Abstract
The formation of biogenic selenium nanoparticles (SeNPs) through microbial activities is a promising technique that can contribute to the development of reliable, non-toxic and environmentally friendly synthesis methods. Among these, under optimal conditions, myconanotechnology confers particular characteristics due to the generation of bioactive fungal metabolites with various bioactivities. The formed SeNPs are known to be stabilized by the biomolecules of the microorganism, forming a so-called bio-corona or capping structure. The composition of this bio-corona greatly impacts the SeNPs activity, but investigations have been limited to date. The SeNPs produced by Trichoderma sp. have potential applications in crops and environmental management, as both selenium and Trichoderma are known to benefit cultivated plants and phytoremediation. This review summarizes the biosynthesis of SeNPs by Trichoderma sp. and contextualizes the possible correlations between SeNPs and biomolecules produced by Trichoderma; it also provides a missing analysis that could help understand and optimize this process. Biosynthesis methods and probable mechanisms are briefly discussed as well as the role and applications of trichogenic SeNPs as plant protectants, plant biostimulants, and safe biofortifying agents. The knowledge gaps related to mechanisms of trichogenic SeNPs biosynthesis, the control of the desired characteristics for a specific agricultural function, and technology scale-up are discussed in connection with the needed future research directions.
1. Introduction
Selenium (Se) is an essential microelement for organisms from several phyla, wherein there was an evolutionary change from sulfur to selenium (i.e., cysteine with selenocysteine) as the active site of several proteins [1,2], which was associated with selenocysteine (Sec) incorporation machinery during protein synthesis [3,4]. Such replacement generates better redox activity and more resistance to permanent oxidation [2,5].
Due to its biological characteristics (resulting from its physical and chemical properties), Se is regarded as an element of great potential in various scientific fields, such as biomedical, food, agricultural, and environmental [6,7,8,9]. Selenium has not been classified as an essential element for terrestrial plants [10,11] or fungi [12,13,14]. Selenium is an essential toxin [15]. Selenium deficiency is linked to several human chronic diseases [16,17], including Alzheimer disease [18,19], as well as animal diseases [20]. Se deficiency was linked to mass extinctions in Phanerozoic oceans [21]. Selenium excess affects ecosystem functions, including animals from the top of food chains, due to bioaccumulation and biomagnification [22,23].
In areas with selenium deficiencies in the soil, selenium was applied to cultivated plants to increase Se levels in the food chain via biofortification [8,24,25,26]. Biofortification is considered to produce a better effect on human and animal health than direct supplementation, especially in the case of selenium, which is well known to have a narrow physiological window [16,27,28]. Biofortification treatments on cultivated plants produced effects similar to biostimulants [29,30]. Several effects similar to plant biostimulants were confirmed in the last decade: (i) enhancement of nutrients uptake and nutrients use efficiency [31], especially of nitrogen [32] and essential micronutrients [33,34]; (ii) increased tolerance to abiotics [35,36,37], including potentially toxic elements [38,39,40] and biotic stress [41,42,43]; (iii) and improved crop quality traits [44,45,46,47,48,49].
In addition to these effects, similar to plant biostimulants, Se application on cultivated plants increases photosynthesis and enhances seed germination [50,51]. Due to its multiple effects on plants, it was recently proposed that selenium should be considered essential for terrestrial plants [50].
One of the selenium biotechnological applications is the biosynthesis of selenium-enriched yeast as Se dietary supplements [52], although Saccharomyces cerevisiae does not have the Sec incorporation machinery [53]. Yeasts accumulate organic and inorganic selenium and detoxify it through various mechanisms [54], including by forming less toxic Se nanoparticles (SeNPs) [55,56].
Nanoparticles (NPs) are believed to have superior reactive surfaces due to their high area-to-volume ratio [57,58,59]. Nanoformulations can enhance absorption, sensitivity, stability and resistance [60,61]. NPs are also highly biocompatible and bioavailable [62]. In addition to the aforementioned valuable features, selenium nanoparticles (SeNPs) have lower toxicity compared to other forms of Se due to the zero-valent state of oxidation of nano-selenium [63].
SeNPs have applications in various fields. Comprehensive reviews were published on biomedical [6,64,65], agronomical [51,66,67], and environmental applications [57,68] of SeNPs. Selenium nanoparticles were used for various (bio)sensing devices [69,70], including wearables electronics [71].
The most common means of obtaining SeNPs have been chemical methods. These approaches are based on reducing selenium using reducing agents. The reactions in aqueous media often occur in the presence of certain emulsifiers or surfactants, which create stable colloidal systems that will finally contain the SeNPs. Examples of such stabilizing agents are quaternary ammonium compounds [72], polysaccharides, arabic gum [73], chondroitin sulfate [74], chitosan [75,76], carrageenan [77], lignosulfonate [78]; proteins/peptides, zein [79], beta-lactoglobulin [80], chymotrypsin [81], peanut meal peptides [82], tilapia peptides [83], and polyvinyl alcohol [84]. Physical methods of SeNPs synthesis mainly involve the exposure of selenium precursors to different radiation sources. In this regard, photoablation or different non-ionizing radiations (UV radiation) or ionizing radiation (gamma radiations) are used to target selenium compounds, finally leading to SeNPs formation. Apart from being expensive and time-consuming, the harsh conditions of the aforementioned techniques can create a toxic processing environment as well as non-sustainable waste and can interfere in the use of SeNPs in biological systems [64,85,86]. Biological synthesis methods are considered safe, eco-friendly, cost-effective, non-toxic and waste minimizing [87,88,89].
Biogenic selenium nanoparticles are synthesized using microorganisms or plants. SeNP biosynthesis can occur either intracellularly or extracellularly. Specific biomolecules of the physiological apparatus of microorganisms can carry out reducing and stabilizing functions, which are vital for SeNPs production [90]. As they provide just the right conditions for this process, microorganisms acquired a reputation as nano-factories. Their great potential also lies in the possibility of modifying their cellular machinery to facilitate SeNP synthesis [91].
Fungi are thought to be among the most efficient microorganisms regarding biogenic NP synthesis [92]. In addition to their capacity to biosynthesize high quantities of NPs, the stabilizing enzymes and metabolites, fungi are manageable microorganisms with accelerated growth rates. An interesting and resourceful trait of fungi is their ability to tolerate metals, even in high concentrations [93]. Compared to bacteria, fungi are almost ideal biocatalysts for the biosynthesis of NPs, since they are known to produce higher levels of bioactive compounds, making them more suitable for large-scale production [94].
Nanoparticles synthesized by fungi have been successfully studied in a wide range of research areas like agroecosystems, plant science, and eco-friendly formulations with protective potential for agricultural crops. Abd-Elsalam (2022) predicts that large-scale myconanoparticles strategies in agriculture will be increasingly introduced in the upcoming years due to the continuous reports of their newly discovered uses in agri-food fields over the last few years [95].
All things considered, the fungal-mediated synthesis of SeNPs is a resourceful research direction. The main principle of this biosynthesis relies on using fungal biomass as a host for the reduction of Se precursors such as selenate (SeVIO42−) and selenite (SeIVO32−) oxyanions to the less toxic Se0. SeNPs synthesis using fungal biomass has been successfully performed using various strains: Gliocladium roseum [96], Aspergillus terreus [97], Alternaria alternata [98], Aspergillus oryzae [99], Penicillium chrysogenum [100], and Trichoderma sp., the latter being detailed below.
The last few years have witnessed an unprecedented interest in SeNPs and its applications—especially biogenic ones. Several reviews that approach this subject are available [48,51,66,88,101,102,103], including special reviews on the applications of SeNPs in agriculture [51,102,103]. Therefore, the purpose of this work is not an in-depth review of the general aspects of SeNPs but rather a critical and focused analysis of the probably most relevant type of SeNPs for agriculture, the so-called trichogenic SeNPs., i.e., SeNPs produced by Trichoderma sp.
Trichoderma (Hypocrea) sp. is a specific species of interest for the production of SeNPs. Trichogenic SeNPs have a wide variety of potential applications, especially in agriculture, as the benefits of Trichoderma sp. uses on plants are well known. Trichoderma colonizes wood and herbaceous plant materials, showing a high level of genetic diversity. The adaptability of Trichoderma sp. to various substrates and its tolerance to toxic compounds (e.g., from fungicides, herbicides or pollutants) make these fungi excellent soil biocontrol agents [104]. Therefore, using this fungus for SeNP synthesis could potentiate its properties and those of the SeNPs themselves, leading to novel products of commercial and ecological interest [105].
Despite the significant importance of Trichoderma sp. and the SeNPs it can produce, a thorough review focusing on trichogenic SeNPs is still necessary. The current reviews either present various nanoparticles and nanomaterials produced by Trichoderma sp., with a relatively brief mention of trichogenic SeNPs, or focus on the general fungal biosynthesis of SeNPs. Moreover, probably one of the most critical aspects for agriculture applications, i.e., the nature of the capping molecules forming the so-called bio-corona of trichogenic SeNPs, has not been emphasized enough. Therefore, we considered it necessary to bring the main aspects of the trichogenic biosynthesis of SeNPs to the forefront and place them within a wider context. In particular, we address the following issues important for understanding trichogenic SeNPs and for future developments: different types of methods for obtaining trichogenic SeNPs, the putative mechanism of trichogenic SeNPs biosynthesis based on the information from other organisms, methods that were/could be used for trichogenic SeNP characterization and manipulation, aspects on the ecological safety and environmental impact that should be investigated, applications in the agri-food sectors, and future research directions.
2. Methods of Trichogenic SeNPs Biosynthesis
Trichoderma-derived SeNPs are currently in the developmental stage. Several Trichoderma species have been employed in nanotechnology to obtain NPs of metallic origin: titanium, gold, zinc, silver, etc., as recently reviewed in [106]. In particular, some strains have substantial potential in the biosynthesis of nanoparticles at an industrial level [94,107].
There is limited study on the Trichoderma sp.-mediated synthesis of SeNPs. The biosynthesis methods are influenced by the fungal strain, the selenium precursor concentration, and the culture medium where it grows and develops the metabolites important for nanoparticles formation. For the production of mycelial biomass involved in biosynthesis, the most common growth media used in studies for Trichoderma strains were Czapek–Dox agar (CDA) [108], potato dextrose broth (PDB) [87,109,110], AP1 agar supplemented with nutrients [111], and Martin-modified broth (MMB) medium [93,112].
The first report describing the biosynthesis methodology based on Trichoderma sp. along with other filamentous fungi (Aspergillus funiculosus, Aspergillus niger, Coriolus versicolor, Rhizopus arrhizus etc.) and yeasts (Saccharomyces cerevisiae, Candida glabrata, etc.) was by Gharieb et al. (1994) [108]. The study was based on understanding the capacity of strains to reduce selenite to Se0. Based on this screening, the authors demonstrated that T. reesei can reduce selenite to elemental selenium. The first hint of the reduction is the change in the red color of the growth medium (due to the presence of amorphous elemental selenium).
Later, more strains of Trichoderma were tested together with other methodologies of SeNPs biosynthesis, i.e., in vitro, by using cell lysate, culture filtrate and/or cell walls. Nandini et al. (2017) studied downy mildew control in pearl millet, synthetizing SeNPs by way of T. asperellum, T. harzianum, T. atroviride, T. virens, T. longibrachiatum and T. brevicompactum strains, using three forms of fungal culture (culture filtrate, cell lysate, and crude cell wall) [109]. A similar method was performed on T. atroviride [113]. In another study, eight Trichoderma strains were investigated to obtain SeNPs with bioactive metabolite support with applications against phytopathogens and mycotoxins [110]. Trichoderma sp. WL-Go was subjected to several conditions to identify the optimal ones [112]. T. harzianum appears to be the most studied strain in SeNPs formation, which was identified as a strain with auspicious biosynthesis potential [110,111,114].
Considering the previous studies, the biogenic synthesis based on Trichoderma sp. has several benefits in terms of efficiency and the generation of diverse metabolites under optimal surroundings [92]. The inoculation conditions serve as key points right from the beginning of the experiment. As a first step, Trichoderma sp. needs to be inoculated on a solid growth medium. Potato dextrose agar (PDA) is the classic culture medium in fungal isolation and culture, and it is also valid for Trichoderma sp. [109,110]. The malt extract agar (MEA) is used as well [111]. The development of the mycelial mass in the liquid growth medium takes place in the dark under static [109,113] or under stirring conditions [87,111]. The growth phase of microorganisms is an important step [90].
Most of the studies investigating trichogenic SeNPs used culture filtrate (extracellular content), lysate (intracellular content), and/or cell walls from Trichoderma sp. to perform in vitro bio-assisted SeNPs synthesis. To collect the metabolites for the in vitro bio-assisted synthesis of trichogenic SeNPs, a mycelial mat is subjected to several processes such as ground, lysis through sonication, and centrifugation. Then, an aqueous solution of Se precursor is added. Sodium selenite (Na2SeO3), a well-known bioactive chemical, has commonly been used as a precursor in trichogenic SeNPs in vitro and in vivo biosynthesis, but its concentration varies from study to study [87,108,109,110,111,113,114]. The reaction mixture is usually kept in the dark until a red sediment is present, because light induces Se reduction and could induce the formation of other Se forms [87]. Nevertheless, light is a parameter that needs to be tested with respect to the biological effects of the trichogenic SeNPs. A previous study on SeNPs produced by a culture filtrate of Penicillium crustosum reported enhanced antimicrobial, anticancer, and catalytic activity in the presence of light [93].
In vivo biosynthesis, reported in fewer studies than in vitro biosynthesis, can result in either intracellular, extracellular or both types of SeNPs, the extracellular ones either by exporting them from the cells or by direct extracellular SeNPs formation [108,111,112]. A brief presentation of the biosynthesis methods in relation to the obtained SeNPs is presented in Table 1.

Table 1.
Parameters of the biosynthesis of trichogenic selenium nanoparticles.
There is not much information about the yield and efficiency of SeNPs biosynthesized by Trichoderma. Quantitative data are available for one study that used in vitro bio-assisted synthesis [113] and two studies featuring in vivo biosynthesis [111,112]. From the data provided by Joshi et al. [113], we calculated a 19% approx. yield of SeNPs produced by the metabolites of a T. atroviride strain, which could be in fact lower, as the quantification of the resulting SeNPs was performed by weighing the resulting nanoparticles without accounting for the bio-corona. Liang et al. determined by ICP-MS a higher apparent trichogenic SeNPs yield, 42%, by in vivo biosynthesis using a T. harzianum strain and sodium selenite [111], and Diko et al. obtained an even higher yield, 84.73% after supernatant filtration, using an isolated, unidentified Trichoderma sp. WL-Go strain and 2 mM SeO2. It is not clear yet what parameters influence the yield mostly, but it is possible that besides the specificity of each Trichoderma strain, the in vivo biosynthesis might be more efficient than the in vitro one.
Understanding the parameters influencing the yield and quality of trichogenic SeNPs biosynthesis would help optimize these outputs. For example, pH seems to play an important role regarding some aspects of trichogenic SeNPs. Diko et al. concluded that the pH is a critical parameter for trichogenic SeNPs synthesis [112]. It is assumed that the alkaline medium prevents the agglomeration of nanoparticles and promotes the stabilization of capping agents from fungi on the NP surfaces, such as proteins and other biomolecules, forming the so-called “bio-corona” [93]. The nature of capping biomolecules has a high impact on the stability of SeNPs with respect to aggregation. An alkaline pH induced an increase in the permeability of fungal cell membrane, which resulted in extracellular SeNPs [90]. The optimum pH for the extracellular synthesis of SeNPs from Trichoderma sp. was found to be 8 when performed in vivo [112] and was reported in vitro at pH 8–12 in one study [87]. Both in vitro and in vivo biosynthesis of fungal SeNPs, including trichogenic SeNPs, seem to have advantages and disadvantages. In vitro biosynthesis enables an easier manipulation of SeNPs by avoiding the necessity of mycelium lysis for their recovery, which is helpful as lysis could result in alterations to their properties. In vivo biosynthesis enables generating specific metabolites and other biomolecules triggered by the interaction with Se. These metabolites might have superior biological properties to the metabolites produced in the absence of Se, and these properties would be transferred to SeNPs as well if these metabolites are part of the capping bio-corona. In-depth studies comparing the two types of biosynthesis with the same Trichoderma strains would give more information in this respect.
3. Putative Mechanism of Trichogenic SeNPs Biosynthesis
The exact mechanism of SeNPs formation by Trichoderma sp. has not been investigated, but some clues can be obtained from the available studies of other nanoparticles and/or fungal and other microorganism species. Most of the studies reported that the synthesis of NPs in fungi might be a defense mechanism for reducing the toxicity of different elements that they encounter. It is already recognized that fungi have great tolerance to metal and non-metal ions [63,101,116]. One of the fungi coping mechanisms for this kind of abiotic stress involves their ability to reduce the metal/non-metal ions to lower the states of oxidation and, implicitly, synthesize nanoparticles which are usually less toxic (e.g., reducing Se4+ and Se6+ from selenite (SeO32−) and, respectively, selenate (SeO42−) to zero valent selenium—creating SeNPs). This process could be viewed as a detoxifying mechanism [116,117,118].
Although the accumulation of nanoparticles can cause physiological changes in microorganisms, fungi can tolerate impressive amounts of NP accumulation. Therefore, fungi continue to develop even after the biosynthesis of nanoparticles [101,119]. The synthesis of NPs in fungal biomass can occur inside the cytoplasm (absorption), within the periplasm or outside the cell membrane (adsorption). The intracellular synthesis of NP avoids the formation of aggregated, clustered, and large-sized nanoparticles. In comparison to intracellular processes, extracellular synthesis offers the advantage of obtaining large quantities of NPs in a relatively pure state, free from other cellular fungal biomolecules or microbial cells, thus making the downstream process easier [120]. Additionally, fungi release extracellular reductive proteins that can be employed in later stages of the process [94,113]—Figure 1.
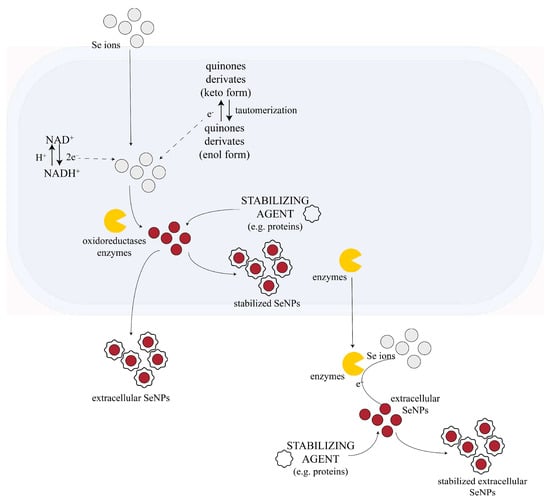
Figure 1.
