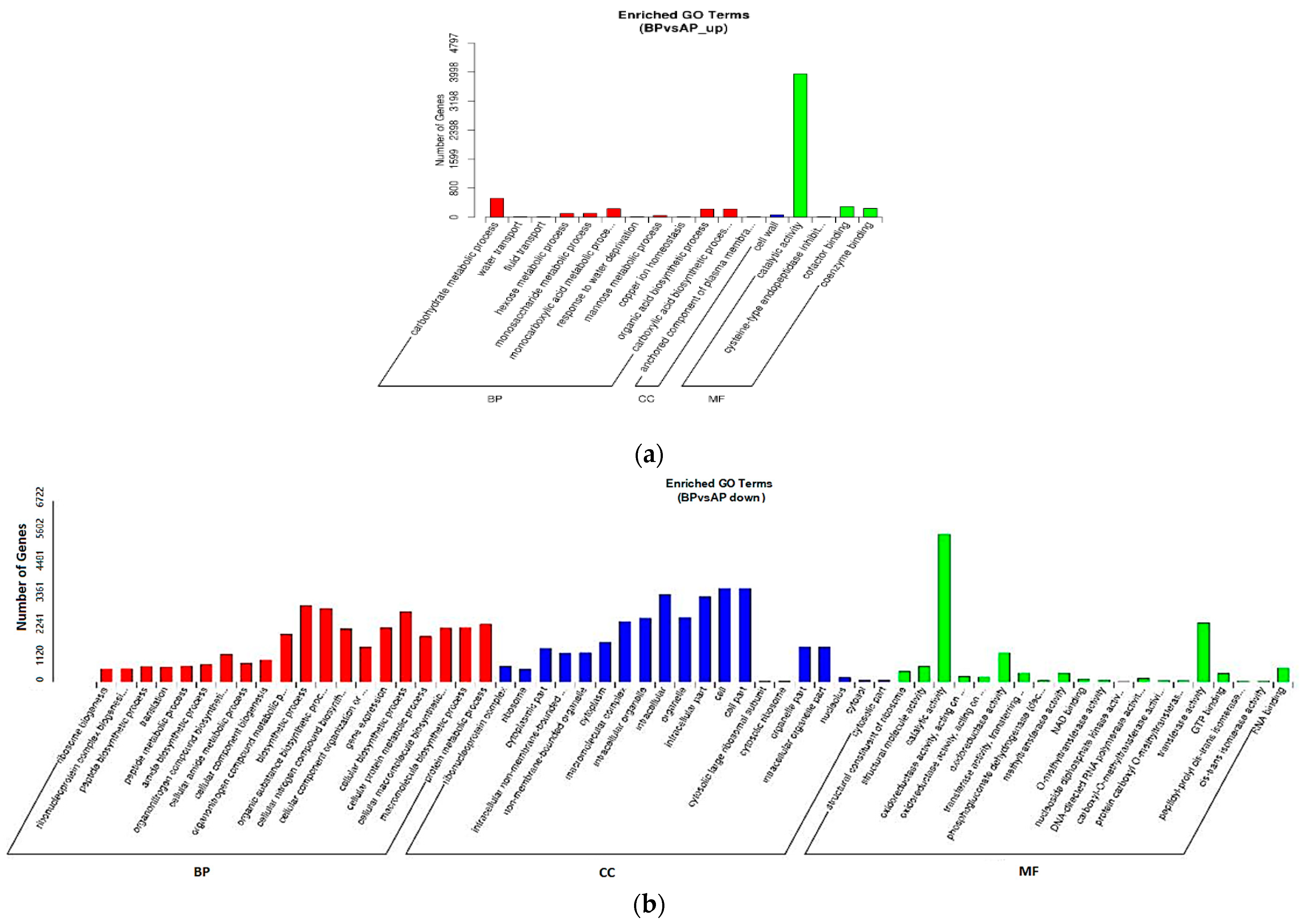
3.4. GO and KEGG of DEGs Annotation and Enrichment Analysis
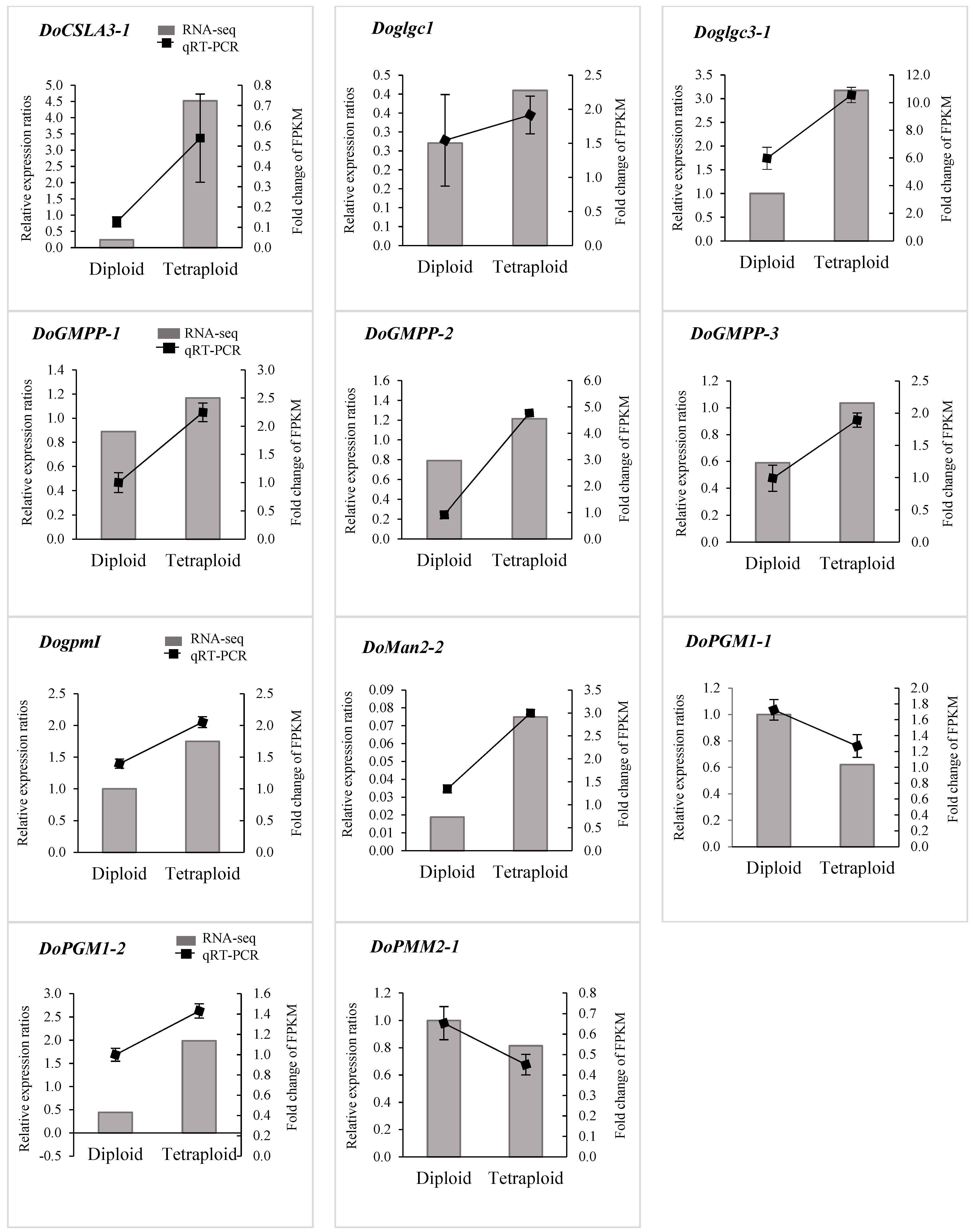
3.5. qRT-PCR Validation for Genes Involved in Polysaccharide Biosynthesis between Diploid and Tetraploid D. officinale
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohammed, A.S.A.; Naveed, M.; Jost, N. Polysaccharides; classification, chemical properties, and future perspective applications in fields of pharmacology and biological medicine (A review of current applications and upcoming potentialities). J. Polym. Environ. 2021, 29, 2359–2371. [Google Scholar] [CrossRef] [PubMed]
- Claus-Desbonnet, H.; Nikly, E.; Nalbantova, V.; Karcheva-Bahchevanska, D.; Ivanova, S.; Pierre, G.; Benbassat, N.; Katsarov, P.; Michaud, P.; Lukova, P.; et al. Polysaccharides and their derivatives as potential antiviral molecules. Viruses 2022, 14, 426. [Google Scholar] [CrossRef] [PubMed]
- Tzianabos, A.O. Polysaccharide immunomodulators as therapeutic agents: Structural aspects and biological function. Clin. Microbiol. Rev. 2000, 13, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Xie, L.; Chen, Y.; Zhang, H. Purification, characterization, and anticoagulant activity of the polysaccharides from green tea. Carbohydr. Polym. 2013, 92, 1086–1090. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Synytsya, A.; Kim, H.B.; Choi, D.J.; Lee, S.; Lee, J.; Kim, W.J.; Jang, S.J.; Park, Y.I. Purification, characterization and immunomodulating activity of a pectic polysaccharide isolated from Korean mulberry fruit Oddi (Morus alba L.). Int. Immunopharmacol. 2013, 17, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.; Morris, G.A. The anti-diabetic potential of polysaccharides extracted from members of the cucurbit family: A review. Bioact. Carbohydr. Diet. Fibre 2014, 3, 106–114. [Google Scholar] [CrossRef]
- Xie, S.Z.; Liu, B.; Zhang, D.D.; Zha, X.Q.; Luo, J.P. Intestinal immunomodulating activity and structural characterization of a new polysaccharide from stems of Dendrobium officinale. Food Funct. 2016, 7, 2789–2799. [Google Scholar] [CrossRef]