Possible mechanism of fungal-mediated SeNPs synthesis. NADPH-dependent oxidoreductases mediate the reduction of selenium ions from precursors to SeNPs along with quinone derivates, which can be electron shuttles in the oxidoreduction process. Capping agents (e.g., proteins from fungal biomass) can surround the SeNPs, acting as NP stabilizers. This process can occur either intracellularly (after the absorption of the Se ions into the cell) or extracellularly. Some information about membrane transport processes and other metabolites involved in the fungal synthesis of NP are not fully understood yet. The figure is adapted and based on the information from [116,121].
Although exceedingly researched, these superpowers of fungi have not been fully explained yet; the complete mechanism of fungi-mediated NPs, and especially SeNPs synthesis, remain unknown. However, studies suggest that the NADPH-dependent reductases (nitrate (NO3−) and nitrite (NO2−) reductases) from the cell wall or plasmatic membrane of the cell play a major role in the fungal biosynthesis of some NPs, probably including the trichogenic synthesis of SeNPs. These reductases are thought to be catalysts for the reaction between the 2e− donor—NADPH—and inorganic ions [101,116,119,122,123,124]. However, Li et al. (2012) showed that NADPH alone is not enough to form these kinds of nanoparticles [124]. Their experiments demonstrate that fungal NP biosynthesis is not possible using just NADPH and the NP precursor alone. Moreover, when fungal biomass was added to NADPH and the NP precursor, NPs were formed. Therefore, other molecules besides NADPH and oxidoreductases are necessary for nanoparticle synthesis by fungi [124]. For example, in selenite reduction, biomolecules like phenazine-1-carboxylic acid and glutathione might be involved in selenite reduction [85]. Some research revealed that there are certain fungal metabolites that contribute to biogenic NP formation. Compounds like penitric acid, chrysogine, chrysogenin, fungisporin, roquefortines or non-enzyme proteins like phytochelatins and methalothenin are believed to play a role in the process. In addition to their reduction properties, most of the molecules involved in the fungal-mediated biosynthesis of NPs act also as capping agents that stabilize the NPs [101,111,116,119,123]. Other hypotheses propose that some of these metabolites might be the quinine derivates of anthraquinones and naphthoquinones [111], but this hypothesis requires experimental confirmation. These molecules act as electron shuttles. Quinone shuttles were shown to be necessary for NP production in some cases, as they might act as redox centers and/or electron carriers in the oxidoreduction reactions. There are studies that confirm that the quinones of T. harzianum have great reducing properties [125]. Liang et al. reported for the first time in 2019 the formation of not only elemental Se but also Se oxide by the interaction of sodium selenite and selenate with a T. harzianum strain, which indicates a more complex system than generally assumed [111]. The mechanism generated in vivo could be completely differently than the one in vitro using lysate or filtrate, as most probably Trichoderma produces additional and specific metabolites when in contact with toxic concentrations of Se salts. Unfortunately, mechanistic insights into the in vivo biosynthesis of SeNPs by Trichoderma and even fungi in general, at the molecular level, are not available yet, and it represents a vast unexplored field. In contrast to fungi, the biosynthesis of SeNPs by bacteria has been mechanistically studied more in depth, as reviewed by Tugarova et al. [126]. In the case of bacteria, there were two obligatory stages established, which are involved in both intracellular and extracellular biosynthesis: (1) the redox reaction and (2) the assembly of Se0 to form SeNPs. Two other stages are involved in the case of in vivo intracellular biosynthesis: (3) the transport of Se oxyanions (in)to the cell and (4) the export of Se0 nuclei and SeNPs to the extracellular environment. These four stages could be extrapolated to intracellular biosynthesis by yeast and fungi, including Trichoderma as well. Another common mechanism between bacteria and fungi could be related to the transporter responsible for Se oxyanions accumulation in the cell. In the case of bacteria, no specific selenate or selenite uptake system has been identified. Selenate, which is structurally similar to sulfate, can be transported via sulfate permeases and other non-specific transport systems, such as the sulfate transport complex ABC (ATP-binding cassette). This has been shown for yeast as well, as reviewed by Kieliszek et al. [54]. A similar route of sulfate permease was proposed to function in filamentous fungi such as Neurospora crassa and Hypocrea jecorina (anamorph T. reesei) [127]. Moreover, the response of H. jecorina to selenate depends on the C source (inducer or not of cellulase) and light/dark conditions. The response to selenium toxicity was proposed to involve the regulatory machinery of sulfur metabolism, SCF–ubiquitin–ligase complex SCF(Met30) and transcription factor Met4, as in the case of cadmium and other heavy metals [127], but the experimental proof is lacking. In the case of bacteria and yeast, there seems to be at least two types of transport systems, a low- and high-affinity/efficiency, respectively, whereas much less is known in the case of filamentous fungi. Electrostatic interactions are believed to participate as well in fungal NP synthesis. The negatively charged cellular membrane surface interacts electrostatically with the inorganic ions, mediating the transportation of ions throughout the cell membrane and promoting the complexation of ions with ligands [116,121]. In the case of selenium, these types of interactions are less likely to occur, as the selenite and selenate ions are negatively charged.
Regarding the redox reaction, several mechanisms have been proposed for bacteria, including nitrate/nitrite or even selenate/selenite reductases; thiol groups of peptides/proteins—glutathione, glutathione reductase, thioredoxin, and thioredoxin reductase; chaperons; translocases; elongation factors; and oxidoreductases [126]. If and how these pieces fit together in a puzzle or whether they are independent pathways is not known. As described above, some of these mechanisms were proposed also for fungi.
Recently, Xu et al. found 1075 differentially expressed proteins of the macromyceta Ganoderma lucidum in the presence of 200 ppm Na2SeO3 compared to the control without selenite, with 601 and 474 upregulated and downregulated proteins, respectively [128]. Most of these proteins were located in the cytoplasm, mitochondria and nucleus, with some in the plasma membrane. Most of the molecular functions of these proteins involved oxidoreductase and transmembrane transporter activity. It is worth mentioning that ABC transporters were among the upregulated proteins by selenite salt both in G. lucidum and in Pleorotus citrinopileatus reported in another study [129], which indicates similarities with bacteria. Some of the regulated genes are related to the response of the microorganism to the oxidative stress induced by the high concentration of selenium salt, which generates reactive oxygen species (ROS). These include genes coding for proteins involved in xenobiotic metabolism and defense metabolism, such as antioxidant enzymes. Enzymes involved in producing various forms of reduced, less toxic forms of selenium (Se0, methylated and volatile Se compounds, organic thiol–selenium compounds) were upregulated. Among these, thioredoxin, NADPH-dependent thioredoxin reductase, and glutathione reductase are probably involved in SeNPs formation and feature common players and pathways between bacteria and macromycetes. Other enzymes upregulated by selenite, including alcohol dehydrogenase and propanol-preferring enzymes (AdhP), have been proposed to be involved in controlling the size of SeNPs produced by bacteria [130]. In addition, genes coding for proteins involved in the phospholipid metabolism, chitinase, and cytoskeletal proteins were regulated as well, indicating involvement in SeNPs transport within and outside the cells. Considering the similarities between the studied macromycetes and bacteria, some of these players and mechanisms are probably common to Trichoderma as well, but future experimental reports alone will be able to confirm this.
Clearly, there are more pieces that need to be put together in this metabolic puzzle of fungal-mediated NPs synthesis, and current research on the topic will continue to provide answers to questions regarding this process.
4. Characterization and Manipulation Issues of SeNPs
SeNPs, formed from zero-valent selenium, are lyophobic colloids that tend to aggregate in aqueous systems without stabilizing agents [131]. As we mentioned in the case of chemically synthesized SeNPs, various emulsifiers or surfactants were used to make these nanostructures thermodynamically stable. Hydrophobic and electrostatic interactions stabilized the SeNPs in the aqueous systems [82,83]. Different stabilizers determine different biological and physicochemical characteristics. SeNPs stabilized by chitosan (CS-SeNPs) had higher storage stability but tended to aggregate at higher ionic strength or alkaline pH. SeNPs stabilized with carrageenan (Cg-SeNPs) were less cytotoxic than CS-SeNPs. Gum arabic-stabilized SeNPs demonstrated a higher thermal stability due to the branched structure of gum arabic biopolymers [132].
The chemically synthesized SeNPs form a protein bio-corona in contact with the biological systems. SeNPs stabilized with different surfactants, such as sodium dodecyl sulfate (SDS), cetyltrimethylammonium bromide (CTAB), and brij-58, form bio-coronas with serum proteins [133]. The formation of a protein bio-corona is driven mainly by electrostatic forces. Two types of bio-corona were highlighted—a “soft” bio-corona, loosely bound, that undergoes changes, and a “hard” bio-corona firmly bound to SeNPs surfaces [133].
The biogenic SeNPs are stabilized by the proteins and the metabolites from the biological (based) system, which featured a significant contribution of antioxidant groups, e.g., thiols, to selenium oxyanion bio-reduction and stabilization [134]. The biomacromolecules prevent zero-selenium crystallization and maintain a shell of amorphous selenium, which is bioavailable for conversion into seleno-amino acids [135].
A precise description of SeNP characteristics should define their properties in correlations with the applications. The shape, size, stability and chemical structure of the bio-corona are some of the parameters that were frequently reported in studies of SeNPs. These NP characteristics are often analyzed in correlation with their stabilizing biomolecules, which are mainly proteins and amino acids [93,109,126,136,137] or proteins and polysaccharides [110,112,138]. The main techniques used for NP characterization are inductively coupled plasma–mass spectrometry (ICP-MS), inductively coupled plasma–optical emission spectrometry (ICP-OES) and colorimetric methods for quantification, single-particle mode ICP-MS (SP-ICP-MS), UV-Vis spectroscopy, transmission electron microscopy (TEM), scanning electron microscopy (SEM), X-ray fluorescence analysis (XFA), electron energy-loss spectroscopy (EELS) for selenium detection, X-ray absorption (XANES and EXAFS) for oxidation state, X-ray diffraction (XRD) in small and wide-angle scattering (SAXS and WAXS), X-ray photoelectron spectroscopy (XPS), dynamic light scattering (DLS) and zeta potential, analytical centrifugation methods such as differential centrifugal sedimentation (DCS) [139], isothermal titration calorimetry, surface tension and contact angle (which can provide information on the hydrophobicity/hydrophilicity of the system and the interaction of SeNPs with the solvent), Fourier-transform infrared (FTIR) and Raman spectroscopy, sodium dodecyl sulfate–polyacrylamide gel (SDS-PAGE) electrophoresis for the identification of proteins capping the NPs, and mass spectrometry coupled with chromatography for the identification of biomolecules from bio-corona. Another useful technique that can separate nanoparticles based on size is asymmetric flow-field flow fractionation (AF4) coupled with various detectors such as ICP-MS, multi-angle light scattering (MALS), or refractive index detector (RID) [56,140], but this technique is less reported in the literature, at least concerning SeNPs. ICP-MS and SP-ICP-MS, along with other techniques, are powerful instruments for the Se speciation of both inorganic and organic forms of Se [48]. Other particular methods can be found in recent reviews [56,141].
One of the most suggestive properties of SeNPs is their color. The post-incubation changes of color in the culture medium to orange or red can be used to confirm the SeNPs formation [85,93]. The intensity of the color gradually increases with time, as the selenium precursor is being reduced, from colorless to ruby red color [93]. The red color is characteristic of other nanoparticles as well, such as AuNP, AgNPs and CuNPs. The nanoparticles adsorb light in the violet, blue, and green wavelengths (200–550 nm) and scatter the complementary yellow, orange, and red wavelengths through the surface plasmon resonance (SPR) phenomenon [142,143,144,145,146], meaning the collective oscillations of nanoparticle conduction electrons excited by the optical electric field [143,147,148]. The color change upon the formation of SeNPs is a helpful indication, as an intense red color can correspond to a high potential of the fungal strain to produce this kind of NP. Color change can also evidence the end of the reduction reaction when no further color change is observed: for example after 24 h incubation at room temperature of Na2SeO3 in the presence of active metabolites secreted by the endophytic fungal strain Penicillium crustosum EP-1 [93]. In a 72 h color variation study of SeNPs trichogenesis using the crude cell wall (CCW), cell lysate (CL), and culture filtrate (CF) of T. asperellum, the main color was achieved after 24 h, which was orange-red for SeNPs with CF, dark yellow for SeNPs with CL and dark brown for SeNPs with CCW. The red SeNPs with CF showed the highest anti-mildew activity, with the minimum inhibitory concentration of 150 ppm, compared with 350 ppm for SeNPs with CL and 500 ppm for SeNPs with CCW [109]. Similarly, after 24 h of Na2SeO3 incubation at 23 ± 2 °C with culture lysate (CL), cell wall debris (CW), and culture filtrate (CF) from Trichoderma atroviride (Tri_AtJSB2) sp., the reduction was considered completed: the most intense brick-red color was obtained with CL, which was followed by orange SeNPs reduced with CW and pale yellow SeNPs reduced with CF [113]. In the mentioned study, although the SeNPs reduced with CF were the least red colored, they were arbitrary chosen for further characterization using FTIR, XRD, SEM, TEM, UV-Vis, XPS and antifungal properties, whereas only the DLS size distribution and zeta potential were used to compare the SeNPs reduced with CL, CW and CF [113]. Considering only the color aspect of reduced SeNPs in the mentioned study [113], the T. atroviride culture lysate (CL) might have the highest content of reducing agents that led to red SeNPs suspension, which was followed by orange SeNPs reduced by cell wall debris (CW) and pale yellow SeNPs from culture filtrate (CF). The intensity of the color can be further quantified by UV-Vis spectrophotometric analysis. The UV-Vis spectra of trichogenic SeNPs evidenced that the wavelengths at which these kinds of NP absorb light vary greatly, depending on the Trichoderma strain, and on the nano/microparticles sizes. Previous analysis indicated absorption peaks around 280 nm for SeNPs reduced with six Trichoderma species (T. asperellum, T. harzianum, T. atroviride, T. virens, T. longibrachiatum and T. brevicompactum) [109], at 259 nm for SeNPs reduced with an unknown Trichoderma species [87], at 260 nm for SeNPs reduced with T. atroviride [113] or at 270 nm for SeNPs reduced with T. harzianum [114]. The wavelength showing the maximum absorption can be used for a rough estimation of the size of nanoparticles based on the localized surface plasmon resonance (LSPR) peak, smaller nanoparticles having the maximum absorbance at lower wavelengths [141,142]. The quantification of SeNPs can be obtained by an assay which uses Na2S for the dissolution of SeNPs [149].
Transmission electron microscopy (TEM) coupled with energy-dispersive X-ray spectroscopy (EDS) is one of the most popular methods for observing the morphology and composition of biogenic SeNPs, along with scanning electron microscopy (SEM). These microscopy techniques offer information about the shape, size and intracellular or extracellular distribution of SeNPs. Notwithstanding, some studies point out that an optimum imaging can be achieved especially when the samples that are being subjected to this assay are highly diluted [94]. The surface morphology and size of SeNPs from Trichoderma sp. were observed as hexagonal, spherical or pseudo-spherical and irregular shapes [87,109,112,113]. The size range of trichogenic nanoparticles observed through TEM is rather broad, depending on the fungal strain. Overall, studies reported measurements from 26.45 nm (T. harzianum) to 312.5 nm (T. virens) [109,110,112,113,114]. In addition to TEM analysis, an EDS spectrum can offer information about the degree of purity of the biosynthesized SeNPs [150].
The scattering phenomenon of X-rays by sample electrons at specific diffraction angles is a frequently employed method to determine the crystallographic particularities of biogenic nanoparticles. Depending on the X-ray technique, different morphological particularities can be deduced, like crystal lattice, crystallinity, and the nanoparticles’ size distribution and shape. Considering Bragg’s law, the wide-angle X-ray scattering (WAXS) offers information about the scattering angles induced by sub-nanometric structures (0.1–1 nm), and the small-angle X-ray scattering (SAXS) covers the particle sizes from 1 to 100 nm, whereas the ultra-small-angle X-ray scattering (USAXS) extends the analysis range up to 1000 nm [151]. Complementary techniques that are usually discussed together with SAXS and USAXS are the small-angle neutron scattering (SANS) and ultra-small-angle neutron scattering (USANS), respectively, in which the incoming neutrons interact with the sample nuclei and the neutron scattering is a quantum-mechanical effect [152]. The WAXS technique (generally identified as XRD) is usually employed to determine the diffraction patterns of the (bio)synthesized nanoparticles often in comparison with the crystalline date of the initial reactants to evidence the bio(reduction).
The WAXS technique was used to investigate the reduction of selenium with SeNPs formation and the crystallographic structure of SeNPs. A detailed structural analysis can be provided by XRD assay for biogenic SeNPs as well. Several XRD assays confirmed that biogenic SeNPs in general and trichogenic SeNPs in particular appear as amorphous structures [87,109,110,111,150], but there were also reports of a crystalline nature [112,113]. The crystalline nature of SeNPs is probably most influenced by the characteristics of the bio-corona, especially its size, but the studies performed previously on trichogenic SeNPs did not investigate this aspect. The XRD studies of trichogenic SeNPs have been performed only in the wide-angle X-ray scattering (WAXS) mode, which gives information on the crystalline nature of a material. A relevant XRD analysis of (nano)materials in general and trichogenic SeNPs in particular should provide crystallographic patterns of raw materials in comparison with the intermediate (where mechanistically relevant) and final (nano)materials, focusing on the 2θ peak position, peak relative intensity, diffraction planes, crystallite size and crystallinity degree (in the case of semi-crystalline nanomaterials). Surprisingly, it is rather hard to find relevant X-ray diffraction details, even for Na2SeO3 as a raw material.
The XRD-WAXS patterns of SeNPs biosynthesized using the fungi Aureobasidium pullulans, Mortierella humilis, Trichoderma harzianum and Phoma glomerata were compared with the diffraction patterns of elemental Se and SeO2 without peak details [111]. All strains produced Se with peaks around 22°, 30° and 45°, and only Trichoderma harzianum produced both SeNPs and SeO2, with the main SeO2 characteristic peaks around 15°, 21°, 24°, 28°, 31°, 34°, 36° and 47°. Additional XRD peaks were assigned to organic impurities or capping agents. SeNPs obtained classically by the reduction with ascorbic acid and stabilized with gum arabic were compared with trichogenic SeNPs bioreduced with T. harzianum metabolites. The XRD patterns were similar: predominantly amorphous with two broad peaks around 25° and 55° [110].
Crystalline trichogenic SeNPs obtained with Na2SeO3 and the culture filtrate of T. atroviride showed diffraction peaks at 23.80°, 29.99°, 41.63°, 43.95°, 45.66° and 51.99° [113]. Meanwhile, other crystalline trichogenic SeNPs obtained with SeO2 and Trichoderma sp. WL-Go culture broth [112] showed a multitude of diffraction peaks. Out of these, only the peaks at 23.68°, 31.36° and 44.14° were assigned to SeNPs. In contrast, the strong peaks around 21°, 24°, 28°, 30°, 32°, 34°, 36°, 38°, and 46° remained unassigned but probably corresponded to the unreacted SeO2, which had similar characteristic peaks [111].
For other crystalline SeNPs derived from Na2SeO3 reduced with T. harzianum extract, only the diffraction planes of SeNPs assigned to oval crystalline SeNPs were described [114], while the apparent 2θ angles assigned to trichogenic SeNPs appeared around 22°, 26°, 41°, 43°, 45°, 54°. Additionally, strong peaks around 24°, 25°, 32°, 33°, 37°, and 38° were generally assigned to bioactive compounds from T. harzianum extract and background noise [114], although it is more probably related to unreacted Na2SeO3, which presented similar peaks [138,153]. The necessity of analytic comparison of the biosynthesized SeNPs with the raw materials is therefore essential.
SeNPs synthesized using Trichoderma fungus filtrate were reported in one study to be amorphous, although the XRD pattern appears to be 100% crystalline without a characteristic broad amorphous peak and instead showing nine sharp peaks. Of these, the strongest four peaks appeared at 2θ angles: 23.49°, 29.79°, 43.77° and 45.53° [87]. These peaks were considered similar to the crystalline SeNPs obtained with an ethanol extract of bee propolis, which had four main XRD peaks at 23.2°, 29.5°, 31° and 45.5° [154]. A high crystallinity of SeNPs might suggest a weak bio-corona or an incomplete reduction reaction, which could correlate also with a large particle size (137 nm) and with the high Se weight and low C and O content determined by SEM-EDX, i.e., 87.98% Se in the mentioned study [87]. These crystalline SeNPs had biocide properties against Spodoptera litura larvae.
XRD patterns for Na2SeO3 and the corresponding trichogenic SeNPs are provided in [109]. Sodium selenite had a multitude of peaks between 17 and 70°, without a particular mention of the main 2θ angles, but these were visually appearing to be around 18°, 20°, 22°, 24°, 26°, 28°, 32°, 33°, 37°, 38°, 45° and 53°. The SeNPs appeared to be semi-crystalline with an amorphous peak centered around 25° and additional crystalline peaks comparable with the main peaks of Na2SeO3. A strong amorphous peak was also found around the 2θ angle of 25.70° in SeNPs reduced with sea buckthorn leaf extract, which was semi-crystalline but predominantly amorphous due to a bio-corona with a 36% crystallinity degree [138]. This peak was accompanied by small crystalline peaks at 19.18°, 21.48°, and 23.30°, which were quite different than the main Na2SeO3 peaks at 17.72°, 20.16°, 22.00°, 24.20°, 26.02°, 31.64°, 32.46°, 36.60°, 37.64°, and 44.92°, suggesting a strong reduction of initial sodium selenite. The crystallite sizes of these SeNPs were evaluated by WAXS to be around 3.5, 8.9, 6.7 and 12.3 nm with a clear pattern toward a 3 nm core crystallite, which was confirmed by a similar pattern in SAXS with SeNPs crystallites clusters around 9, 12, 18, 24, 27 and 54 nm [138], as further detailed.
The SAXS technique uses diffraction patterns generated by the particle electron density at small diffraction angles, usually at 2θ < 2°, to gather information about the nanoparticle size distribution, nanoparticle shape or nanopores [155]. SAXS has a number of advantages over other analytic techniques used for studying nanoparticles like TEM and DLS. For instance, SAXS offers an overall view on the bulk size distribution of particle diameters between 1 and 100 nm, whereas the small grid size of TEM and the reduced concentration of nanoparticles necessary for a good dispersion, together with the natural propensity toward acquiring aesthetic images, make TEM less relevant for a global picture of the nanoparticle size distribution. In addition, SAXS is a non-destructive method that allows the unaltered recovery of the sample after analysis. Compared to DLS, SAXS has a clear advantage in analyzing opaque, colored or semi-translucent colloidal suspensions, and the data reliability of SAX for nanoparticles between 1 and 30 nm is superior to that of DLS. Additionally, SAXS allows the morphological determination of size, shape, size distribution, organization and surface structure of the sample electron density differences in their native (bio)synthesized state, liquid, sol–gel or colloidal, and even in situ synthesis progression [155]. Actually, the first SAXS study was performed on a “colloidal gold” in 1951 before the term “nanoparticle” was defined [155,156].
The literature of SeNPs characterization using SAXS is scarce, suggesting an available innovative niche at the crossroads of these two directions. In a first example from our group, SAXS performed on SeNPs phytosynthesized using sea buckthorn leaf extract evidenced a polydisperse distribution of SeNPs with four main population between 9 and 60 nm and also suggested, in correlation with WAXS analysis, that the SeNPs nanograins are clusters of 3 nm crystallites, with SAXS peaks in the scattering wavevector q values ranges of 0.1–0.7 nm−1 (2θ between 0.2 and 1°) corresponding to 9, 12, 18, 24, 27 and 54 nm nanoparticle diameters [138]. In another study, spherical red SeNPs with a mean size of 45 nm were obtained by the reduction of Na2SeO3 with L-cysteine and further stabilized with 29 kDa polyvinylpyrrolidone [157]. STEM images evidenced relative uniform spherical nanoparticles with a polymeric shell and diameters between 20 and 60 nm that agglomerated after solvent drying on the STEM grid, which is a known inconvenience of the TEM/STEM analysis. SAXS analysis performed between 2θ angles 0 and 1° evidenced a high background noise at 2θ > 0.2°, and the experimental curve was fitted with a Gamma distribution model for particles with a 45 nm mean diameter [157]. These SeNPs were used in combination with a boric acid-based fungicide to protect the wood products against the brown-rot Serpula lacrymans. SAXS was used to characterize SeNPs obtained by the reduction of Na2SeO3 with ascorbic acid and stabilized with 200 kDa chitosan as 3% suspension in 2% succinic acid [76]. SAXS performed between the scattering wavevector (q) values from 0 to 3 nm−1 evidenced a bimodal particle size distribution centered around 2.5 nm and 37 nm in a volumetric ratio of 1:2 [76]. These SeNPs were intended for the development of adaptogenic drugs due to their biocompatibility and antioxidant properties. There are no available data of SAXS analysis performed on trichogenic SeNPs, to the best of our knowledge.
XPS assays of trichogenic SeNPs confirmed the presence of elemental Se by a peak at the binding energy of elemental Se, 55.6 eV [109]. These data confirm the reduction of Se from its precursors and implicitly the formation of SeNPs [90]. XPS is a powerful technique that can give additional information on the bio-corona composition and the types and energy of Se interactions, e.g., via 3d orbital electrons, with the molecules in the bio-corona [158,159], but no in-depth analysis is available in the case of trichogenic SeNPs.
Dynamic light scattering (DLS) and SP-ICP-MS techniques can be used to estimate the size of hydrated SeNPs in solution, including the bio-corona [87]. Whereas TEM gives information mainly on the size of the selenium core, DLS gives information on the hydrodynamic radius of the entire structure. SP-ICP-MS can also determine the size of NPs [111]. The zeta potential indicates the net charge of a particle and is a good indicator of NP stability in terms of colloidal dispersion and tendency for aggregation. The closer the value is to zero, the higher the chance that the particles will aggregate due to attractive van der Waals interactions [160]. Values higher than ±30 mV are considered to be highly stable. But because the zeta potential does not offer information on the strength of these van der Waals interactions, there are cases of unstable NP suspensions despite high zeta potential and vice versa [161]. The zeta potential is determined by the characteristics of the biomolecules capping the Se core of the NP and forming the bio-corona. The determinations of zeta potential of biogenic SeNPs indicated that these kinds of structures are in general rather stable. To the best of our knowledge, there are only a few studies that determined the zeta potential of trichogenic SeNPs [109,112,113,114]. An intriguing aspect is that the zeta potential seems to have significant variations across Trichoderma species and also on the fraction of the culture used, in the case of in vitro bio-assisted synthesis, varying from +11 mV to as negative as −200 mV [109]. This observation deserves further more in-depth studies, including correlations between the capping biomolecules and zeta potential. Most of the zeta potentials indicating stable colloidal suspensions of SeNPs had negative values, whereas the positive values were in the range indicating unstable systems. A comparison of the zeta values of washed and unwashed SeNPs showed that washing and resuspending SeNPs in distilled water can enhance their stability [112,113,161].
Another technique that can give valuable information on the compounds forming the bio-corona is FTIR. This technique has been applied in almost all studies involving trichogenic SeNPs. Most of the FTIR studies show the existence of hydrogen bonds in the diagnostic region of 3400–3000 cm−1 and protein bands, e.g., amide bands I, II, and III in the fingerprint region 1600–1200 cm−1 [87,109,110,112,113,114], although in some cases, their presence is not properly discussed. A comparison between the FTIR spectra of trichogenic SeNPs obtained with the sterile filtrate and control SeNPs with PDB medium showed spectral differences that indicated a protein-dominant and a saccharide-dominant bio-corona in the trichogenic SeNPs and control SeNPs, respectively [110]. The FTIR spectra of T. harzianum extract and SeNPs indicated as well that proteins from the trichogenic biomass are probably capping the SeNPs [114]. In comparison, FTIR analysis evidenced the characteristic absorption bands of Se-O and Se=O in Na2SeO3 around 880 cm−1 and 714 cm−1 [136,138] and further selenite reduction by phytosynthesis with sea buckthorn leaf extract [138] or by pollen–Kombucha fermentation [136]. Here, the SeNPs bio-corona was stabilized by multiple hydrogen bonds of amino acids, proteins and oligo-saccharides. FTIR analysis is the most suitable method to evaluate the hydrogen bonding in SeNPs as the first molecular layer around Se0, the disruption of the hydrogen bonds environment with strong polar solvents like NaOH and dimethylsulfoxide leading to selenium aggregation and precipitation [162].
As FTIR data indicate that proteins contribute significantly to the capping and stabilization of trichogenic SeNPs, protein SDS-PAGE electrophoresis can be a valuable tool that should be used to characterize these proteins. Protein electrophoresis can provide hints on the abundance of capping proteins, as well as their molecular weight, and, in combination with LC-MS/MS, the proteins can be identified. There are almost no studies including an electrophoresis analysis of proteins involved in trichogenic SeNP formation. We found only one study in which the authors found two protein bands at 15 and 19 kDa proposed to be part of the capping bio-corona [112], but the bands were not identified.
Using liquid chromatography and mass spectrometry (TripleTOF LC-MS), one research group identified 35 various metabolites in the aqueous solution of SeNPs produced by the sterile filtrate of a T. harzianum strain. Among them, there were organic acids, amino acids, sugars, and some intermediates from the carbohydrate metabolism; 27 compounds showed potent antifungal activity against phytopathogens [110]. This result indicates that the bio-corona can be in fact highly heterogeneous, and it partially explains the significant differences in zeta potential of SeNPs among Trichoderma species. It additionally highlights the usefulness of combining various complementary techniques to better understand the properties and role of the bio-corona of trichogenic SeNPs; however, this approach is currently missing.
Table 2 summarizes the information available from the literature with respect to the composition and the induced physicochemical characteristics of the bio-corona capping trichogenic SeNPs.

Table 2.
Composition and physicochemical characteristics of bio-corona capping trichogenic SeNPs.
Two bottleneck issues in manipulating fungal SeNPs are, firstly, its separation from the fungal biomass in the case of in vivo biosynthesis and especially for intracellularly formed SeNPs, and secondly, obtaining sterile, pure SeNPs, with an intact bio-corona, to be biologically tested at the laboratory level. The first issue requires the disruption of the fungal mycelium, which could influence the characteristics of SeNPs. Moreover, the intracellular SeNPs could have different properties compared with the ones released extracellularly in the culture medium during incubation. These aspects are currently still unexplored and should be investigated in the future.
The bio-corona manipulation of trichogenic SeNPs should be targeted to enhance the specific SeNPs properties—for example, antimicrobial activities. The antimicrobial activities of the chemically synthesized SeNPs stabilized with cetyltrimethylammonium chloride (CTAC) against Escherichia coli, Micrococcus luteus, and Mucor were significantly higher than those of the initial constituents, including selenious acid [163]. It was reported that the surface chemistry has the highest influence on the antimicrobial activities of chemically synthesized SeNPs [164]. The SeNPs stabilized with bovine serum albumin effectively inhibited monomicrobial and dual-species biofilm development [164].
The trichogenic SeNPs, prepared with filtrate-containing metabolites of T. harzianum JG309, exhibited a threefold antimicrobial effect compared to chemically synthesized SeNPs. The biocontrol efficacy of trichogenic SeNPs was more evident than that of chemically synthesized SeNPs in reducing mycotoxin production by Alternaria and Fusarium toxigenic strains. The reduction ranged from 63% in the case of fumonisins B1 to 83% in the case of tenuazonic acid. The proposed mechanisms involve the “recognition metabolites” from the SeNP bio-corona that drive SeNPs action on the expression of key biosynthetic genes of mycotoxins [110].
Bio-coronas are complex chemical systems with emergent properties that derive from the interactions of the components [165]. The complexity of the bio-corona is a significant challenge for its manipulation directed toward enhancing a specific activity relevant to a given application. More studies are needed to understand the relationship between the biological activities of various Trichoderma proteins and metabolites, their probability of being included in the “soft” or “hard” bio-corona, and the biological activities of the resulting SeNPs. The probability of being included in the bio-corona results from the thermodynamics of the interactions.
Isothermal titration calorimetry (ITC) is used to study the thermodynamic profile of interactions (binding and dissociation) in solutions/suspensions [166]. In the last few years, this technique was used to characterize the interactions between the surface of the SeNPs and the bio-corona [82,83,167,168]. In the ITC experiments, a highly sensitive microcalorimeter continuously records the calorimetric curve of interactions between the NP surface and a bio-corona component. The ITC experiment provides thermodynamic and kinetic information of the biomolecular interaction—the enthalpy variation in the system (ΔH), entropy change (ΔS), binding free energy (ΔG), the binding constant (Ka), and the stoichiometry, e.g., the number of binding sites (N) [166]. To the best of our knowledge, ITC has not been used yet to investigate the formation and stabilization of trichogenic nanoparticles.
5. Applications of Trichogenic SeNPs
Improvements in crop yield, the management of nutrient levels in soil and plants, insect control, and the management of environmental factors are priorities for the agricultural sector [120]. Nanoparticles could be one of the solutions to satisfy these requirements, and it is considered a research direction with great innovative potential. Trichoderma sp. can be used for the biogenic synthesis of NPs with powerful properties needed for the agriculture sector. The use of these species might exhibit its potential to act in synergism with the NP that it produces, providing new leads in creating biotechnologies that might enhance crop quality and resistance against phytopathogens [92,169].
One of the most prevalent bioactivities of trichogenic SeNPs is their antifungal activity [66,92,169,170]. This nanotechnology has been proved to suppress the sporulation of Sclerospora graminicola, which is the pathogen that causes downy mildew in pearl millet [109]. Biogenic SeNPs synthesized using T. atroviridae strains have been proved to have great zoosporicidal and antifungal activities for Phytophthora infestans in tomato, which is one of the most devastating pathogens in tomato [113]. The antifungal mechanism could be attributed to the united and synergistic effect of SeNPs and diverse fungal metabolites, including multiple organic acids and its derivates [110]. In addition, SeNPs synthesized by Trichoderma sp. have larvicidal and antifeedant activity, which is a main component of crop protection [87]. Recently, Helmy et al. showed a higher antifungal activity of trichogenic SeNPs against Fusarium oxysporum and a higher reduction of Fusarium wilt infection of tomato plant compared with selenite [171].
Constant mycotoxins contamination is a serious challenge of crop production, food safety animal and human health. Trichoderma sp.-derived SeNPs might address these needs by providing a functional biocontrol of mycotoxins in agricultural and food safety procedures [110]. These NPs could represent affordable, environmentally beneficial and non-toxic solutions [113]. A test conducted by our group on Vigna radiata seeds which germinated in trichogenic SeNPs aqueous solutions validates the non-toxic potential of the nanoparticles [172].
Several biotests have revealed the biostimulant effects of biogenic SeNPs. Zogra et al. (2021) highlighted that in addition to beneficial microorganisms, the use of biogenic SeNPs is known as an environmentally friendly approach to enhance crop production by alleviating biotic and abiotic stresses (alleviating drought, salinity, heavy metal, heat stresses, and bacterial and fungal diseases in plants) [173]. Joshi et al. demonstrated enhanced resistance of tomato to late blight disease by priming mechanisms [115]. The application of biogenic SeNPs in plants can improve crop value. Moreover, the treatment of crops with biogenic SeNPs could lead to the production of edible foods fortified with Se, which might be of great importance in managing the deficiencies of this micronutrient [66]. Table 3 summarizes the previously reported biological effects of trichogenic SeNPs that have potential applications in agriculture and the food industry.

Table 3.
Biological effects of trichogenic SeNPs with applications in agriculture and the food industry.
For a proper optimization of the trichogenic SeNPs bioactivity and use in agriculture, the mechanism behind the biological effects needs to be understood in depth, such as the priming mechanisms mentioned above, as well as the properties that correlate with its bioactivity. The first step is to establish these correlations; in this sense, only a handful of studies provide some information with respect to trichogenic SeNPs. Nandini et al. found an inversely proportional correlation, but with a modest correlation coefficient, between the growth and proliferation inhibition of S. graminicola and the size of the SeNPs produced by various fractions of Trichoderma spp. Cultures [109]. The reason for this type of correlation is not straightforward, as the sizes of SeNPs were in general larger than 50 nm, reaching microparticle regime (>100 nm). The translocation of SeNPs of this size into the cells would be rather difficult, although the smallest nanoparticles present are probably translocated. Other explanations might contribute more, such as a different rate of selenium disproportionation or differences in the biomolecules forming the bio-corona. It would have been interesting to perform a correlation analysis with the zeta potential as well, which varied largely in their study, depending on the Trichoderma strain and culture fraction used, as we mentioned above.