- Chen, W.H.; Wu, J.J.; Li, X.F.; Lu, J.M.; Wu, W.; Sun, Y.Q.; Zhu, B. Isolation, structural properties, bioactivities of polysaccharides from Dendrobium officinale Kimura et. Migo: A review. Int. J. Biol. Macromol. 2021, 184, 1000–1013. [Google Scholar] [CrossRef]
- Zhu, S.; Niu, Z.; Xue, Q.; Wang, H.; Xie, X.; Ding, X. Accurate authentication of Dendrobium officinale and its closely related species by comparative analysis of complete plastomes. Acta Pharm. Sin. B 2018, 8, 969–980. [Google Scholar] [CrossRef]
- Xu, J.; Han, Q.B.; Li, S.L.; Chen, X.J.; Wang, X.N.; Zhao, Z.Z.; Chen, H.B. Chemistry, bioactivity and quality control of Dendrobium, a commonly used tonic herb in traditional Chinese medicine. Phytochem. Rev. 2013, 12, 341–367. [Google Scholar] [CrossRef]
- Niu, Z.; Zhu, F.; Fan, Y.; Li, C.; Zhang, B.; Zhu, S.; Hou, Z.; Wang, M.; Yang, J.; Xue, Q.; et al. The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm. Sin. B. 2021, 11, 2080–2092. [Google Scholar] [CrossRef] [PubMed]
- Kamemoto, H.; Amore, T.D.; Kuehnle, A.R. Breeding Dendrobium Orchids in Hawaii; University of Hawaii Press: Honolulu, HI, USA, 1999; 176p. [Google Scholar]
- Zhang, X.; Gao, J. In vitro tetraploid induction from multigenotype protocorms and tetraploid regeneration in Dendrobium officinale. Plant Cell Tissue Organ Cult. 2020, 141, 289–298. [Google Scholar] [CrossRef]
- Teixeira da Silva, J.A.; Zeng, S.; Galdiano, R.F., Jr.; Dobra’nszki, J.; Cardoso, J.C.; Vendrame, W.A. In vitro conservation of Dendrobium germplasm. Plant Cell Rep. 2014, 33, 1413–1423. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.M.; Kant, R.; Van, P.T.; Winarto, B.; Zeng, S.; Teixera da Silva, J.A. The application of biotechnology to orchids. Crit. Rev. Plant Sci. 2013, 32, 69–139. [Google Scholar] [CrossRef]
- Manzoor, A.; Ahmad, T.; Bashir, M.A.; Hafiz, I.A.; Silvestri, C. Studies on colchicine induced chromosome doubling for enhancement of quality traits in ornamental plants. Plants 2019, 8, 194. [Google Scholar] [CrossRef]
- Atichart, P. Polyploid induction by colchicine treatments and plant regeneration of Dendrobium chrysotoxum. Thai J. Agric. Sci. 2013, 46, 59–63. [Google Scholar]