In another study, the antifeedant activity of trichogenic SeNPs against Spodoptera litura was directly proportional to the nanoparticle concentration [87]. Other correlations between trichogenic SeNPs activity and their properties are not available according to our survey. Questions remain open on if and how the capping bio-corona variation, in relation to the SeNP crystallinity, affects the bioactivity, considering the differences between the Trichoderma species observed. There is no systematic study available to assign any particular capping composition to a specific effect. Moreover, few investigations studied whether the effects observed are due to the capping biomolecules, the released soluble Se species or small trichogenic SeNPs that could translocate into the cells, or whether there is a synergism/antagonism between these contributions and to what extent. As mentioned in Section 4, the work of Hu et al. showed that the trichogenic SeNPs significantly enhanced the antifungal activity against phytopathogens compared with standard SeNPs [110]. The enhanced activity was despite a lower absorption of selenium from trichogenic SeNPs compared with standard SeNPs, as determined by ICP-MS, which indicates that it was mainly due to metabolites such as organic acids forming the bio-corona. An analysis of speciation would have helped to estimate the absorbed elemental Se from other Se species. Moreover, we believe that the constituents of the NP could influence each other. For example, the capping bio-corona might influence the Se disproportionation reactions, and the disproportionation could influence the bio-corona’s stability. For example, the protein corona was proposed to reduce the oxidation of trichogenic SeNPs [171]. Therefore, investigations of the stability of SeNPs should consider not only colloidal aspects but also the oxidation state of Se and the bio-corona’s integrity. Bacterial proteins were shown to influence the size of SeNPs [137], and this is probably true for fungal proteins as well.
The plant biostimulant effects of trichogenic SeNPs on seed germination have not been yet investigated. The application of Trichoderma strains on seeds promotes germination and seedling development [66,174,175]. The chemically synthetized SeNPs, stabilized with didecyldimethylammonium chloride, improved the germinability and germination of barley seeds (Hordéum vulgáre L.) with impaired morphofunctional characteristics due to 10 years of storage [176]. The trichogenic SeNPs should have a better effect as biostimulants of seed germination due to the combined effect of SeNPs and the bio-corona consisting of proteins and metabolites from biostimulant Trichoderma strains.
All in all, the relatively limited information available indicates that specifically targeted effects could be obtained by manipulating the in situ capping of SeNPs with the most suitable metabolites able to form stable complexes with the nanoparticles. That is why it is important that future studies include an in-depth characterization of the trichogenic bio-corona mentioned in Section 4. The biotests of in vitro particular designs of specific bio-corona combinations with Trichoderma metabolites and proteins would help optimize the outcome.
The strains from Trichoderma are plant-beneficial fungi with multiple agronomical functions: plant protection against plant pathogens [177], fungi, [178,179], bacteria [180], viruses [181], nematodes [182,183] and insects [184,185]; plant biostimulants [186], increasing nutrient uptake and nutrients use efficiency [122], enhancing plant tolerance to abiotic stress [187], and improving crop quality traits [188]; biofortification enhancers [189]; and agents for bioremediation [190]/phyto(rhizo)remediation [191,192].
Selenium nanoparticles demonstrated similar agronomical functions as biostimulant [193], plant protection [115], biofortification [194] and phytoremediation agents [195].
Trichoderma agronomical functions are mediated by various categories of bioactive compounds: proteins, lytic exo-enzymes [196], expansin-like proteins [197], hydrophobins [198], other protein elicitors [199], and secondary metabolites [200] such as 6-pentyl-pyrones [201], trichothecenes [202], peptaibols [203], siderophores [204], and butenolide [205]. The involvement of these bioactive compounds in the bio-corona of the trichogenic SeNPs should enhance SeNPs’ agronomical function—as it is possible to share activities among the biological systems.
In addition, for agricultural applications in open fields, one has to consider the effects of soil microbiota and composition on SeNPs. Microorganisms, including fungi, can induce not only a reduction but also oxidation and methylation of selenium species, which could influence the Se disproportionation and the relative abundance of various states of Se [111,206].
Potential Environmental Impacts and Safety Considerations of SeNPs Use in Agriculture
Quite a few studies describe the use of selenium and selenium-based products in agriculture, especially for plant biofortification and phytoremediation. These practices aim to manage selenium deficiencies in humans and animals [8,32,207,208,209,210,211,212,213,214]. The available reports acknowledge this novel biotechnology could be a safe, sustainable and simple alternative to other forms of selenium in agricultural applications. The main advantages of SeNPs reported so far referred to the higher biological activity and bioavailability as well as the lower toxicity of nano-selenium [8,25,103,193,215,216,217,218,219,220].
A complete understanding of putative secondary effects and the environmental impact of products used in crop management is mandatory for preserving both food safety and sustainability in agricultural practices. The trichogenic SeNPs have not been investigated in terms of ecological safety and/or environmental impact yet. Therefore, we present here general aspects concluded from studies of other types of SeNPs as well as aspects that should be established for trichogenic SeNPs in future research.
In addition to the production of Se-enriched edible crops, which might prevent and counteract selenium deficiencies, the environmental impact of SeNPs agricultural uses seems to be mostly beneficial, considering the fact that SeNPs act as efficient heavy metal decontaminants for soil and waters [57,221]. Additionally, the antimicrobial activity of SeNPs could enhance soil quality and preserve plant health by inhibiting pathogen growth [51].
The progress achieved through industrialization has come at the cost of heavy metal pollution, which has become a pressing ecological issue over the last two centuries. This phenomenon was proved to be health threatening, as exposure to heavy metals might cause DNA damage, neurotoxicity, cell membrane damage and protein-related metabolic disorders in humans [222,223]. A wide range of studies described SeNPs, particularly of biogenic nature, to be great decontaminants for heavy metal-polluted waters and soils. SeNPs have proved to be excellent tools in the nano-bioremediation of these biospheres due to their capacity to adsorb heavy metals [57,66,221,224,225,226,227,228,229]. In addition to their ability to reduce heavy metals (directly or post-adsorption) to lower, less toxic, valence states, SeNPs possess some additional properties which ensure an efficient adsorption of contaminants. Precisely, the interaction forces between SeNPs and heavy metals are enhanced by the amorphous surface of the nanoparticles, which consequently increases absorptivity rate. Even more, nanoparticles have a large and active surface area and possess numerous active sites, which increases the reaction yield of these adsorbents with the contaminant substrates [229,230,231]. The bio-corona that caps the biogenic SeNPs promotes the formation of electrostatic and chemical interactions with the contaminants, facilitating heavy metal entrapment on the SeNPs surface. Therefore, SeNPs entering vital ecosystems such as soils and water through agricultural uses might bring additional detoxifying benefits to a metal-polluted environment [227].
The antimicrobial properties, described in Section 5 of this review, bring out the benefits of agricultural uses of SeNPs in plant defense against phytopathogens. However, these benefits could be extended by improving the soil ecosystems in terms of rhizosphere microbiome. Not only could SeNPs prevent and combat plant diseases, they could also be efficient antimicrobial agents for soils and simultaneously preserve the probiotic activity [51,232,233,234]. Moreover, SeNPs seem to directly enhance soil quality by improving its enzymatic content and ensuring an optimal level of signaling molecules, which might trigger plant defense mechanisms. SeNPs might also improve the soil quality by preserving and stimulating beneficial organic substances which are necessary for plant growth [51,67,235].
The ecological safety of SeNPs has not been extensively analyzed yet. However, there are some studies that stated the adverse impact of SeNPs on the environment. It has been established that the toxic behavior of SeNPs varies greatly among plant and animal species and it is mainly dose-dependent. The time of exposure and type of biogenic coating are also important factors which might determine the degree of toxicity and stability of these compounds. The accumulation of selenium, which might be ecologically hazardous, is influenced by similar factors [67,85,236,237,238]. Therefore, although SeNPs are generally considered much safer than other forms of selenium, it is tremendously important to take into consideration that any potential application of this resourceful and innovative bionanotechnology should be adapted and optimized especially in terms of dosage and time of exposure.
6. Future Research Directions
As mentioned in the Introduction, the biosynthesis of SeNPs by Trichoderma spp. is still an insufficiently explored subject, despite the fact that the first study was reported in 1995. Still, interest has grown in the last few years, which is due in part to the development of plant biostimulants and plant protection bioproducts. The future directions should fill the knowledge gaps that limit the exploitation of the agronomical functions of trichogenic selenium nanoparticles. In particular, future works should address the significant research gaps that remain at present. These include deciphering the complete mechanism of trichogenic SeNPs biosynthesis as well as providing a more in-depth characterization of in vivo versus in vitro (intracellular and extracellular in both cases) produced SeNPs for the same Trichoderma strains, especially with respect to the bio-corona and the dissociation of biomolecules from it. Other research avenues include investigating its biological activity, optimizing the production yield, processing SeNPs, manipulating the bioactivity of SeNPs, finding new applications of SeNPs, assessing their safety, and determining the environmental impact of trichogenic SeNP use in agriculture.
In the case of characterization, using as many as possible of the methods described in Section 4 on the same system will help build a more complete picture of the system, correlating physicochemical characteristics to the biological activities. In particular, the composition and characteristics of the bio-corona should be characterized in depth, using techniques such as DLS, zeta potential, analytical ultracentrifugation, surface tension and contact angle, FTIR, SDS-PAGE, proteomics, LC-MS, ITC, etc. An important aspect is that the characterizations should be performed additionally under relevant conditions related to the targeted applications. The modifications and transformation of SeNPs under these conditions should be established. The stability of the bio-corona and the oxidation state of Se should be considered complementary to the colloidal stability of trichogenic SeNPs.
For optimizing the yield and quality of SeNPs biosynthesis, design of experiments (DoE) such as response surface methodology (RSM) combined with high-throughput screening (HTS) are useful techniques that should be taken into consideration for an efficient output. These approaches are useful for reducing the number of experiments, producing optimal results and providing information on the interactions between the independent parameters. The main problem in applying these techniques lies in the manipulation of SeNPs post-synthesis, which is usually time consuming and most often performed manually. This could be solved by using multi-well plates, in combination with multichannel pipettes, multiscreen filter plates and, if available, robot systems. Another issue that could arise is the need for high-force centrifugation in order to sediment the SeNPs. This cannot be completed using multi-well plates; other methods would need to be developed. Up to now, this type of optimization has not been reported for the biosynthesis of SeNPs to the best of our knowledge. In the case of in vivo intracellular SeNPs, optimal methods for mycelium disruption should be developed in the future.
Another focus of future research should be the optimization of biosynthesis parameters in order to maximally reduce the aggregation of trichogenic SeNPs produced in vivo. This would facilitate obtaining pure, sterile-filtered SeNPs that could be properly characterized, biologically tested and formulated. Comparison with in vitro-produced SeNPs would be facilitated and should be performed. For agronomical applications, non-sterile trichogenic SeNPs can be applied as well, but the contribution of individual components to the biological effects should be known.
Regarding individual components, the contribution of individual SeNP parameters such as capping composition and properties, size, crystallinity, etc. on the biological effects observed should be established. The biological effects should be further assigned to individual contributions such as bio-corona molecules, released soluble selenium species, intracellular translocated SeNPs, etc., and eventual relations of synergism/antagonism between them should be established. Knowing how these individual parameters of SeNPs influence each other will be beneficial for the best optimization of the final formulation for the specific application considered.
A systemic approach is needed to select the optimal Trichoderma strain and parameters for the formation of such trichogenic SeNPs sharing activities with the biological system in question—see Figure 2.
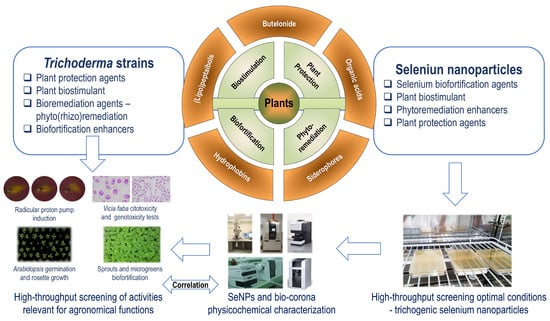
Figure 2.
The systemic investigation to select Trichoderma strains and the optimal conditions producing trichogenic SeNPs that share activities with the biological systems in question.
The approach proposed in Figure 2 follows the CESM (Composition, Environment, Structure and Mechanism) model [239]. The composition of the bio-corona (proteins, metabolites) results from the peculiarities of each Trichoderma strain and is investigated by the methods presented in Section 4. The environment provides the component of the “soft” and “hard” bio-corona and is characterized by (optimal) parameters to form and to stabilize the SeNPs—temperature, pH, and light intensity. The structure notion refers to the interaction between the bio-corona components. The mechanisms represent the processes involved in the slow release of the inorganic selenium species and bio-corona components from trichogenic SeNPs and in the common/enhanced biological activity. The CESM model was used as a systemic approach to explain the emergent properties of biological systems, including plant ecology [240]. This systemic approach is intended to further develop trichogenic SeNPs as complex chemical systems that contain complex elements relevant to biological activities and agronomical functions with the biological systems in question. The plant-beneficial strains from Trichoderma genera were selected due to their multifaceted role in cultivated plant management and similar agronomical functions.
The research and development activities needed to upscale the Trichoderma-mediated SeNPs biosynthesis for commercial applications are summarized in Figure 3. These activities are related to the upscaling of the selected conditions for producing trichogenic SeNPs that share activities with the biological systems in question, creating formulations specific for the intended agronomical application, and developing the precise application systems.
Figure 3.
The research and development activities needed to upscale the Trichoderma-mediated SeNPs biosynthesis for commercial applications.
The production of extracellular SeNPs should benefit from the emergent fungal biofilm bioreactors [241]. Selection of the most appropriate material used as support for biofilm development is necessary since the physicochemical surface properties of these materials modulate biofilm physiology and the production of proteins and metabolites [242]. In the case of the intracellular biosynthesized SeNPs, a critical upscaling scale is cellular lysis [243]. Mild energy/shearing force should be used to reduce the probability of bio-corona destabilization. Adaptation of the ultrafiltration techniques to SeNPs is needed for an efficient separation from cell debris [244].
Reduction in the water activity for long-term storage, requested for technology upscaling to the commercial level, should be combined with developing various formulations (water-dispersible powder/granules, concentrated solution, film-forming formulations), specific for agronomical applications—seed coating, foliar spraying, soil drenching, injection in the drip irrigation systems [245].
Efficient SeNPs applications bearing fruit from SeNPs bioenhanced agronomical functions require the development of adapted precision application systems [246,247]. The innovation ecosystems that promote such developments are the Living Labs, wherein the scientists and farmers share knowledge, providing user-centric ideas and solutions [248]. The context-related, holistic knowledge of the farmers will support the sustainable exploitation of the agronomical functions of trichogenic SeNPs sharing activities with the biological systems in question [249,250]. In the innovative ecosystem of Living Labs, new directions for trichogenic SeNPs applications, i.e., sprout and microgreen biofortification, food packaging, and the control of food pathogens, could be developed and upscaled.
7. Conclusions
Selenium nanoparticles (SeNPs) have been intensely studied over the last decade due to their extraordinary properties and their various biological activities. The present review confirms that the biosynthesis of selenium nanoparticles involving Trichoderma sp. reveal promising potential in an eco-friendly manner. The SeNPs obtained through trichogenic biosynthesis are non-toxic and have enhanced biological activity compared with chemically synthesized SeNPs. The current study described as well several documented hypotheses on the fungal-mediated NP biosynthesis that could explain the synthesis mechanism of trichogenic SeNPs. Biogenic nanoparticles, including the trichogenic SeNPs, can be characterized by a wide range of analysis methods, depending on the aim of their final use. Many pieces are still missing in the puzzle, and more complementary physicochemical and biological investigations should be performed as well as correlation analysis for optimizing the biological activity. Applications of trichogenic selenium nanoparticles target mainly the agri-food sector, aiming to improve crop quality.
Author Contributions
Conceptualization, F.O. and D.C.-A.; writing—original draft preparation, L.T.C., V.B. and Ș.-O.D.; writing—review and editing, I.C.F., D.C.-A. and F.O.; visualization, D.C.-A. and F.O.; supervision, I.C.F. and F.O.; project administration, D.C.-A.; funding acquisition, D.C.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministry of Research, Innovation and Digitization, CNCS/CCCDI—UEFISCDI, project number 107PCE/2021, within PNCDI III.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Tsuji, P.A.; Santesmasses, D.; Lee, B.J.; Gladyshev, V.N.; Hatfield, D.L. Historical Roles of Selenium and Selenoproteins in Health and Development: The Good, the Bad and the Ugly. Int. J. Mol. Sci. 2022, 23, 5. [Google Scholar] [CrossRef]
- Reich, H.J.; Hondal, R.J. Why nature chose selenium. ACS Chem. Biol. 2016, 11, 821–841. [Google Scholar] [CrossRef]
- Serrão, V.H.B.; Scortecci, J.F. Why Selenocysteine Is Unique? Front. Mol. Biosci. 2020, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.Z.; Krahn, N. The selenocysteine toolbox: A guide to studying the 21st amino acid. Arch. Biochem. Biophys. 2022, 730, 109421. [Google Scholar] [CrossRef] [PubMed]
- Maroney, M.J.; Hondal, R.J. Selenium versus sulfur: Reversibility of chemical reactions and resistance to permanent oxidation in proteins and nucleic acids. Free Radic. Biol. Med. 2018, 127, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Nayak, V.; Singh, K.R.; Singh, A.K.; Singh, R.P. Potentialities of selenium nanoparticles in biomedical science. New J. Chem. 2021, 45, 2849–2878. [Google Scholar] [CrossRef]
- Xiao, D.; Li, T.; Huang, X.; Zhu, K.; Li, Z.; Dong, Y.; Wang, L.; Huang, J. Advances in the Study of Selenium-Enriched Probiotics: From the Inorganic Se into Se Nanoparticles. Mol. Nutr. Food Res. 2023, 67, 2300432. [Google Scholar] [CrossRef]
- Schiavon, M.; Nardi, S.; Dalla Vecchia, F.; Ertani, A. Selenium biofortification in the 21st century: Status and challenges for healthy human nutrition. Plant Soil 2020, 453, 245–270. [Google Scholar] [CrossRef]
- Trippe III, R.C.; Pilon-Smits, E.A. Selenium transport and metabolism in plants: Phytoremediation and biofortification implications. J. Hazard. Mater. 2021, 404, 124178. [Google Scholar] [CrossRef]
- Schiavon, M.; Pilon-Smits, E.A. The fascinating facets of plant selenium accumulation–biochemistry, physiology, evolution and ecology. New Phytol. 2017, 213, 1582–1596. [Google Scholar] [CrossRef]
- Chauhan, R.; Awasthi, S.; Srivastava, S.; Dwivedi, S.; Pilon-Smits, E.A.H.; Dhankher, O.P.; Tripathi, R.D. Understanding selenium metabolism in plants and its role as a beneficial element. Crit. Rev. Environ. Sci. Technol. 2019, 49, 1937–1958. [Google Scholar] [CrossRef]
- Lobanov, A.V.; Hatfield, D.L.; Gladyshev, V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2009, 1790, 1424–1428. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Ni, J.; Liu, Q. Evolution of selenoproteins in the metazoan. BMC Genom. 2012, 13, 446. [Google Scholar] [CrossRef]
- Wells, M.; Basu, P.; Stolz, J.F. The physiology and evolution of microbial selenium metabolism. Metallomics 2021, 13, mfab024. [Google Scholar] [CrossRef] [PubMed]
- Lenz, M.; Lens, P.N.L. The essential toxin: The changing perception of selenium in environmental sciences. Sci. Total Environ. 2009, 407, 3620–3633. [Google Scholar] [CrossRef] [PubMed]
- Rocourt, C.R.; Cheng, W.H. Selenium supranutrition: Are the potential benefits of chemoprevention outweighed by the promotion of diabetes and insulin resistance? Nutrients 2013, 5, 1349–1365. [Google Scholar] [CrossRef]
- Shimada, B.K.; Alfulaij, N.; Seale, L.A. The Impact of Selenium Deficiency on Cardiovascular Function. Int. J. Mol. Sci. 2021, 22, 10713. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Chen, C.; Jia, S.-Z.; Cao, X.-C.; Liu, M.; Tian, J.; Hoffmann, P.R.; Xu, H.-X.; Ni, J.-Z.; Song, G.-L. Selenium restores synaptic deficits by modulating NMDA receptors and selenoprotein K in an alzheimer’s disease model. Antioxid. Redox Signal. 2021, 35, 863–884. [Google Scholar] [CrossRef]
- Pereira, M.E.; Souza, J.V.; Galiciolli, M.E.A.; Sare, F.; Vieira, G.S.; Kruk, I.L.; Oliveira, C.S. Effects of Selenium supplementation in patients with mild cognitive impairment or alzheimer’s disease: A systematic review and meta-analysis. Nutrients 2022, 14, 3205. [Google Scholar] [CrossRef]
- Hosnedlova, B.; Kepinska, M.; Skalickova, S.; Fernandez, C.; Ruttkay-Nedecky, B.; Malevu, T.D.; Sochor, J.; Baron, M.; Melcova, M.; Zidkova, J. A summary of new findings on the biological effects of selenium in selected animal species—A critical review. Int. J. Mol. Sci. 2017, 18, 2209. [Google Scholar] [CrossRef]
- Long, J.A.; Large, R.R.; Lee, M.S.; Benton, M.J.; Danyushevsky, L.V.; Chiappe, L.M.; Halpin, J.A.; Cantrill, D.; Lottermoser, B. Severe selenium depletion in the Phanerozoic oceans as a factor in three global mass extinction events. Gondwana Res. 2016, 36, 209–218. [Google Scholar] [CrossRef]
- Etteieb, S.; Magdouli, S.; Zolfaghari, M.; Brar, S. Monitoring and analysis of selenium as an emerging contaminant in mining industry: A critical review. Sci. Total Environ. 2020, 698, 134339. [Google Scholar] [CrossRef] [PubMed]
- English, S.G.; Hess, H.; Bishop, C.A.; Porter, E.; Cheng, K.M.; Elliott, J.E. Bioaccumulation and effects of selenium from surface coal mining in an aquatic songbird. Environ. Res. 2022, 208, 112702. [Google Scholar] [CrossRef] [PubMed]
- Oancea, F.; Szabolcs, L.; Oancea, A.-O.; Lăcătuşu, R.; Abraham, B.; Stanciu-Burileanu, M.M.; Meszaros, A.; Lungu, M. Selenium biofortification biotechnologies of wheat grain in south-eastern part of Romania for a better human health. Stud. Univ. Vasile Goldis Ser. Stiintele Vietii (Life Sci. Ser.) 2014, 24, 47–56. [Google Scholar]
- Danso, O.P.; Asante-Badu, B.; Zhang, Z.; Song, J.; Wang, Z.; Yin, X.; Zhu, R. Selenium Biofortification: Strategies, Progress and Challenges. Agriculture 2023, 13, 416. [Google Scholar] [CrossRef]
- Monika, G.; Kim, S.R.M.; Kumar, P.S.; Gayathri, K.V.; Rangasamy, G.; Saravanan, A. Biofortification: A long-term solution to improve global health—A review. Chemosphere 2023, 314, 137713. [Google Scholar] [CrossRef]
- Genchi, G.; Lauria, G.; Catalano, A.; Sinicropi, M.S.; Carocci, A. Biological Activity of Selenium and Its Impact on Human Health. Int. J. Mol. Sci. 2023, 24, 2633. [Google Scholar] [CrossRef]
- Banerjee, M.; Chakravarty, D.; Kalwani, P.; Ballal, A. Voyage of selenium from environment to life: Beneficial or toxic? J. Biochem. Mol. Toxicol. 2022, 36, e23195. [Google Scholar] [CrossRef]
- Oancea, A.; Gaspar, A.; Seciu, A.; Ștefan, L.; Crăciunescu, O.; Georgescu, F.; Lăcătușu, R. Development of a new technology for protective biofortification with selenium of Brassica crops. AgroLife Sci. J. 2015, 4, 80–85. [Google Scholar]
- Mimmo, T.; Tiziani, R.; Valentinuzzi, F.; Lucini, L.; Nicoletto, C.; Sambo, P.; Scampicchio, M.; Pii, Y.; Cesco, S. Selenium Biofortification in Fragaria × ananassa: Implications on Strawberry Fruits Quality, Content of Bioactive Health Beneficial Compounds and Metabolomic Profile. Front. Plant Sci. 2017, 8, 1887. [Google Scholar] [CrossRef]
- Singhal, R.K.; Fahad, S.; Kumar, P.; Choyal, P.; Javed, T.; Jinger, D.; Singh, P.; Saha, D.; Md, P.; Bose, B.; et al. Beneficial elements: New Players in improving nutrient use efficiency and abiotic stress tolerance. Plant Growth Regul. 2023, 100, 237–265. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, J.; Kronzucker, H.J.; Shi, W. Selenium biofortification and interaction with other elements in plants: A review. Front. Plant Sci. 2020, 11, 586421. [Google Scholar] [CrossRef]
- Gui, J.-Y.; Rao, S.; Huang, X.; Liu, X.; Cheng, S.; Xu, F. Interaction between selenium and essential micronutrient elements in plants: A systematic review. Sci. Total Environ. 2022, 853, 158673. [Google Scholar] [CrossRef] [PubMed]
- Poblaciones, M.J.; Broadley, M.R. Foliar selenium biofortification of broccolini: Effects on plant growth and mineral accumulation. J. Hortic. Sci. Biotechnol. 2022, 97, 730–738. [Google Scholar] [CrossRef]
- Liu, H.D.; Xiao, C.M.; Qiu, T.C.; Deng, J.; Cheng, H.; Cong, X.; Cheng, S.Y.; Rao, S.; Zhang, Y. Selenium Regulates Antioxidant, Photosynthesis, and Cell Permeability in Plants under Various Abiotic Stresses: A Review. Plants 2023, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Nazir, F.; Kumari, S.; Mahajan, M.; Khan, M.I.R. An explicit story of plant abiotic stress resilience: Overtone of selenium, plant hormones and other signaling molecules. Plant Soil 2023, 486, 135–163. [Google Scholar] [CrossRef]
- Ahmad, R.; Waraich, E.A.; Nawaz, F.; Ashraf, M.Y.; Khalid, M. Selenium (Se) improves drought tolerance in crop plants–a myth or fact? J. Sci. Food Agric. 2016, 96, 372–380. [Google Scholar] [CrossRef]
- Jiang, H.; Lin, W.; Jiao, H.; Liu, J.; Chan, L.; Liu, X.; Wang, R.; Chen, T. Uptake, transport, and metabolism of selenium and its protective effects against toxic metals in plants: A review. Metallomics 2021, 13, mfab040. [Google Scholar] [CrossRef]
- Lai, X.; Yang, X.; Rao, S.; Zhu, Z.; Cong, X.; Ye, J.; Zhang, W.; Liao, Y.; Cheng, S.; Xu, F. Advances in physiological mechanisms of selenium to improve heavy metal stress tolerance in plants. Plant Biol. 2022, 24, 913–919. [Google Scholar] [CrossRef]
- Feng, R.; Zhao, P.; Zhu, Y.; Yang, J.; Wei, X.; Yang, L.; Liu, H.; Rensing, C.; Ding, Y. Application of inorganic selenium to reduce accumulation and toxicity of heavy metals (metalloids) in plants: The main mechanisms, concerns, and risks. Sci. Total Environ. 2021, 771, 144776. [Google Scholar] [CrossRef]
- So, J.; Choe, D.H.; Rust, M.K.; Trumble, J.T.; Lee, C.Y. The impact of selenium on insects. J. Econ. Entomol. 2023, 116, 1041–1062. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.R.; Xian, L.M.; Yuan, L.X.; Lin, Z.Q.; Chen, X.R.; Wang, J.J.; Li, T. The use of selenium for controlling plant fungal diseases and insect pests. Front. Plant Sci. 2023, 14, 1102594. [Google Scholar] [CrossRef] [PubMed]
- Somalraju, A.; McCallum, J.L.; Main, D.; Peters, R.D.; Fofana, B. Foliar selenium application reduces late blight severity and incidence in potato and acts as a pathogen growth inhibitor and elicitor of induced plant defence. Can. J. Plant Pathol. 2022, 44, 39–55. [Google Scholar] [CrossRef]
- Xu, X.; Wang, J.H.; Wu, H.H.; Yuan, Q.H.; Wang, J.H.; Cui, J.; Lin, A.J. Effects of selenium fertilizer application and tomato varieties on tomato fruit quality: A meta-analysis. Sci. Hortic. 2022, 304, 111242. [Google Scholar] [CrossRef]
- Dima, S.-O.; Neamțu, C.; Desliu-Avram, M.; Ghiurea, M.; Capra, L.; Radu, E.; Stoica, R.; Faraon, V.-A.; Zamfiropol-Cristea, V.; Constantinescu-Aruxandei, D.; et al. Plant Biostimulant Effects of Baker’s Yeast Vinasse and Selenium on Tomatoes through Foliar Fertilization. Agronomy 2020, 10, 133. [Google Scholar] [CrossRef]
- Dima, Ș.-O.; Constantinescu-Aruxandei, D.; Tritean, N.; Ghiurea, M.; Capră, L.; Nicolae, C.-A.; Faraon, V.; Neamțu, C.; Oancea, F. Spectroscopic Analyses Highlight Plant Biostimulant Effects of Baker’s Yeast Vinasse and Selenium on Cabbage through Foliar Fertilization. Plants 2023, 12, 3016. [Google Scholar] [CrossRef]
- Sita, K.; Sehgal, A.; Bhardwaj, A.; Bhandari, K.; Jha, U.; Prasad, P.V.V.; Singh, S.; Kumar, S.; Siddique, K.H.M.; Nayyar, H. Selenium supplementation to lentil (Lens culinaris Medik.) under combined heat and drought stress improves photosynthetic ability, antioxidant systems, reproductive function and yield traits. Plant Soil 2023, 486, 7–23. [Google Scholar] [CrossRef]
- Constantinescu-Aruxandei, D.; Frîncu, R.M.; Capră, L.; Oancea, F. Selenium Analysis and Speciation in Dietary Supplements Based on Next-Generation Selenium Ingredients. Nutrients 2018, 10, 1466. [Google Scholar] [CrossRef]
- Cheng, B.X.; Wang, C.X.; Yue, L.; Chen, F.R.; Cao, X.S.; Lan, Q.Q.; Liu, T.X.; Wang, Z.Y. Selenium nanomaterials improve the quality of lettuce (Lactuca sativa L.) by modulating root growth, nutrient availability, and photosynthesis. Nanoimpact 2023, 29, 100449. [Google Scholar] [CrossRef]
- Khan, Z.; Thounaojam, T.C.; Chowdhury, D.; Upadhyaya, H. The role of selenium and nano selenium on physiological responses in plant: A review. Plant Growth Regul. 2023, 100, 409–433. [Google Scholar] [CrossRef]
- Song, J.; Yu, S.; Yang, R.; Xiao, J.; Liu, J. Opportunities for the use of selenium nanoparticles in agriculture. NanoImpact 2023, 31, 100478. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. The use of high-selenium yeast to raise selenium status: How does it measure up? Br. J. Nutr. 2004, 92, 557–573. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, M.; Salinas, G.; Gabaldón, T.; Gladyshev, V.N. Utilization of selenocysteine in early-branching fungal phyla. Nat. Microbiol. 2019, 4, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Blazejak, S.; Gientka, I.; Bzducha-Wróbel, A. Accumulation and metabolism of selenium by yeast cells. Appl. Microbiol. Biotechnol. 2015, 99, 5373–5382. [Google Scholar] [CrossRef]
- Nie, X.; Yang, X.; He, J.; Liu, P.; Shi, H.; Wang, T.; Zhang, D. Bioconversion of inorganic selenium to less toxic selenium forms by microbes: A review. Front. Bioeng. Biotechnol. 2023, 11, 1167123. [Google Scholar] [CrossRef]
- Fuentes-Cervantes, A.; Ruiz Allica, J.; Calderón Celis, F.; Costa-Fernández, J.M.; Ruiz Encinar, J. The Potential of ICP-MS as a Complementary Tool in Nanoparticle–Protein Corona Analysis. Nanomaterials 2023, 13, 1132. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, K.S. Role of nano-selenium in health and environment. J. Biotechnol. 2021, 325, 152–163. [Google Scholar] [CrossRef]
- Devi, M.S.; Srinivasan, S.; Muthuvel, A. Selenium nanomaterial is a promising nanotechnology for biomedical and environmental remediation: A detailed review. Biocatal. Agric. Biotechnol. 2023, 51, 102766. [Google Scholar] [CrossRef]
- Manjunatha, C.; Rao, P.P.; Bhardwaj, P.; Raju, H.; Ranganath, D. New insight into the synthesis, morphological architectures and biomedical applications of elemental selenium nanostructures. Biomed. Mater. 2021, 16, 022010. [Google Scholar] [CrossRef]
- Hadrup, N.; Ravn-Haren, G. Absorption, distribution, metabolism and excretion (ADME) of oral selenium from organic and inorganic sources: A review. J. Trace Elem. Med. Biol. 2021, 67, 126801. [Google Scholar] [CrossRef]
- Gu, X.; Gao, C.Q. New horizons for selenium in animal nutrition and functional foods. Anim. Nutr. 2022, 11, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, L.; Cattaneo, D.; Abbate, R.; Manoni, M.; Ottoboni, M.; Luciano, A.; von Holst, C.; Pinotti, L. Advances in selenium supplementation: From selenium-enriched yeast to potential selenium-enriched insects, and selenium nanoparticles. Anim. Nutr. 2023, 14, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Parvin, S.; Khan, S.; Alam, P.; Khan, T.H.; Khataibeh, M.; Khan, M.A.; Samad, A.; Baker, A.; Mansoor, S. A Review on Potentialities of Selenium Nanoparticles and Its Application Using Air Borne Fungus. Appl. Ecol. Environ. Sci. 2021, 9, 607–612. [Google Scholar] [CrossRef]
- Bisht, N.; Phalswal, P.; Khanna, P.K. Selenium nanoparticles: A review on synthesis and biomedical applications. Mater. Adv. 2022, 3, 1415–1431. [Google Scholar] [CrossRef]
- Zambonino, M.C.; Quizhpe, E.M.; Mouheb, L.; Rahman, A.; Agathos, S.N.; Dahoumane, S.A. Biogenic Selenium Nanoparticles in Biomedical Sciences: Properties, Current Trends, Novel Opportunities and Emerging Challenges in Theranostic Nanomedicine. Nanomaterials 2023, 13, 424. [Google Scholar] [CrossRef]