- Li, H.; Zheng, X.S.; Long, C.L. Induction of polyploid of Dendrobium devonianum. Acta Bot. Yunnanica 2005, 27, 552–556, (In Chinese with English abstract). [Google Scholar]
- Vichiato, M.R.M.; Vichiato, M.; Pasqual, M.; Rodrigues, F.A.; Castro, D.M. Morphological effects of induced polyploidy in Dendrobium nobile Lindl. (Orchidaceae). Crop Breed. Appl. Biotechnol. 2014, 14, 154–159. [Google Scholar] [CrossRef]
- Wang, A.H.; Wu, Q.Q.; Yang, L.; Xu, H.J.; Chen, Z.L. Study on polyploid of Dendrobium ochreatum induced by colchicine. J. Southwest Univ. 2017, 39, 55–60, (In Chinese with English abstract). [Google Scholar]
- Zhan, Z.G.; Cheng, X.U. Study on colchiploid of Dendrobium officinale induced by colchicines. J. Zhejiang Univ. 2011, 38, 321–325, (In Chinese with English abstract). [Google Scholar]
- Liu, Y.; Duan, S.-D.; Jia, Y.; Hao, L.-H.; Xiang, D.-Y.; Chen, D.-F.; Niu, S.-C. Polyploid Induction and Karyotype Analysis of Dendrobium officinale. Horticulturae 2023, 9, 329. [Google Scholar] [CrossRef]
- Bulpitt, C.J.; Li, Y.; Bulpitt, P.F.; Wang, J. The use of orchids in Chinese medicine. J. R. Soc. Med. 2007, 100, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.M.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 2015, 8, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.X.; Zhao, T.W.; Sheng, Y.J.; Zheng, T.; Fu, L.Z.; Zhang, Y.S. Dendrobium officinale Kimura et Migo: A review on its ethnopharmacology, phytochemistry, pharmacology, and industrialization. Evid. Based Complement. Altern. Med. 2017, 2017, 7436259. [Google Scholar] [CrossRef] [PubMed]
- Mai, Y.; Yang, Z.; Ji, X.; An, W.; Huang, Y.; Liu, S.; He, L.; Lai, X.; Huang, S.; Zheng, X. Comparative analysis of transcriptome and metabolome uncovers the metabolic differences between Dendrobium officinale protocorms and mature stems. All Life 2020, 13, 346–359. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, W.; Liu, Y.; Meng, X.; Su, X.; Cao, M.; Wu, L.; Yu, N.; Xing, S.; Peng, D. Putative genes in alkaloid biosynthesis identified in Dendrobium officinale by correlating the contents of major bioactive metabolites with genes expression between protocorm-like bodies and leaves. BMC Genom. 2021, 22, 579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, X.; Zhu, X.; Hua, Y. Chemical constituents, bioactivities, and pharmacological mechanisms of Dendrobium officinale: A review of the past decade. J. Agric. Food Chem. 2023, 71, 14870–14889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yu, Z.; Zeng, D.; Si, C.; Zhao, C.; Wang, H.; Li, C.; He, C.; Duan, J. Transcriptome and metabolome reveal salt-stress responses of leaf tissues from Dendrobium officinale. Biomolecules 2021, 11, 736. [Google Scholar] [CrossRef]
- Xing, X.H.; Cui, S.W.; Nie, S.P.; Phillips, G.O.; Goff, H.D.; Wang, Q. Study on Dendrobium officinale O-acetyl-glucomannan (Dendronan R): Part II. Fine structures of O-acetylated residues. Carbohydr. Polym. 2015, 117, 422–433. [Google Scholar] [CrossRef]
- Madani, H.; Escrich, A.; Hosseini, B.; Sanchez-Muñoz, R.; Khojasteh, A.; Palazon, J. Effect of polyploidy induction on natural metabolite production in medicinal plants. Biomolecules 2021, 11, 899. [Google Scholar] [CrossRef]
- Yadav, A.K.; Singh, S.; Yadav, S.; Dhyani, D.; Bhardwaj, G.; Sharma, A.; Singh, B. Induction and morpho-chemical characterization of Stevia rebaudiana colchiploids. Indian J. Agric. Sci. 2013, 83, 159–165. [Google Scholar]
- Wallaart, T.; Pras, N.; Quax, W.J. Seasonal variations of artemisinin and its biosynthetic precursors in tetraploid Artemisia annua plants compared with the diploid wild-type. Planta Med. 1999, 65, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhou, Y.; Zhang, J.; Lu, X.; Zhang, F.; Shen, Q.; Wu, S.; Chen, Y.; Wang, T.; Tang, K. Enhancement of artemisinin content in tetraploid Artemisia annua plants by modulating the expression of genes in artemisinin biosynthetic pathway. Biotechnol. Appl. Biochem. 2011, 58, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.L.; Zhu, D.N.; Cai, Z.H.; Xu, D.R. Autotetraploid plants from colchicine treated bud culture of Salvia miltiorrhiza Bge. Plant Cell Tissue Organ Cult. 1996, 47, 73–77. [Google Scholar] [CrossRef]