- Garza-García, J.J.; Hernández-Díaz, J.A.; Zamudio-Ojeda, A.; León-Morales, J.M.; Guerrero-Guzmán, A.; Sánchez-Chiprés, D.R.; López-Velázquez, J.C.; García-Morales, S. The role of selenium nanoparticles in agriculture and food technology. Biol. Trace Elem. Res. 2021, 200, 1–21. [Google Scholar] [CrossRef]
- Bano, I.; Skalickova, S.; Sajjad, H.; Skladanka, J.; Horky, P. Uses of selenium nanoparticles in the plant production. Agronomy 2021, 11, 2229. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Rehman, M.Z.U.; Rinklebe, J.; Tsang, D.C.; Tack, F.M.; Abbasi, G.H.; Hussain, A.; Igalavithana, A.D.; Lee, B.C. Effects of selenium on the uptake of toxic trace elements by crop plants: A review. Crit. Rev. Environ. Sci. Technol. 2021, 51, 2531–2566. [Google Scholar] [CrossRef]
- Prasad, K.S.; Vaghasiya, J.V.; Soni, S.S.; Patel, J.; Patel, R.; Kumari, M.; Jasmani, F.; Selvaraj, K. Microbial selenium nanoparticles (SeNPs) and their application as a sensitive hydrogen peroxide biosensor. Appl. Biochem. Biotechnol. 2015, 177, 1386–1393. [Google Scholar] [CrossRef]
- Ahmed, F.; Dwivedi, S.; Shaalan, N.M.; Kumar, S.; Arshi, N.; Alshoaibi, A.; Husain, F.M. Development of selenium nanoparticle based agriculture sensor for heavy metal toxicity detection. Agriculture 2020, 10, 610. [Google Scholar] [CrossRef]
- Dutta, D.; Reddy, K.S.; Badhulika, S. Exfoliated Se nanoparticles decorated on MoS2/paper-based heterojunction as a flexible, self-powered and highly responsive photodetector. Mater. Sci. Semicond. Process. 2023, 164, 107610. [Google Scholar] [CrossRef]
- Blinov, A.; Maglakelidze, D.; Yasnaya, M.; Gvozdenko, A.; Blinova, A.; Golik, A.; Slyadneva, K.; Pirogov, M. Synthesis of selenium nanoparticles stabilized by quaternary ammonium compounds. Russ. J. Gen. Chem. 2022, 92, 424–429. [Google Scholar] [CrossRef]
- Kong, H.; Yang, J.; Zhang, Y.; Fang, Y.; Nishinari, K.; Phillips, G.O. Synthesis and antioxidant properties of gum arabic-stabilized selenium nanoparticles. Int. J. Biol. Macromol. 2014, 65, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Guo, X.; Lei, Y.; Dennis, B.S.; Wu, S.; Wu, C. Synthesis and characterization of selenium–chondroitin sulfate nanoparticles. Carbohydr. Polym. 2012, 90, 122–126. [Google Scholar] [CrossRef]
- Zhai, X.; Zhang, C.; Zhao, G.; Stoll, S.; Ren, F.; Leng, X. Antioxidant capacities of the selenium nanoparticles stabilized by chitosan. J. Nanobiotechnol. 2017, 15, 1–12. [Google Scholar] [CrossRef]
- Apryatina, K.V.; Murach, E.I.; Amarantov, S.V.; Erlykina, E.I.; Veselov, V.S.; Smirnova, L.A. Synthesis of a Bioactive Composition of Chitosan-Selenium Nanoparticles. Appl. Biochem. Microbiol. 2022, 58, 126–131. [Google Scholar] [CrossRef]
- Lesnichaya, M.; Shendrik, R.; Titov, E.; Sukhov, B. Synthesis and comparative assessment of antiradical activity, toxicity, and biodistribution of κ-carrageenan-capped selenium nanoparticles of different size: In vivo and in vitro study. IET Nanobiotechnol. 2020, 14, 519–526. [Google Scholar] [CrossRef]
- Modrzejewska-Sikorska, A.; Konował, E.; Klapiszewski, Ł.; Nowaczyk, G.; Jurga, S.; Jesionowski, T.; Milczarek, G. Lignosulfonate-stabilized selenium nanoparticles and their deposition on spherical silica. Int. J. Biol. Macromol. 2017, 103, 403–408. [Google Scholar] [CrossRef]
- Wang, H.; Xu, M.-Z.; Liang, X.-Y.; Nag, A.; Zeng, Q.-Z.; Yuan, Y. Fabrication of food grade zein-dispersed selenium dual-nanoparticles with controllable size, cell friendliness and oral bioavailability. Food Chem. 2023, 398, 133878. [Google Scholar] [CrossRef]
- Zhang, J.; Teng, Z.; Yuan, Y.; Zeng, Q.-Z.; Lou, Z.; Lee, S.-H.; Wang, Q. Development, physicochemical characterization and cytotoxicity of selenium nanoparticles stabilized by beta-lactoglobulin. Int. J. Biol. Macromol. 2018, 107, 1406–1413. [Google Scholar] [CrossRef]
- Borovikova, L.; Titova, A.; Matveeva, N.; Pisarev, O. Stabilizing selenium nanoparticles with chymotrypsin: The effect of pH and nanoparticle-enzyme concentration ratios on the stability of nanocomplexes. Russ. J. Phys. Chem. A 2013, 87, 998–1001. [Google Scholar] [CrossRef]
- Ye, M.-J.; Xu, Q.-L.; Tang, H.-Y.; Jiang, W.-Y.; Su, D.-X.; He, S.; Zeng, Q.-Z.; Yuan, Y. Development and stability of novel selenium colloidal particles complex with peanut meal peptides. LWT 2020, 126, 109280. [Google Scholar] [CrossRef]
- Tang, H.-Y.; Huang, Q.; Wang, Y.-L.; Yang, X.-Q.; Su, D.-X.; He, S.; Tan, J.-C.; Zeng, Q.-Z.; Yuan, Y. Development, structure characterization and stability of food grade selenium nanoparticles stabilized by tilapia polypeptides. J. Food Eng. 2020, 275, 109878. [Google Scholar] [CrossRef]
- Hashemi-Firouzi, N.; Afshar, S.; Asl, S.S.; Samzadeh-Kermani, A.; Gholamigeravand, B.; Amiri, K.; Majidi, M.; Shahidi, S. The effects of polyvinyl alcohol-coated selenium nanoparticles on memory impairment in rats. Metab. Brain Dis. 2022, 37, 3011–3021. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S.; Chen, J.; Divya, M.; Durán-Lara, E.F.; Prasannakumar, M.; Vaseeharan, B. A Review on Biogenic Synthesis of Selenium Nanoparticles and Its Biological Applications. J. Inorg. Organomet. Polym. Mater. 2022, 32, 2355–2370. [Google Scholar] [CrossRef]
- Arunthirumeni, M.; Veerammal, V.; Shivakumar, M.S. Biocontrol Efficacy of Mycosynthesized Selenium Nanoparticle Using Trichoderma sp. on Insect Pest Spodoptera litura. J. Clust. Sci. 2022, 33, 1645–1653. [Google Scholar] [CrossRef]
- Reddy, B.; Bandi, R. Synthesis of selenium nanoparticles by using microorganisms and agri-based products. In Agri-Waste and Microbes for Production of Sustainable Nanomaterials; Elsevier: Amsterdam, The Netherlands, 2022; pp. 655–683. [Google Scholar]
- Mates, I.; Antoniac, I.; Laslo, V.; Vicas, S.; Brocks, M.; Fritea, L.; Milea, C.; Mohan, A.; Cavalu, S. Selenium nanoparticles: Production, characterization and possible applications in biomedicine and food science. Sci. Bull. B Chem. Mater. Sci. UPB 2019, 81, 205–216. [Google Scholar]
- Zhang, H.; Zhou, H.; Bai, J.; Li, Y.; Yang, J.; Ma, Q.; Qu, Y. Biosynthesis of selenium nanoparticles mediated by fungus Mariannaea sp. HJ and their characterization. Colloids Surf. A Physicochem. Eng. Asp. 2019, 571, 9–16. [Google Scholar] [CrossRef]
- Ghosh, S.; Ahmad, R.; Zeyaullah, M.; Khare, S.K. Microbial nano-factories: Synthesis and biomedical applications. Front. Chem. 2021, 9, 626834. [Google Scholar] [CrossRef]
- Alghuthaymi, M.A.; Abd-Elsalam, K.A.; AboDalam, H.M.; Ahmed, F.K.; Ravichandran, M.; Kalia, A.; Rai, M. Trichoderma: An Eco-Friendly Source of Nanomaterials for Sustainable Agroecosystems. J. Fungi 2022, 8, 367. [Google Scholar] [CrossRef] [PubMed]
- Fouda, A.; Hassan, S.E.-D.; Eid, A.M.; Abdel-Rahman, M.A.; Hamza, M.F. Light enhanced the antimicrobial, anticancer, and catalytic activities of selenium nanoparticles fabricated by endophytic fungal strain, Penicillium crustosum EP-1. Sci. Rep. 2022, 12, 11834. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, A.; Klimek-Ochab, M. Fungal synthesis of size-defined nanoparticles. Adv. Nat. Sci. Nanosci. Nanotechnol. 2017, 8, 043001. [Google Scholar] [CrossRef]
- Abd-Elsalam, K.A. Fungal Cell Factories for Sustainable Nanomaterials Productions and Agricultural Applications; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Srivastava, N.; Mukhopadhyay, M. Biosynthesis and structural characterization of selenium nanoparticles using Gliocladium roseum. J. Clust. Sci. 2015, 26, 1473–1482. [Google Scholar] [CrossRef]
- Zare, B.; Babaie, S.; Setayesh, N.; Shahverdi, A.R. Isolation and characterization of a fungus for extracellular synthesis of small selenium nanoparticles. Nanomed. J. 2013, 1, 13–19. [Google Scholar]
- Sarkar, J.; Dey, P.; Saha, S.; Acharya, K. Mycosynthesis of selenium nanoparticles. Micro Nano Lett. 2011, 6, 599–602. [Google Scholar] [CrossRef]
- Mosallam, F.M.; El-Sayyad, G.S.; Fathy, R.M.; El-Batal, A.I. Biomolecules-mediated synthesis of selenium nanoparticles using Aspergillus oryzae fermented Lupin extract and gamma radiation for hindering the growth of some multidrug-resistant bacteria and pathogenic fungi. Microb. Pathog. 2018, 122, 108–116. [Google Scholar] [CrossRef]
- Amin, M.A.; Ismail, M.A.; Badawy, A.A.; Awad, M.A.; Hamza, M.F.; Awad, M.F.; Fouda, A. The Potency of fungal-fabricated selenium nanoparticles to improve the growth performance of Helianthus annuus L. and control of cutworm Agrotis ipsilon. Catalysts 2021, 11, 1551. [Google Scholar] [CrossRef]
- Sarkar, J.; Mridha, D.; Davoodbasha, M.A.; Banerjee, J.; Chanda, S.; Ray, K.; Roychowdhury, T.; Acharya, K.; Sarkar, J. A State-of-the-Art Systemic Review on Selenium Nanoparticles: Mechanisms and Factors Influencing Biogenesis and Its Potential Applications. Biol. Trace Elem. Res. 2023, 201, 5000–5036. [Google Scholar] [CrossRef]
- Hashem, A.H.; Abdelaziz, A.M.; Attia, M.S.; Salem, S.S. Selenium and Nano-Selenium-Mediated Biotic Stress Tolerance in Plants. In Selenium and Nano-Selenium in Environmental Stress Management and Crop Quality Improvement; Hossain, M.A., Ahammed, G.J., Kolbert, Z., El-Ramady, H., Islam, T., Schiavon, M., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 209–226. [Google Scholar] [CrossRef]
- Samynathan, R.; Venkidasamy, B.; Ramya, K.; Muthuramalingam, P.; Shin, H.; Kumari, P.S.; Thangavel, S.; Sivanesan, I. A Recent Update on the Impact of Nano-Selenium on Plant Growth, Metabolism, and Stress Tolerance. Plants 2023, 12, 853. [Google Scholar] [CrossRef]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The Current Status of Its Application in Agriculture for the Biocontrol of Fungal Phytopathogens and Stimulation of Plant Growth. Int. J. Mol. Sci. 2022, 23, 2329. [Google Scholar] [CrossRef] [PubMed]
- Ogunleye, G.E.; Adebayo-Tayo, B.C.; Oyinlola, K.A. Biological evaluation of extracellular mycosynthesized silver nanoparticles by Trichoderma asperellum. BioMetals 2023, 36, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Castro-Longoria, E. Biosynthesis of Metal-Based NanoparticlesNanoparticles by TrichodermaTrichodermaand Its Potential Applications. In Advances in Trichoderma Biology for Agricultural Applications; Amaresan, N., Sankaranarayanan, A., Dwivedi, M.K., Druzhinina, I.S., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 433–463. [Google Scholar] [CrossRef]
- Khabat, V.; Mansoori, G.A.; Karimi, S. Biosynthesis of silver nanoparticles by fungus Trichoderma reesei. Insci. J. 2011, 1, 65–79. [Google Scholar]
- Gharieb, M.; Wilkinson, S.; Gadd, G. Reduction of selenium oxyanions by unicellular, polymorphic and filamentous fungi: Cellular location of reduced selenium and implications for tolerance. J. Ind. Microbiol. 1995, 14, 300–311. [Google Scholar] [CrossRef]
- Nandini, B.; Hariprasad, P.; Prakash, H.S.; Shetty, H.S.; Geetha, N. Trichogenic-selenium nanoparticles enhance disease suppressive ability of Trichoderma against downy mildew disease caused by Sclerospora graminicola in pearl millet. Sci. Rep. 2017, 7, 2612. [Google Scholar] [CrossRef]
- Hu, D.; Yu, S.; Yu, D.; Liu, N.; Tang, Y.; Fan, Y.; Wang, C.; Wu, A. Biogenic Trichoderma harzianum-derived selenium nanoparticles with control functionalities originating from diverse recognition metabolites against phytopathogens and mycotoxins. Food Control 2019, 106, 106748. [Google Scholar] [CrossRef]
- Liang, X.; Perez, M.A.M.-J.; Nwoko, K.C.; Egbers, P.; Feldmann, J.; Csetenyi, L.; Gadd, G.M. Fungal formation of selenium and tellurium nanoparticles. Appl. Microbiol. Biotechnol. 2019, 103, 7241–7259. [Google Scholar] [CrossRef]
- Diko, C.S.; Zhang, H.; Lian, S.; Fan, S.; Li, Z.; Qu, Y. Optimal synthesis conditions and characterization of selenium nanoparticles in Trichoderma sp. WL-Go culture broth. Mater. Chem. Phys. 2020, 246, 122583. [Google Scholar] [CrossRef]
- Joshi, S.M.; De Britto, S.; Jogaiah, S.; Ito, S.-i. Mycogenic Selenium Nanoparticles as Potential New Generation Broad Spectrum Antifungal Molecules. Biomolecules 2019, 9, 419. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Sathiyaseelan, A.; Zhang, X.; Park, S.; Wang, M.-H. Purinoceptor targeted cytotoxicity of adenosine triphosphate-conjugated biogenic selenium nanoparticles in human colon cancer cells. Pharmaceuticals 2022, 15, 582. [Google Scholar] [CrossRef]
- Joshi, S.M.; De Britto, S.; Jogaiah, S. Myco-engineered selenium nanoparticles elicit resistance against tomato late blight disease by regulating differential expression of cellular, biochemical and defense responsive genes. J. Biotechnol. 2021, 325, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Lee, S.Y. Biosynthesis of inorganic nanomaterials using microbial cells and bacteriophages. Nat. Rev. Chem. 2020, 4, 638–656. [Google Scholar] [CrossRef] [PubMed]
- Shoeibi, S.; Mozdziak, P.; Golkar-Narenji, A. Biogenesis of Selenium Nanoparticles Using Green Chemistry. Top. Curr. Chem. 2017, 375, 88. [Google Scholar] [CrossRef]
- Zambonino, M.C.; Quizhpe, E.M.; Jaramillo, F.E.; Rahman, A.; Santiago Vispo, N.; Jeffryes, C.; Dahoumane, S.A. Green synthesis of selenium and tellurium nanoparticles: Current trends, biological properties and biomedical applications. Int. J. Mol. Sci. 2021, 22, 989. [Google Scholar] [CrossRef] [PubMed]
- Tsivileva, O.; Pozdnyakov, A.; Ivanova, A. Polymer nanocomposites of selenium biofabricated using fungi. Molecules 2021, 26, 3657. [Google Scholar] [CrossRef]
- Annamalai, J.; Ganesan, S.; Murugan, K.; Janjaroen, D. Recent breakthroughs set by fungal enzymes in the biosynthesis of nanoparticles. In Fungal Cell Factories for Sustainable Nanomaterials Productions and Agricultural Applications; Elsevier: Amsterdam, The Netherlands, 2023; pp. 131–162. [Google Scholar]
- Bahrulolum, H.; Nooraei, S.; Javanshir, N.; Tarrahimofrad, H.; Mirbagheri, V.S.; Easton, A.J.; Ahmadian, G. Green synthesis of metal nanoparticles using microorganisms and their application in the agrifood sector. J. Nanobiotechnol. 2021, 19, 1–26. [Google Scholar] [CrossRef]
- Dhillon, G.S.; Brar, S.K.; Kaur, S.; Verma, M. Green approach for nanoparticle biosynthesis by fungi: Current trends and applications. Crit. Rev. Biotechnol. 2012, 32, 49–73. [Google Scholar] [CrossRef]
- Khandel, P.; Shahi, S.K. Mycogenic nanoparticles and their bio-prospective applications: Current status and future challenges. J. Nanostruct. Chem. 2018, 8, 369–391. [Google Scholar] [CrossRef]
- Li, G.; He, D.; Qian, Y.; Guan, B.; Gao, S.; Cui, Y.; Yokoyama, K.; Wang, L. Fungus-mediated green synthesis of silver nanoparticles using Aspergillus terreus. Int. J. Mol. Sci. 2012, 13, 466–476. [Google Scholar] [CrossRef]
- Liu, S.-Y.; Lo, C.-T.; Chen, C.; Liu, M.-Y.; Chen, J.-H.; Peng, K.-C. Efficient isolation of anthraquinone-derivatives from Trichoderma harzianum ETS 323. J. Biochem. Biophys. Methods 2007, 70, 391–395. [Google Scholar] [CrossRef]
- Tugarova, A.V.; Kamnev, A.A. Proteins in microbial synthesis of selenium nanoparticles. Talanta 2017, 174, 539–547. [Google Scholar] [CrossRef]
- Gremel, G.; Dorrer, M.; Schmoll, M. Sulphur metabolism and cellulase gene expression are connected processes in the filamentous fungus Hypocrea jecorina (anamorph Trichoderma reesei). BMC Microbiol. 2008, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhu, S.; Wang, Q.; Chen, L.; Li, Y.; Xu, S.; Gu, Z.; Shi, G.; Ding, Z. Pivotal biological processes and proteins for selenite reduction and methylation in Ganoderma lucidum. J. Hazard. Mater. 2023, 444, 130409. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, J.; Chen, N.; Bai, H.; Qu, L.; Yang, Y.; Ye, H.; Xiao, H.; Yan, H.; Zhang, T. Antioxidant Activity and Transcriptomic Analysis of Se-Enriched Golden Oyster Mushroom Pleurotus citrinopileatus (Agaricomycetes). Int. J. Med. Mushrooms 2020, 22, 755–762. [Google Scholar] [CrossRef]
- Pinel-Cabello, M.; Chapon, V.; Ruiz-Fresneda, M.A.; Alpha-Bazin, B.; Berthomieu, C.; Armengaud, J.; Merroun, M.L. Delineation of cellular stages and identification of key proteins for reduction and biotransformation of Se(IV) by Stenotrophomonas bentonitica BII-R7. J. Hazard. Mater. 2021, 418, 126150. [Google Scholar] [CrossRef] [PubMed]
- Valueva, S.; Titova, A.; Borovikova, L. Selenium-containing nanosystems based on biocompatible polymer stabilizers: Kinetics, morphology, and thermodynamics. Russ. J. Phys. Chem. A 2015, 89, 1633–1637. [Google Scholar] [CrossRef]
- Song, X.; Chen, Y.; Sun, H.; Liu, X.; Leng, X. Physicochemical stability and functional properties of selenium nanoparticles stabilized by chitosan, carrageenan, and gum Arabic. Carbohydr. Polym. 2021, 255, 117379. [Google Scholar] [CrossRef]
- Chakraborty, D.; Chauhan, P.; Alex, S.A.; Chaudhary, S.; Ethiraj, K.R.; Chandrasekaran, N.; Mukherjee, A. Comprehensive study on biocorona formation on functionalized selenium nanoparticle and its biological implications. J. Mol. Liq. 2018, 268, 335–342. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Ferrante, F.; Cavallaro, G.; Alduina, R.; Chillura Martino, D.F. Biogenic Selenium Nanoparticles: A Fine Characterization to Unveil Their Thermodynamic Stability. Nanomaterials 2021, 11, 1195. [Google Scholar] [CrossRef]
- Li, K.; Li, J.; Zhang, S.; Zhang, J.; Xu, Q.; Xu, Z.; Guo, Y. Amorphous structure and crystal stability determine the bioavailability of selenium nanoparticles. J. Hazard. Mater. 2024, 465, 133287. [Google Scholar] [CrossRef]
- Tritean, N.; Dima, Ș.-O.; Trica, B.; Stoica, R.; Ghiurea, M.; Moraru, I.; Cimpean, A.; Oancea, F.; Constantinescu-Aruxandei, D. Selenium-Fortified Kombucha-Pollen Beverage by In Situ Biosynthesized Selenium Nanoparticles with High Biocompatibility and Antioxidant Activity. Antioxidants 2023, 12, 1711. [Google Scholar] [CrossRef] [PubMed]
- Dobias, J.; Suvorova, E.I.; Bernier-Latmani, R. Role of proteins in controlling selenium nanoparticle size. Nanotechnology 2011, 22, 195605. [Google Scholar] [CrossRef] [PubMed]
- Tritean, N.; Dimitriu, L.; Dima, Ș.-O.; Stoica, R.; Trică, B.; Ghiurea, M.; Moraru, I.; Cimpean, A.; Oancea, F.; Constantinescu-Aruxandei, D. Cytocompatibility, Antimicrobial and Antioxidant Activity of a Mucoadhesive Biopolymeric Hydrogel Embedding Selenium Nanoparticles Phytosynthesized by Sea Buckthorn Leaf Extract. Pharmaceuticals 2024, 17, 23. [Google Scholar] [CrossRef]
- Perez-Potti, A.; Lopez, H.; Pelaz, B.; Abdelmonem, A.; Soliman, M.G.; Schoen, I.; Kelly, P.M.; Dawson, K.A.; Parak, W.J.; Krpetic, Z.; et al. In depth characterisation of the biomolecular coronas of polymer coated inorganic nanoparticles with differential centrifugal sedimentation. Sci. Rep. 2021, 11, 6443. [Google Scholar] [CrossRef]
- Nwoko, K.C.; Liang, X.; Perez, M.A.M.J.; Krupp, E.; Gadd, G.M.; Feldmann, J. Characterisation of selenium and tellurium nanoparticles produced by Aureobasidium pullulans using a multi-method approach. J. Chromatogr. A 2021, 1642, 462022. [Google Scholar] [CrossRef] [PubMed]
- Lapresta-Fernández, A.; Salinas-Castillo, A.; de la Llana, S.A.; Costa-Fernández, J.M.; Domínguez-Meister, S.; Cecchini, R.; Capitán-Vallvey, L.F.; Moreno-Bondi, M.C.; Marco, M.P.; Sánchez-López, J.C.; et al. A General Perspective of the Characterization and Quantification of Nanoparticles: Imaging, Spectroscopic, and Separation Techniques. Crit. Rev. Solid State Mater. Sci. 2014, 39, 423–458. [Google Scholar] [CrossRef]
- Nikam, P.B.; Salunkhe, J.D.; Minkina, T.; Rajput, V.D.; Kim, B.S.; Patil, S.V. A review on green synthesis and recent applications of red nano Selenium. Results Chem. 2022, 4, 100581. [Google Scholar] [CrossRef]
- Maity, M.; Bera, K.; Pal, U.; Khamaru, K.; Maiti, N.C. Sensing of Iron(III) Ion via Modulation of Redox Potential on Biliverdin Protected Silver Nanosurface. ACS Appl. Nano Mater. 2018, 1, 6099–6111. [Google Scholar] [CrossRef]
- Kesharwani, P.; Ma, R.Y.; Sang, L.; Fatima, M.; Sheikh, A.; Abourehab, M.A.S.; Gupta, N.; Chen, Z.S.; Zhou, Y. Gold nanoparticles and gold nanorods in the landscape of cancer therapy. Mol. Cancer 2023, 22, 31. [Google Scholar] [CrossRef]
- Oliveira, A.E.F.; Pereira, A.C.; Resende, M.A.C.; Ferreira, L.F. Gold Nanoparticles: A Didactic Step-by-Step of the Synthesis Using the Turkevich Method, Mechanisms, and Characterizations. Analytica 2023, 4, 250–263. [Google Scholar] [CrossRef]
- Akar, N.; Asar, B.; Dizge, N.; Koyuncu, I. Investigation of characterization and biofouling properties of PES membrane containing selenium and copper nanoparticles. J. Membr. Sci. 2013, 437, 216–226. [Google Scholar] [CrossRef]
- Wiley, B.J.; Im, S.H.; Li, Z.Y.; McLellan, J.; Siekkinen, A.; Xia, Y.N. Maneuvering the surface plasmon resonance of silver nanostructures through shape-controlled synthesis. J. Phys. Chem. B 2006, 110, 15666–15675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.P.; He, J.F.; Wang, Y.M.; Liu, F.F. Terahertz beat oscillation of plasmonic electrons interacting with femtosecond light pulses. Sci. Rep. 2016, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.C.; Barton, L.L.; Tsui, W.L.; Shuman, K.; Gillespie, J.; Eze, C.S. A novel method for the measurement of elemental selenium produced by bacterial reduction of selenite. J. Microbiol. Methods 2011, 86, 140–144. [Google Scholar] [CrossRef] [PubMed]
- El-Sayyad, G.S.; El-Bastawisy, H.S.; Gobara, M.; El-Batal, A.I. Gentamicin-assisted mycogenic selenium nanoparticles synthesized under gamma irradiation for robust reluctance of resistant urinary tract infection-causing pathogens. Biol. Trace Elem. Res. 2020, 195, 323–342. [Google Scholar] [CrossRef]
- Hamley, I.W. Small-Angle Scattering: Theory, Instrumentation, Data and Applications; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2022; p. 278. [Google Scholar]
- Gommes, C.J.; Jaksch, S.; Frielinghaus, H. Small-angle scattering for beginners. J. Appl. Crystallogr. 2021, 54, 1832–1843. [Google Scholar] [CrossRef]
- Helmholdt, R.B.; Sonneveld, E.J.; Schenk, H. The crystal structure of sodium selenite Na2SeO3 from powder diffraction data. Z. Für Krist. 1999, 214, 151–153. [Google Scholar] [CrossRef]
- Shubharani, R.; Mahesh, M.; Yogananda Murthy, V. Biosynthesis and characterization, antioxidant and antimicrobial activities of selenium nanoparticles from ethanol extract of Bee Propolis. J. Nanomed. Nanotechnol. 2019, 10, 1000522. [Google Scholar]
- Li, T.; Senesi, A.J.; Lee, B. Small Angle X-ray Scattering for Nanoparticle Research. Chem. Rev. 2016, 116, 11128–11180. [Google Scholar] [CrossRef]
- Turkevich, J.; Hubbell, H.H. Low Angle X-Ray Diffraction of Colloidal Gold and Carbon Black1a. J. Am. Chem. Soc. 1951, 73, 1–7. [Google Scholar] [CrossRef]
- Gablech, E.; Fohlerova, Z.; Svec, K.; Zales, F.; Benada, O.; Kofronova, O.; Pekarkova, J.; Caha, O.; Gablech, I.; Gabriel, J.; et al. Selenium nanoparticles with boron salt-based compound act synergistically against the brown-rot Serpula lacrymans. Int. Biodeterior. Biodegrad. 2022, 169, 105377. [Google Scholar] [CrossRef]
- Borgatta, J.R.; Lochbaum, C.A.; Elmer, W.H.; White, J.C.; Pedersen, J.A.; Hamers, R.J. Biomolecular corona formation on CuO nanoparticles in plant xylem fluid. Environ. Sci. Nano 2021, 8, 1067–1080. [Google Scholar] [CrossRef]
- Chen, W.; Li, X.; Cheng, H.; Zhan, X.; Xia, W. Synthesis, characterization, and anticancer activity of protamine sulfate stabilized selenium nanoparticles. Food Res. Int. 2023, 164, 112435. [Google Scholar] [CrossRef] [PubMed]
- Dahman, Y. (Ed.) Chapter 5—Nanoparticles. In Nanotechnology and Functional Materials for Engineers; Elsevier: Amsterdam, The Netherlands, 2017; pp. 93–119. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential–what they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Jia, X.W.; Liu, Q.Y.; Zou, S.W.; Xu, X.J.; Zhang, L.N. Construction of selenium nanoparticles/beta-glucan composites for enhancement of the antitumor activity. Carbohydr. Polym. 2015, 117, 434–442. [Google Scholar] [CrossRef]
- Blinova, A.; Blinov, A.; Kravtsov, A.; Nagdalian, A.; Rekhman, Z.; Gvozdenko, A.; Kolodkin, M.; Filippov, D.; Askerova, A.; Golik, A.; et al. Synthesis, Characterization and Potential Antimicrobial Activity of Selenium Nanoparticles Stabilized with Cetyltrimethylammonium Chloride. Nanomaterials 2023, 13, 3128. [Google Scholar] [CrossRef]
- Filipović, N.; Ušjak, D.; Milenković, M.T.; Zheng, K.; Liverani, L.; Boccaccini, A.R.; Stevanović, M.M. Comparative Study of the Antimicrobial Activity of Selenium Nanoparticles With Different Surface Chemistry and Structure. Front. Bioeng. Biotechnol. 2021, 8, 624621. [Google Scholar] [CrossRef]
- Ashkenasy, G.; Hermans, T.M.; Otto, S.; Taylor, A.F. Systems chemistry. Chem. Soc. Rev. 2017, 46, 2543–2554. [Google Scholar] [CrossRef]
- Archer, W.R.; Schulz, M.D. Isothermal titration calorimetry: Practical approaches and current applications in soft matter. Soft Matter 2020, 16, 8760–8774. [Google Scholar] [CrossRef]
- Zhao, M.; Wu, Y.; Zhang, F.; Zheng, S.; Wang, L.; Bai, J.; Yang, Y. Preparation of Ribes nigrum L. polysaccharides-stabilized selenium nanoparticles for enhancement of the anti-glycation and α-glucosidase inhibitory activities. Int. J. Biol. Macromol. 2023, 253, 127122. [Google Scholar] [CrossRef]
- Jiang, W.; He, S.; Su, D.; Ye, M.; Zeng, Q.; Yuan, Y. Synthesis, characterization of tuna polypeptide selenium nanoparticle, and its immunomodulatory and antioxidant effects in vivo. Food Chem. 2022, 383, 132405. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Valdespino, C.A.; Orrantia-Borunda, E. Trichoderma and nanotechnology in sustainable agriculture: A review. Front. Fungal Biol. 2021, 2, 764675. [Google Scholar] [CrossRef]
- Shahbaz, M.; Akram, A.; Raja, N.I.; Mukhtar, T.; Mashwani, Z.U.; Mehak, A.; Fatima, N.; Sarwar, S.; Haq, E.U.; Yousaf, T. Green synthesis and characterization of selenium nanoparticles and its application in plant disease management: A review. Pak. J. Phytopathol. 2022, 34, 189–202. [Google Scholar] [CrossRef]
- Helmy, E.; Salah, R.; El-Shazly, M.M.; Alqhtani, A.; Pokoo-Aikins, A.; Yosri, M. Investigation of the Impact of Mycogenic Titanium and Selenium Nanoparticles on Fusarium Wilt Infection of Tomato Plant. J. Pure Appl. Microbiol. 2023, 17, 1800–1813. [Google Scholar] [CrossRef]
- Bărbieru, O.-G.; Dimitriu, L.; Călin, M.; Răut, I.; Constantinescu-Aruxandei, D.; Oancea, F. Plant Biostimulants Based on Selenium Nanoparticles Biosynthesized by Trichoderma Strains. Proceedings 2019, 29, 95. [Google Scholar]
- Zohra, E.; Ikram, M.; Omar, A.A.; Hussain, M.; Satti, S.H.; Raja, N.I.; Ehsan, M. Potential applications of biogenic selenium nanoparticles in alleviating biotic and abiotic stresses in plants: A comprehensive insight on the mechanistic approach and future perspectives. Green Process. Synth. 2021, 10, 456–475. [Google Scholar] [CrossRef]
- Fernando, D.; Milagrosa, S.; Francisco, C.; Francisco, M. Biostimulant Activity of Trichoderma saturnisporum in Melon (Cucumis melo). Hortscience 2018, 53, 810–815. [Google Scholar] [CrossRef]
- de Sousa, W.N.; Brito, N.F.; Felsemburgh, C.A.; Vieira, T.A.; Lustosa, D.C. Evaluation of Trichoderma spp. Isolates in Cocoa Seed Treatment and Seedling Production. Plants 2021, 10, 1964. [Google Scholar] [CrossRef]
- Nagdalian, A.A.; Blinov, A.V.; Siddiqui, S.A.; Gvozdenko, A.A.; Golik, A.B.; Maglakelidze, D.G.; Rzhepakovsky, I.V.; Kukharuk, M.Y.; Piskov, S.I.; Rebezov, M.B.; et al. Effect of selenium nanoparticles on biological and morphofunctional parameters of barley seeds (Hordéum vulgáre L.). Sci. Rep. 2023, 13, 6453. [Google Scholar] [CrossRef]
- Manzar, N.; Kashyap, A.S.; Goutam, R.S.; Rajawat, M.V.S.; Sharma, P.K.; Sharma, S.K.; Singh, H.V. Trichoderma: Advent of Versatile Biocontrol Agent, Its Secrets and Insights into Mechanism of Biocontrol Potential. Sustainability 2022, 14, 12786. [Google Scholar] [CrossRef]
- Guzmán-Guzmán, P.; Kumar, A.; de los Santos-Villalobos, S.; Parra-Cota, F.I.; Orozco-Mosqueda, M.D.; Fadiji, A.E.; Hyder, S.; Babalola, O.O.; Santoyo, G. Trichoderma Species: Our Best Fungal Allies in the Biocontrol of Plant Diseases—A Review. Plants 2023, 12, 432. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.O.; De Mio, L.L.M.; Soccol, C.R. Trichoderma as a powerful fungal disease control agent for a more sustainable and healthy agriculture: Recent studies and molecular insights. Planta 2023, 257, 31. [Google Scholar] [CrossRef] [PubMed]
- Konappa, N.; Krishnamurthy, S.; Siddaiah, C.N.; Ramachandrappa, N.S.; Chowdappa, S. Evaluation of biological efficacy of Trichoderma asperellum against tomato bacterial wilt caused by Ralstonia solanacearum. Egypt. J. Biol. Pest Control 2018, 28, 1–11. [Google Scholar] [CrossRef]
- Kai, K.; Mine, K.; Akiyama, K.; Ohki, S.; Hayashi, H. Anti-plant viral activity of peptaibols, trichorzins HA II, HA V, and HA VI, isolated from Trichoderma harzianum HK-61. J. Pestic. Sci. 2018, 43, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Sharon, E.; Bar-Eyal, M.; Chet, I.; Herrera-Estrella, A.; Kleifeld, O.; Spiegel, Y. Biological control of the root-knot nematode Meloidogyne javanica by Trichoderma harzianum. Phytopathology 2001, 91, 687–693. [Google Scholar] [CrossRef]
- Sahebani, N.; Hadavi, N. Biological control of the root-knot nematode Meloidogyne javanica by Trichoderma harzianum. Soil Biol. Biochem. 2008, 40, 2016–2020. [Google Scholar] [CrossRef]
- Rodríguez-González, A.; Casquero, P.A.; Suárez-Villanueva, V.; Carro-Huerga, G.; Alvarez-García, S.; Mayo-Prieto, S.; Lorenzana, A.; Cardoza, R.E.; Gutiérrez, S. Effect of trichodiene production by Trichoderma harzianum on Acanthoscelides obtectus. J. Stored Prod. Res. 2018, 77, 231–239. [Google Scholar] [CrossRef]
- Rodriguez-Gonzalez, A.; Guerra, M.; Ramirez-Lozano, D.; Casquero, P.A.; Gutierrez, S. Germination and Agronomic Traits of Phaseolus vulgaris L. Beans Sprayed with Trichoderma Strains and Attacked by Acanthoscelides obtectus. Agronomy 2021, 11, 2130. [Google Scholar] [CrossRef]
- López-Bucio, J.; Pelagio-Flores, R.; Herrera-Estrella, A. Trichoderma as biostimulant: Exploiting the multilevel properties of a plant beneficial fungus. Sci. Hortic. 2015, 196, 109–123. [Google Scholar] [CrossRef]
- Pacheco-Trejo, J.; Aquino-Torres, E.; Reyes-Santamaría, M.I.; Islas-Pelcastre, M.; Pérez-Ríos, S.R.; Madariaga-Navarrete, A.; Saucedo-García, M. Plant Defensive Responses Triggered by Trichoderma spp. as Tools to Face Stressful Conditions. Horticulturae 2022, 8, 1181. [Google Scholar] [CrossRef]
- Şesan, T.E.; Oancea, A.O.; Ştefan, L.M.; Mănoiu, V.S.; Ghiurea, M.; Răut, I.; Constantinescu-Aruxandei, D.; Toma, A.; Savin, S.; Bira, A.F.; et al. Effects of Foliar Treatment with a Trichoderma Plant Biostimulant Consortium on Passiflora caerulea L. Yield and Quality. Microorganisms 2020, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- Marra, R.; Lombardi, N.; Piccolo, A.; Bazghaleh, N.; Prashar, P.; Vandenberg, A.; Woo, S. Mineral Biofortification and Growth Stimulation of Lentil Plants Inoculated with Trichoderma Strains and Metabolites. Microorganisms 2022, 10, 87. [Google Scholar] [CrossRef]