- Gao, S.L.; Chen, B.J.; Zhu, D.N. In vitro production and identification of autotetraploids of Scutellaria baicalensis. Plant Cell Tissue Organ Cult. 2002, 70, 289–293. [Google Scholar] [CrossRef]
- Kim, Y.S.; Hahn, E.J.; Murthy, H.N.; Paek, K.Y. Effect of polyploidy induction on biomass and ginsenoside accumulations in adventitious roots of ginseng. J. Plant Biol. 2004, 47, 356–360. [Google Scholar] [CrossRef]
- Lavania, U.C.; Srivastava, S.; Lavania, S.; Basu, S.; Misra, N.K.; Mukai, Y. Autopolyploidy differentially influences body size in plants, but facilitates enhanced accumulation of secondary metabolites, causing increased cytosine methylation. Plant J. 2012, 71, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.L.; Li, Y.X.; Guo, H.R.; Zeng, R.Z.; Xie, L.; Zhang, Z.S. Changes in morphological characteristics, regeneration ability, and polysaccharide content in tetraploid Dendrobium officinale. Hortscience 2019, 54, 1879–1886. [Google Scholar] [CrossRef]
- He, C.; Zhang, J.; Liu, X.; Zeng, S.; Wu, K.; Yu, Z.; Wang, X.; Teixeira da Silva, J.A.; Lin, Z.; Duan, J. Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol. 2015, 88, 219–231. [Google Scholar] [CrossRef]
- Zhang, J.; He, C.; Wu, K.; Teixeira da Silva, J.A.; Zeng, S.; Zhang, X.; Yu, Z.; Xia, H.; Duan, J. Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci. 2016, 7, 5. [Google Scholar] [CrossRef]
- Shen, C.; Guo, H.; Chen, H.; Shi, Y.; Meng, Y.; Lu, J.; Feng, S.; Wang, H. Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Sci. Rep. 2017, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, B.; Tang, X.; Zhang, J.; Lin, J. Comparative transcriptome analysis of different Dendrobium species reveals active ingredients-related genes and pathways. Int. J. Mol. Sci. 2020, 21, 861. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.Z.; Thompson, A.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Brown, G.R.; Maglott, D.R. NCBI Reference Sequences (RefSeq): Current status, new features, and genome annotation policy. Nucleic Acids Res. 2012, 40, D130–D135. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopaedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Bkatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Midchoud, K.; O’Donovan, C.; Phan, I.; et al. The Swiss-prot protein knowledgebase and its supplement trembl in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, Y.; Adjobi, O.I.; Wang, Y.; Liu, A. Research advances in multi-omics on the traditional Chinese herb Dendrobium officinale. Front. Plant Sci. 2022, 12, 808228. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Pu, T.; Gui, C.; Zhang, X.; Gong, L. Transcriptome analysis reveals biosynthesis of important bioactive constitutents and mechanism of stem formation of Derndrobium huoshanense. Sci. Rep. 2020, 10, 2857. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Shi, M.; Zhang, Q.; Xie, W.; Chu, L.; Qiu, M.; Li, L.; Zeng, Z.; Han, L.; Sun, Z. Transcriptomic and metabolomic analyses reveal differences in flavonoid pathway gene expression profiles between two Dendrobium varieties during vernalization. Int. J. Mol. Sci. 2023, 24, 11039. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Liu, X.; Wang, J.; Teng, S.Y.; Shi, J.Q.; Li, Y.Y.; Huang, M.R. Transcriptome sequencing and development of novel genic SSR markers for Dendrobium officinale. Mol. Breed 2017, 37, 18. [Google Scholar] [CrossRef]
- Han, R.; Xie, D.; Tong, X.; Zhang, W.; Liu, G.; Peng, D.; Yu, N. Transcriptomic landscape of Dendrobium huoshanense and its genes related to polysaccharide biosynthesis. Acta Soc. Bot. Pol. 2018, 87, 3574. [Google Scholar] [CrossRef]
- Yuan, Y.; Yu, M.; Jia, Z.; Song, X.; Liang, Y.; Zhang, J. Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genom. 2018, 19, 978. [Google Scholar] [CrossRef]
- Lei, Z.; Zhou, C.; Ji, X.; Wei, G.; Huang, Y.; Yu, W.; Luo, Y.; Qiu, Y. Transcriptome analysis reveals genes involved in flavonoid biosynthesis and accumulation in Dendrobium catenatum from different locations. Sci. Rep. 2018, 8, 6373–6388. [Google Scholar] [CrossRef]