- Tripathi, P.; Singh, P.C.; Mishra, A.; Chauhan, P.S.; Dwivedi, S.; Bais, R.T.; Tripathi, R.D. Trichoderma: A potential bioremediator for environmental clean up. Clean Technol. Environ. Policy 2013, 15, 541–550. [Google Scholar] [CrossRef]
- Yao, S.; Zhou, B.; Duan, M.; Cao, T.; Wen, Z.; Chen, X.; Wang, H.; Wang, M.; Cheng, W.; Zhu, H.; et al. Combination of Biochar and Trichoderma harzianum Can Improve the Phytoremediation Efficiency of Brassica juncea and the Rhizosphere Micro-Ecology in Cadmium and Arsenic Contaminated Soil. Plants 2023, 12, 2939. [Google Scholar] [CrossRef]
- Teng, Y.; Luo, Y.; Ma, W.; Zhu, L.; Ren, W.; Luo, Y.; Christie, P.; Li, Z. Trichoderma reesei FS10-C enhances phytoremediation of Cd-contaminated soil by Sedum plumbizincicola and associated soil microbial activities. Front. Plant Sci. 2015, 6, 220. [Google Scholar] [CrossRef] [PubMed]
- Garza-García, J.J.O.; Hernández-Díaz, J.A.; León-Morales, J.M.; Velázquez-Juárez, G.; Zamudio-Ojeda, A.; Arratia-Quijada, J.; Reyes-Maldonado, O.K.; López-Velázquez, J.C.; García-Morales, S. Selenium nanoparticles based on Amphipterygium glaucum extract with antibacterial, antioxidant, and plant biostimulant properties. J. Nanobiotechnol. 2023, 21, 252. [Google Scholar] [CrossRef]
- El-Ramady, H.; Abdalla, N.; Taha, H.S.; Alshaal, T.; El-Henawy, A.; Faizy, S.E.D.A.; Shams, M.S.; Youssef, S.M.; Shalaby, T.; Bayoumi, Y.; et al. Selenium and nano-selenium in plant nutrition. Environ. Chem. Lett. 2016, 14, 123–147. [Google Scholar] [CrossRef]
- Sardar, R.; Ahmed, S.; Shah, A.A.; Yasin, N.A. Selenium nanoparticles reduced cadmium uptake, regulated nutritional homeostasis and antioxidative system in Coriandrum sativum grown in cadmium toxic conditions. Chemosphere 2022, 287, 132332. [Google Scholar] [CrossRef]
- Viterbo, A.; Ramot, O.; Chernin, L.; Chet, I. Significance of lytic enzymes from Trichoderma spp. in the biocontrol of fungal plant pathogens. Antonie Van Leeuwenhoek 2002, 81, 549–556. [Google Scholar] [CrossRef]
- Brotman, Y.; Briff, E.; Viterbo, A.; Chet, I. Role of swollenin, an expansin-like protein from Trichoderma, in plant root colonization. Plant Physiol. 2008, 147, 779–789. [Google Scholar] [CrossRef]
- Ruocco, M.; Lanzuise, S.; Lombardi, N.; Woo, S.L.; Vinale, F.; Marra, R.; Varlese, R.; Manganiello, G.; Pascale, A.; Scala, V.; et al. Multiple Roles and Effects of a Novel Trichoderma Hydrophobin. Mol. Plant-Microbe Interact. 2015, 28, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Salwan, R.; Sharma, V. Extracellular proteins of Trichoderma and their role in plant health. S. Afr. J. Bot. 2022, 147, 359–369. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K. Beneficial effects of Trichoderma secondary metabolites on crops. Phytother. Res. 2020, 34, 2835–2842. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P.; Woo, S.L.; Comite, E.; El-Nakhel, C.; Rouphael, Y.; Fusco, G.M.; Borzacchiello, A.; Lanzuise, S.; Vinale, F. Application of Trichoderma harzianum, 6-pentyl-α-pyrone and plant biopolymer formulations modulate plant metabolism and fruit quality of plum tomatoes. Plants 2020, 9, 771. [Google Scholar] [CrossRef]
- Malmierca, M.; Cardoza, R.; Alexander, N.; McCormick, S.; Hermosa, R.; Monte, E.; Gutiérrez, S. Involvement of Trichoderma trichothecenes in the biocontrol activity and induction of plant defense-related genes. Appl. Environ. Microbiol. 2012, 78, 4856–4868. [Google Scholar] [CrossRef]
- Tamandegani, P.R.; Marik, T.; Zafari, D.; Balázs, D.; Vágvölgyi, C.; Szekeres, A.; Kredics, L. Changes in Peptaibol Production of Trichoderma Species during In Vitro Antagonistic Interactions with Fungal Plant Pathogens. Biomolecules 2020, 10, 730. [Google Scholar] [CrossRef]
- Vinale, F.; Nigro, M.; Sivasithamparam, K.; Flematti, G.; Ghisalberti, E.L.; Ruocco, M.; Varlese, R.; Marra, R.; Lanzuise, S.; Eid, A.; et al. Harzianic acid: A novel siderophore from Trichoderma harzianum. Fems Microbiol. Lett. 2013, 347, 123–129. [Google Scholar] [CrossRef]
- Cai, F.; Yu, G.; Wang, P.; Wei, Z.; Fu, L.; Shen, Q.; Chen, W. Harzianolide, a novel plant growth regulator and systemic resistance elicitor from Trichoderma harzianum. Plant Physiol. Biochem. 2013, 73, 106–113. [Google Scholar] [CrossRef]
- Nancharaiah, Y.V.; Lens, P.N.L. Ecology and Biotechnology of Selenium-Respiring Bacteria. Microbiol. Mol. Biol. Rev. 2015, 79, 61–80. [Google Scholar] [CrossRef]
- Wu, Z.; Bañuelos, G.S.; Lin, Z.-Q.; Liu, Y.; Yuan, L.; Yin, X.; Li, M. Biofortification and phytoremediation of selenium in China. Front. Plant Sci. 2015, 6, 136. [Google Scholar] [CrossRef]
- Malagoli, M.; Schiavon, M.; dall’Acqua, S.; Pilon-Smits, E.A.H. Effects of selenium biofortification on crop nutritional quality. Front. Plant Sci. 2015, 6, 280. [Google Scholar] [CrossRef] [PubMed]
- Bañuelos, G.S.; Lin, Z.-Q.; Broadley, M. Selenium Biofortification. In Selenium in plants: Molecular, Physiological, Ecological and Evolutionary Aspects; Pilon-Smits, E.A.H., Winkel, L.H.E., Lin, Z.-Q., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 231–255. [Google Scholar] [CrossRef]
- Hossain, A.; Skalicky, M.; Brestic, M.; Maitra, S.; Sarkar, S.; Ahmad, Z.; Vemuri, H.; Garai, S.; Mondal, M.; Bhatt, R.; et al. Selenium Biofortification: Roles, Mechanisms, Responses and Prospects. Molecules 2021, 26, 881. [Google Scholar] [CrossRef] [PubMed]
- Schiavon, M.; Pilon-Smits, E.A. Selenium Biofortification and Phytoremediation Phytotechnologies: A Review. J. Environ. Qual. 2017, 46, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Faquin, V.; Guilherme, L.; Castro, E.; Ávila, F.; Carvalho, G.; Bastos, C.; Oliveira, C. Selenium biofortification and antioxidant activity in lettuce plants fed with selenate and selenite. Plant Soil Environ. 2010, 56, 584–588. [Google Scholar] [CrossRef]
- Bowen, H.C.; Breward, N.; Broadley, M.R.; Bryson, R.J.; Harriman, M.; Hawkesford, M.J.; Johnson, S.E.; McGrath, S.P.; Meacham, M.C.; Tucker, M.; et al. Biofortification of UK food crops with selenium. Proc. Nutr. Soc. 2006, 65, 169–181. [Google Scholar] [CrossRef]
- Graham, R.; Lyons, G.; Stangoulis, J. High-selenium wheat: Biofortification for better health. Nutr. Res. Rev. 2003, 16, 45–60. [Google Scholar] [CrossRef]
- Abdalla, Z.F.; El-Bassiony, A.E.-M.; El-Ramady, H.; El-Sawy, S.; Shedeed, S. Broccoli Biofortification Using Biological Nano- and Mineral Fertilizers of Selenium: A Comparative Study under Soil Nutrient Deficiency Stress. Egypt. J. Soil Sci. 2023, 63, 57–66. [Google Scholar] [CrossRef]
- El-Ramady, H.; Faizy, S.E.-D.; Abdalla, N.; Taha, H.; Domokos-Szabolcsy, É.; Fari, M.; Elsakhawy, T.; Omara, A.E.-D.; Shalaby, T.; Bayoumi, Y.; et al. Selenium and Nano-Selenium Biofortification for Human Health: Opportunities and Challenges. Soil Syst. 2020, 4, 57. [Google Scholar] [CrossRef]
- El-Bassiony, A.E.-M.M.; El-Ramady, H.; El-Sawy, S.M.; Mahmoud, S.H.; Shedeed, S.I.; Fawzy, Z.F. Onion Biofortification Using Selenium Bionanofertilizer and its Bulk Source under Sandy Soil Conditions. IOP Conf. Ser. Earth Environ. Sci. 2023, 1213, 012043. [Google Scholar] [CrossRef]
- Golubkina, N.A.; Folmanis, G.E.; Tananaev, I.G.; Krivenkov, L.V.; Kosheleva, O.V.; Soldatenko, A.V. Comparative Evaluation of Spinach Biofortification with Selenium Nanoparticles and Ionic Forms of the Element. Nanotechnol. Russ. 2017, 12, 569–576. [Google Scholar] [CrossRef]
- Sun, P.; Wang, Z.; Yuan, N.; Lu, Q.; Sun, L.; Li, Y.; Zhang, J.; Zhang, Y.; Ge, G.; Jia, Y. Evaluation of nano-selenium biofortification characteristics of alfalfa (Medicago sativa L.). Green Process. Synth. 2023, 12, 20228121. [Google Scholar] [CrossRef]
- Mahmoud, S.; Shedeed, S.; El-Ramady, H.; Abdalla, Z.F.; El-Bassiony, A.E.-M.; El-Sawy, S. Biological Nano-Selenium for Eggplant Biofortification under Soil Nutrient Deficiency. Egypt. J. Soil Sci. 2023, 63, 151–162. [Google Scholar] [CrossRef]
- Azeez, L.; Lateef, A.; Olabode, O. An overview of biogenic metallic nanoparticles for water treatment and purification: The state of the art. Water Sci. Technol. 2023, 88, 851–873. [Google Scholar] [CrossRef]
- Jacob, J.M.; Karthik, C.; Saratale, R.G.; Kumar, S.S.; Prabakar, D.; Kadirvelu, K.; Pugazhendhi, A. Biological approaches to tackle heavy metal pollution: A survey of literature. J. Environ. Manag. 2018, 217, 56–70. [Google Scholar] [CrossRef]
- Briffa, J.; Sinagra, E.; Blundell, R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 2020, 6, e04691. [Google Scholar] [CrossRef]
- Bai, Y.; Rong, F.; Wang, H.; Zhou, Y.; Xie, X.; Teng, J. Removal of copper from aqueous solutions by adsorption on elemental selenium nanoparticles. J. Chem. Eng. Data 2011, 56, 2563–2568. [Google Scholar] [CrossRef]
- Jain, R.; Jordan, N.; Schild, D.; van Hullebusch, E.D.; Weiss, S.; Franzen, C.; Farges, F.; Hübner, R.; Lens, P.N.L. Adsorption of zinc by biogenic elemental selenium nanoparticles. Chem. Eng. J. 2015, 260, 855–863. [Google Scholar] [CrossRef]
- Golgoli, T.; Ghanemi, K.; Buazar, F. Cysteine-functionalized selenium nanoparticles for efficient extraction and preconcentration of cadmium, copper, lead, and zinc ions from environmental water samples. J. Mol. Liq. 2024, 393, 123680. [Google Scholar] [CrossRef]
- Hatzikioseyian, A.; Saikia, S.; Lens, P.N.L. Nickel removal by biogenic selenium nanoparticles (SeNPs): Characterization and simulation of non-symmetric breakthrough curves in continuous-flow packed-bed columns. Environ. Nanotechnol. Monit. Manag. 2022, 18, 100729. [Google Scholar] [CrossRef]
- Jain, R. Biogenic Nanoparticles of Elemental Selenium: Synthesis, Characterization and Relevance in Wastewater Treatment. Doctoral Dissertation, l’Université Paris-Est, Paris, France, 2014. [Google Scholar]
- Jain, R.; Dominic, D.; Jordan, N.; Rene, E.R.; Weiss, S.; van Hullebusch, E.D.; Hübner, R.; Lens, P.N. Higher Cd adsorption on biogenic elemental selenium nanoparticles. Environ. Chem. Lett. 2016, 14, 381–386. [Google Scholar] [CrossRef]
- Tsukimura, K.; Miyoshi, Y.; Takagi, T.; Suzuki, M.; Wada, S.-I. Amorphous nanoparticles in clays, soils and marine sediments analyzed with a small angle X-ray scattering (SAXS) method. Sci. Rep. 2021, 11, 6997. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.K.; Ali, D.; Khan, S.H.; Gnanamoorthy, G.; Choudhary, N.; Yadav, K.K.; Thai, V.N.; Hussain, S.A.; Manhrdas, S. Synthesis and Characterization of Amorphous Iron Oxide Nanoparticles by the Sonochemical Method and Their Application for the Remediation of Heavy Metals from Wastewater. Nanomaterials 2020, 10, 1551. [Google Scholar] [CrossRef]
- Liu, J.; Qi, W.-Y.; Chen, H.; Song, C.; Li, Q.; Wang, S.-G. Selenium Nanoparticles as an Innovative Selenium Fertilizer Exert Less Disturbance to Soil Microorganisms. Front. Microbiol. 2021, 12, 746046. [Google Scholar] [CrossRef] [PubMed]
- Nandini, B.; Krishna, L.; Jogigowda, S.C.; Nagaraja, G.; Hadimani, S.; Ali, D.; Sasaki, K.; Jogaiah, S. Significance of Bryophyllum pinnatum (Lam.) for green synthesis of anti-bacterial copper and selenium nanoparticles and their influence on soil microflora. Appl. Nanosci. 2023, 13, 3609–3623. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Z.; Dai, C.; Wang, P.; Fan, S.; Yu, B.; Qu, Y. Antibacterial properties and mechanism of selenium nanoparticles synthesized by Providencia sp. DCX. Environ. Res. 2021, 194, 110630. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Shafeev, G.A.; Glinushkin, A.P.; Shkirin, A.V.; Barmina, E.V.; Rakov, I.I.; Simakin, A.V.; Kislov, A.V.; Astashev, M.E.; Vodeneev, V.A.; et al. Production and Use of Selenium Nanoparticles as Fertilizers. ACS Omega 2020, 5, 17767–17774. [Google Scholar] [CrossRef] [PubMed]
- Selmani, A.; Ulm, L.; Kasemets, K.; Kurvet, I.; Erceg, I.; Barbir, R.; Pem, B.; Santini, P.; Marion, I.D.; Vinković, T.; et al. Stability and toxicity of differently coated selenium nanoparticles under model environmental exposure settings. Chemosphere 2020, 250, 126265. [Google Scholar] [CrossRef]
- Abdelnour, S.A.; Alagawany, M.; Hashem, N.M.; Farag, M.R.; Alghamdi, E.S.; Hassan, F.U.; Bilal, R.M.; Elnesr, S.S.; Dawood, M.A.O.; Nagadi, S.A.; et al. Nanominerals: Fabrication Methods, Benefits and Hazards, and Their Applications in Ruminants with Special Reference to Selenium and Zinc Nanoparticles. Animals 2021, 11, 1916. [Google Scholar] [CrossRef]
- Sarkar, B.; Bhattacharjee, S.; Daware, A.; Tribedi, P.; Krishnani, K.K.; Minhas, P.S. Selenium Nanoparticles for Stress-Resilient Fish and Livestock. Nanoscale Res. Lett. 2015, 10, 371. [Google Scholar] [CrossRef]
- Bunge, M. Emergence and Convergence: Qualitative Novelty and the Unity of Knowledge; University of Toronto Press: Toronto, ON, Canada, 2015. [Google Scholar]
- Wegner, L.H. Formalizing complexity in the life sciences: Systems, emergence, and metafluxes. Theor. Exp. Plant Physiol. 2023, in press. [CrossRef]
- Musoni, M.; Destain, J.; Thonart, P.; Bahama, J.B.; Delvigne, F. Bioreactor design and implementation strategies for the cultivation of filamentous fungi and the production of fungal metabolites: From traditional methods to engineered systems. Biotechnol. Agron. Soc. Environ. 2015, 19, 430–442. [Google Scholar]
- Kakahi, F.B.; Ly, S.; Tarayre, C.; Deschaume, O.; Bartic, C.; Wagner, P.; Compère, P.; Derdelinckx, G.; Blecker, C.; Delvigne, F. Modulation of fungal biofilm physiology and secondary product formation based on physico-chemical surface properties. Bioprocess Biosyst. Eng. 2019, 42, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Das, B.S.; Das, A.; Mishra, A.; Arakha, M. Microbial cells as biological factory for nanoparticle synthesis. Front. Mater. Sci. 2021, 15, 177–191. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Adedeji, I.; Chen, G.; Tang, Y.N. Chemical-Free Recovery of Elemental Selenium from Selenate-Contaminated Water by a System Combining a Biological Reactor, a Bacterium-Nanoparticle Separator, and a Tangential Flow Filter. Environ. Sci. Technol. 2018, 52, 13231–13238. [Google Scholar] [CrossRef] [PubMed]
- Nisha Raj, S.; Anooj, E.S.; Rajendran, K.; Vallinayagam, S. A comprehensive review on regulatory invention of nano pesticides in Agricultural nano formulation and food system. J. Mol. Struct. 2021, 1239, 130517. [Google Scholar] [CrossRef]
- Goyal, V.; Rani, D.; Ritika; Mehrotra, S.; Deng, C.; Wang, Y. Unlocking the Potential of Nano-Enabled Precision Agriculture for Efficient and Sustainable Farming. Plants 2023, 12, 3744. [Google Scholar] [CrossRef]
- Zain, M.; Ma, H.; Ur Rahman, S.; Nuruzzaman, M.; Chaudhary, S.; Azeem, I.; Mehmood, F.; Duan, A.; Sun, C. Nanotechnology in precision agriculture: Advancing towards sustainable crop production. Plant Physiol. Biochem. 2024, 206, 108244. [Google Scholar] [CrossRef]
- Yousefi, M.; Ewert, F. Protocol for a systematic review of living labs in agricultural-related systems. Sustain. Earth Rev. 2023, 6, 11. [Google Scholar] [CrossRef]
- Beaudoin, C.; Joncoux, S.; Jasmin, J.-F.; Berberi, A.; McPhee, C.; Schillo, R.S.; Nguyen, V.M. A research agenda for evaluating living labs as an open innovation model for environmental and agricultural sustainability. Environ. Chall. 2022, 7, 100505. [Google Scholar] [CrossRef]
- Toffolini, Q.; Hannachi, M.; Capitaine, M.; Cerf, M. Ideal-types of experimentation practices in agricultural Living Labs: Various appropriations of an open innovation model. Agric. Syst. 2023, 208, 103661. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).