- Yuan, Y.D.; Zhang, J.C.; Kallman, J.; Liu, X.; Meng, M.J.; Lin, J. Polysaccharide biosynthetic pathway profiling and putative gene mining of Dendrobium moniliforme using RNA-Seq in different tissues. BMC Plant Biol. 2019, 19, 521. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, Y.; Li, C.; Luo, H.; Wang, L.; Qian, J.; Luo, X.; Xiang, L.; Song, J.; Sun, C.; et al. Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene 2013, 527, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Breton, C.; Šnajdrová, L.; Jeanneau, C.; Koča, J.; Imberty, A. Structures and mechanisms of glycosyltransferases. Glycobiology 2006, 16, 29R–37R. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wu, K.; Zhang, J.; Liu, X.; Zeng, S.; Yu, Z.; Zhan, X.; da Silva, J.A.T.; Deng, R.; Tan, J.; et al. Cytochemical localization of polysaccharides in Dendrobium officinale and the involvement of DoCSLA6 in the synthesis of mannan polysaccharides. Front. Plant Sci. 2017, 8, 1599. [Google Scholar] [CrossRef]
- Xing, X.; Cui, S.W.; Nie, S.; Phillips, G.O.; Goff, H.D.; Wang, Q. Study on Dendrobium officinale O-acetyl-glucomannan (Dendronan R): Part I. Extraction, purification, and partial structural characterization. Bioact. Carbohydr. Diet. Fibre 2014, 4, 74–83. [Google Scholar] [CrossRef]
- Goubet, F.; Barton, C.J.; Mortimer, J.C.; Yu, X.; Zhang, Z.; Miles, G.P.; Richens, J.; Liepman, A.H.; Sefen, K.; Dupree, P. Cell wall glucomannan in Arabidopsis is synthesized by CSLA glycosyltransferases and influences the progression of embryogenesis. Plant J. 2009, 60, 527–538. [Google Scholar] [CrossRef]

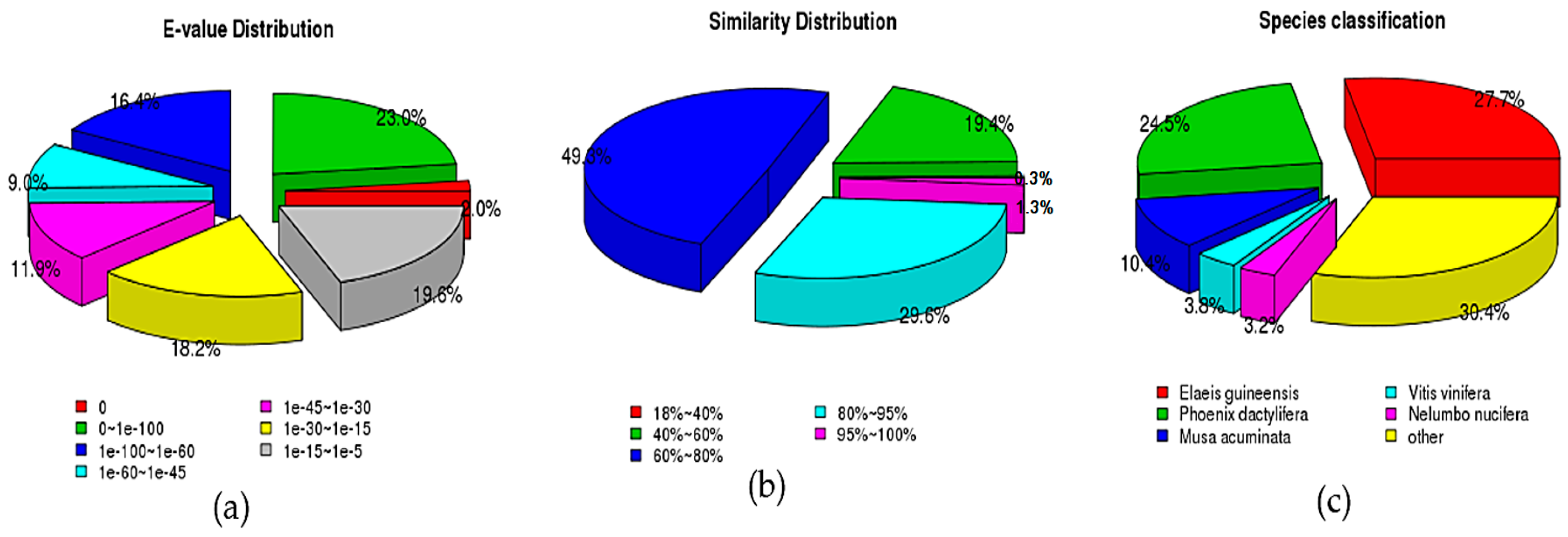
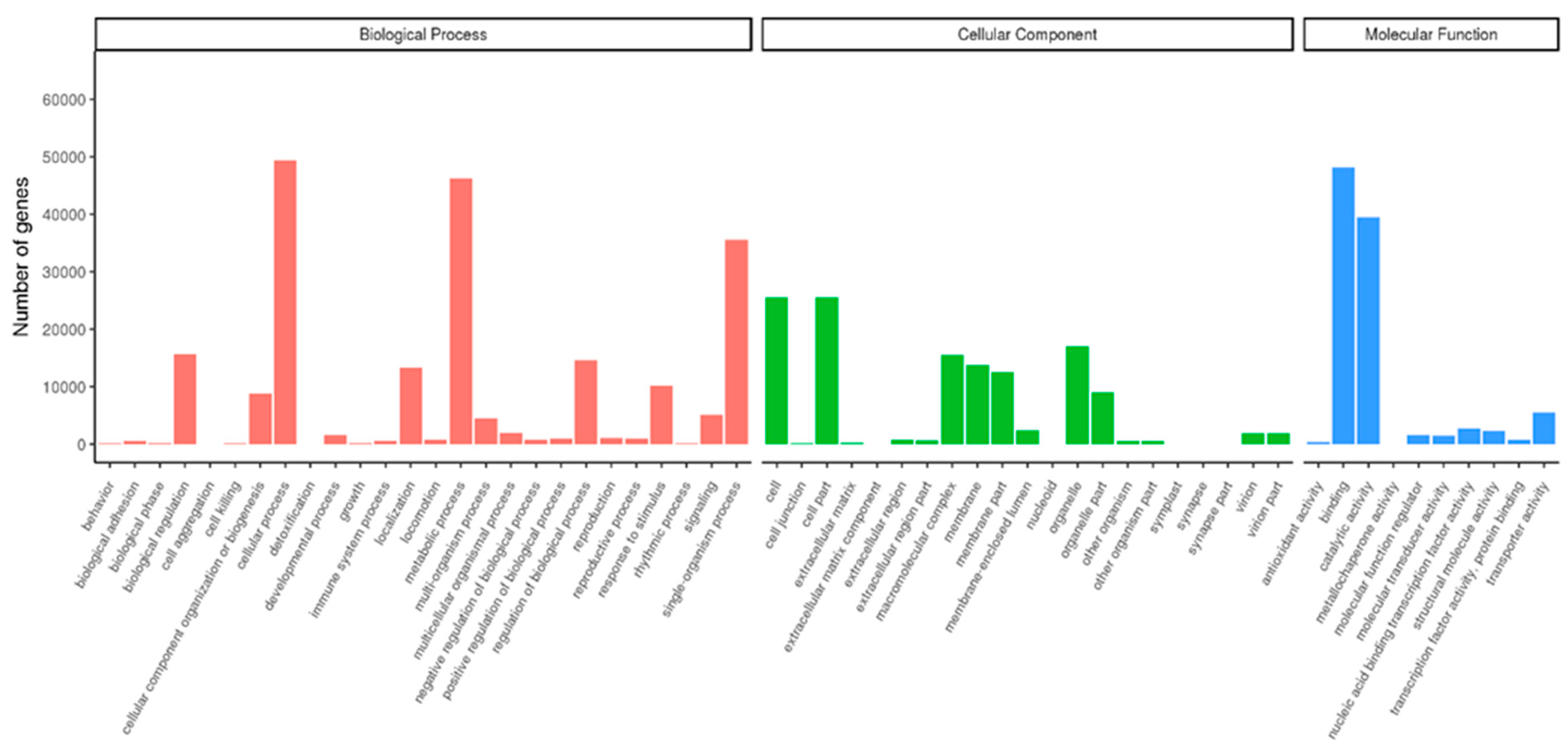
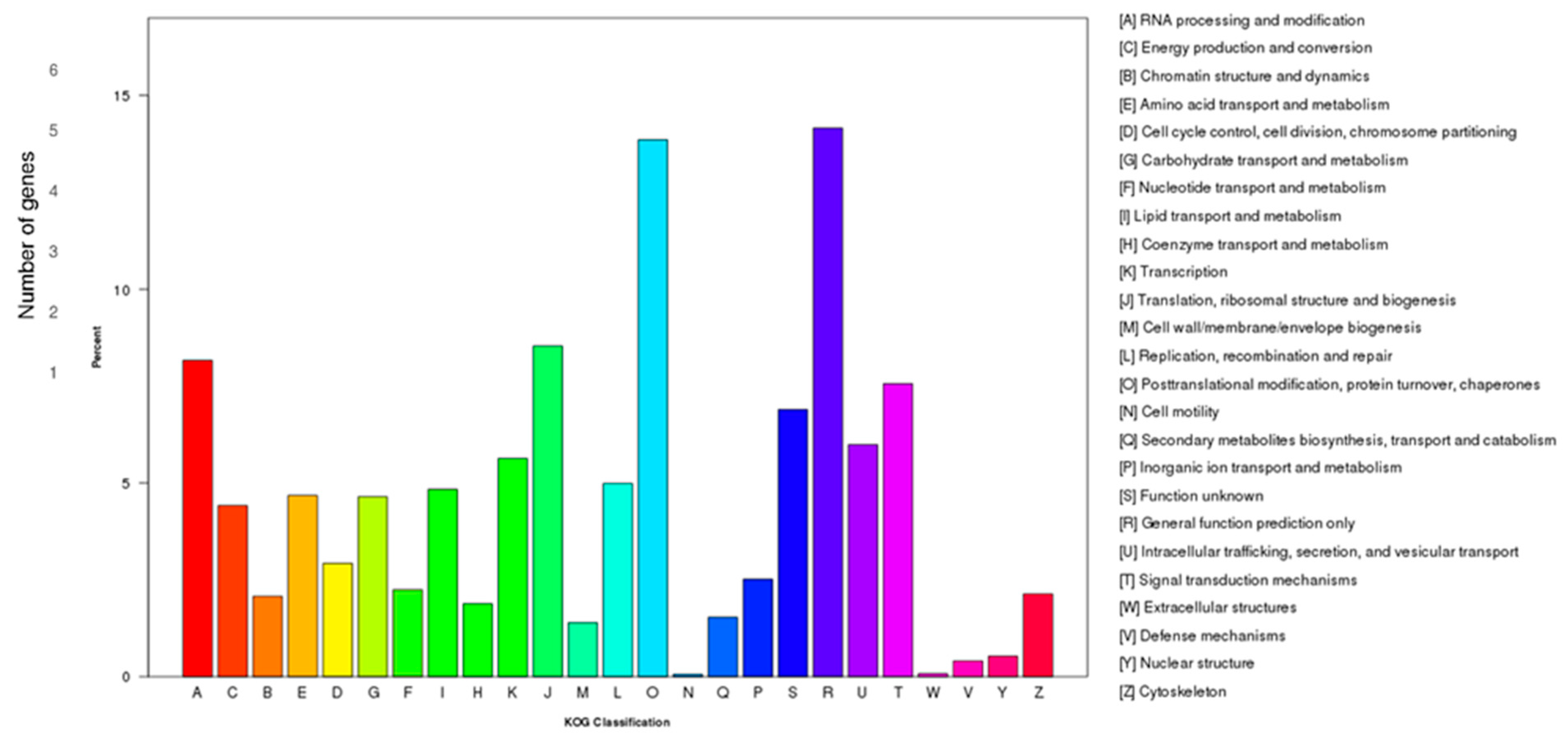
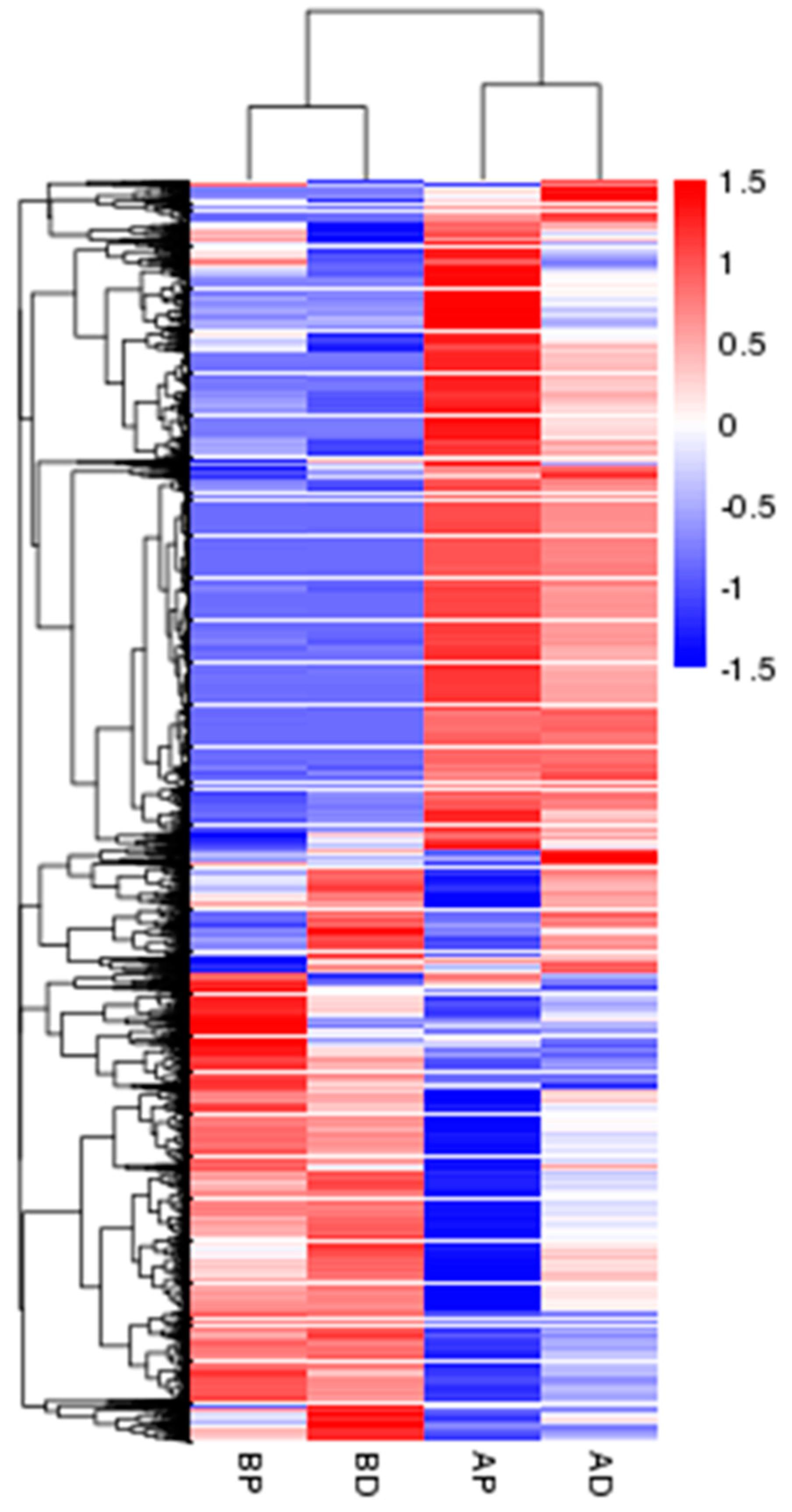
| Samples | Raw Reads | Clean Reads | Clean Bases (Gb) | Error/% | Q20/% | Q30/% | GC/% |
|---|---|---|---|---|---|---|---|
| AP | 56,991,166 | 55,610,478 | 8.34 | 0.02 | 98.07 | 94.4 | 45.98 |
| BP | 55,980,546 | 55,178,130 | 8.28 | 0.03 | 98.01 | 94.24 | 46.50 |
| AD | 60,575,150 | 59,332,712 | 8.90 | 0.02 | 98.04 | 94.31 | 46.45 |
| BD | 61,896,220 | 60,665,298 | 9.10 | 0.03 | 97.93 | 94.10 | 46.82 |
| Total | 235,443,082 | 230,786,618 | 34.62 | ||||
| Average | 98.01 | 94.26 | 46.44 |
| Transcript Length Interval | 200–500 bp | 500 bp–1 kbp | 1–2 kbp | >2 kbp | Total |
|---|---|---|---|---|---|
| Number of transcripts | 235,579 | 90,037 | 63,469 | 27,556 | 416,641 |
| Number of unigenes | 99,607 | 84,270 | 62,998 | 27,528 | 274,403 |
| Component | Number of Unigenes | Percentage (%) |
|---|---|---|
| Annotated in NR | 131,990 | 48.10 |
| Annotated in NT | 183,742 | 66.96 |
| Annotated in KEGG | 45,601 | 16.61 |
| Annotated in Swiss-Prot | 90,095 | 32.83 |
| Annotated in PFAM | 89,223 | 32.51 |
| Annotated in GO | 89,827 | 32.73 |
| Annotated in KOG | 29,056 | 10.58 |
| Annotated in all databases | 17,451 | 6.35 |
| Annotated in at least one database | 203,043 | 73.99 |
| Total unigenes | 274,403 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, P.L.; Le, T.T.C.; Vu, T.T.H.; Nguyen, T.T.; Zhang, Z.-S.; Zeng, R.-Z.; Xie, L.; Nguyen, M.N.; Trang, V.T.H.; Xuan, T.D.; et al. Comparative Transcriptome Analysis and Expression of Genes Associated with Polysaccharide Biosynthesis in Dendrobium officinale Diploid and Tetraploid Plants. Agronomy 2024, 14, 69. https://doi.org/10.3390/agronomy14010069
Pham PL, Le TTC, Vu TTH, Nguyen TT, Zhang Z-S, Zeng R-Z, Xie L, Nguyen MN, Trang VTH, Xuan TD, et al. Comparative Transcriptome Analysis and Expression of Genes Associated with Polysaccharide Biosynthesis in Dendrobium officinale Diploid and Tetraploid Plants. Agronomy. 2024; 14(1):69. https://doi.org/10.3390/agronomy14010069
Chicago/Turabian StylePham, Phu Long, Thi Tuyet Cham Le, Thi Thuy Hang Vu, Thanh Tuan Nguyen, Zhi-Sheng Zhang, Rui-Zhen Zeng, Li Xie, Minh Ngoc Nguyen, Vuong Thi Huyen Trang, Tran Dang Xuan, and et al. 2024. "Comparative Transcriptome Analysis and Expression of Genes Associated with Polysaccharide Biosynthesis in Dendrobium officinale Diploid and Tetraploid Plants" Agronomy 14, no. 1: 69. https://doi.org/10.3390/agronomy14010069
APA StylePham, P. L., Le, T. T. C., Vu, T. T. H., Nguyen, T. T., Zhang, Z.-S., Zeng, R.-Z., Xie, L., Nguyen, M. N., Trang, V. T. H., Xuan, T. D., & Dang Khanh, T. (2024). Comparative Transcriptome Analysis and Expression of Genes Associated with Polysaccharide Biosynthesis in Dendrobium officinale Diploid and Tetraploid Plants. Agronomy, 14(1), 69. https://doi.org/10.3390/agronomy14010069







