Core Germplasm Construction Based on Genetic and Phenotypic Diversity of Buffalograss (Bouteloua dactyloides (Nutt.) Columbus) from the Great Plains of America
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Materials
2.2. Field Phenotype Evaluation
2.3. DNA Extraction and SRAP-PCR
2.4. Ploidy Level Determination Using Flow Cytometry
3. Data Analysis
3.1. Genetic Diversity
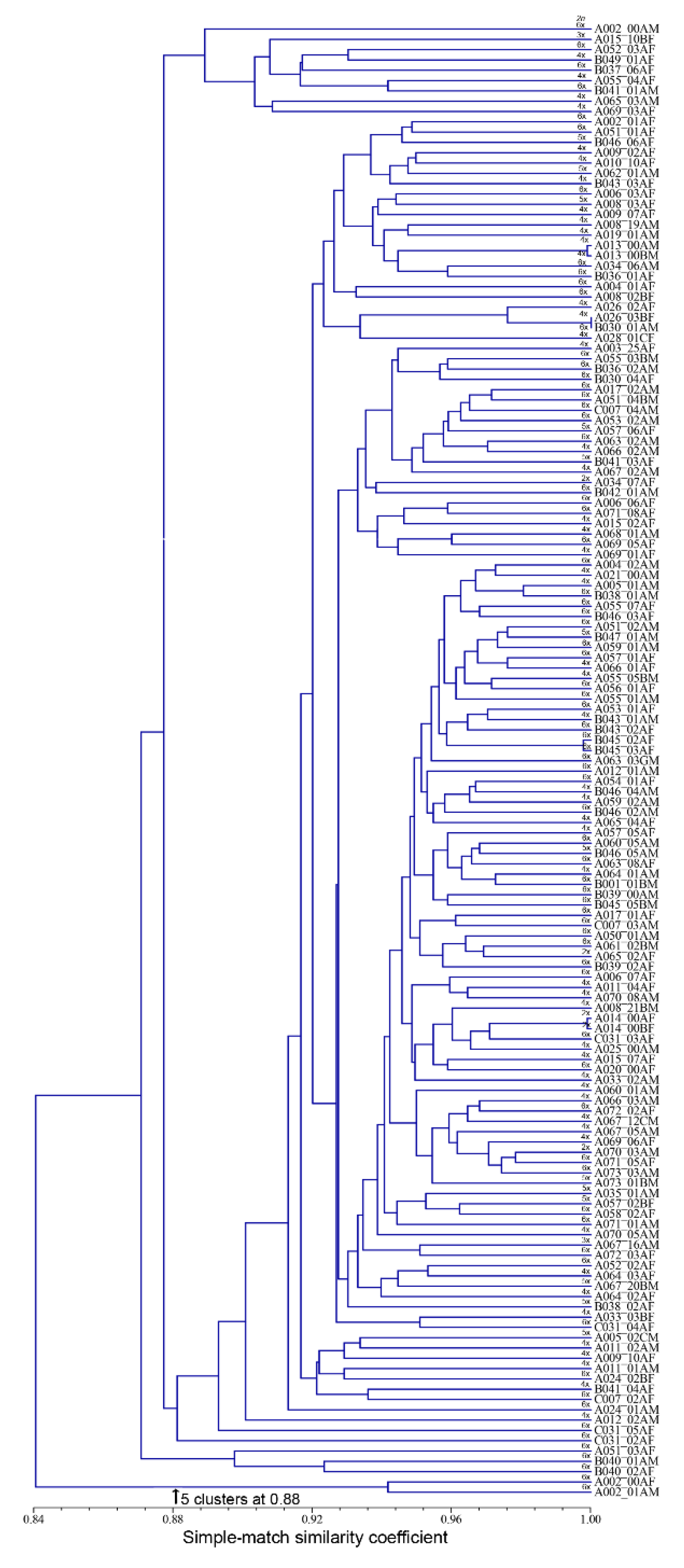
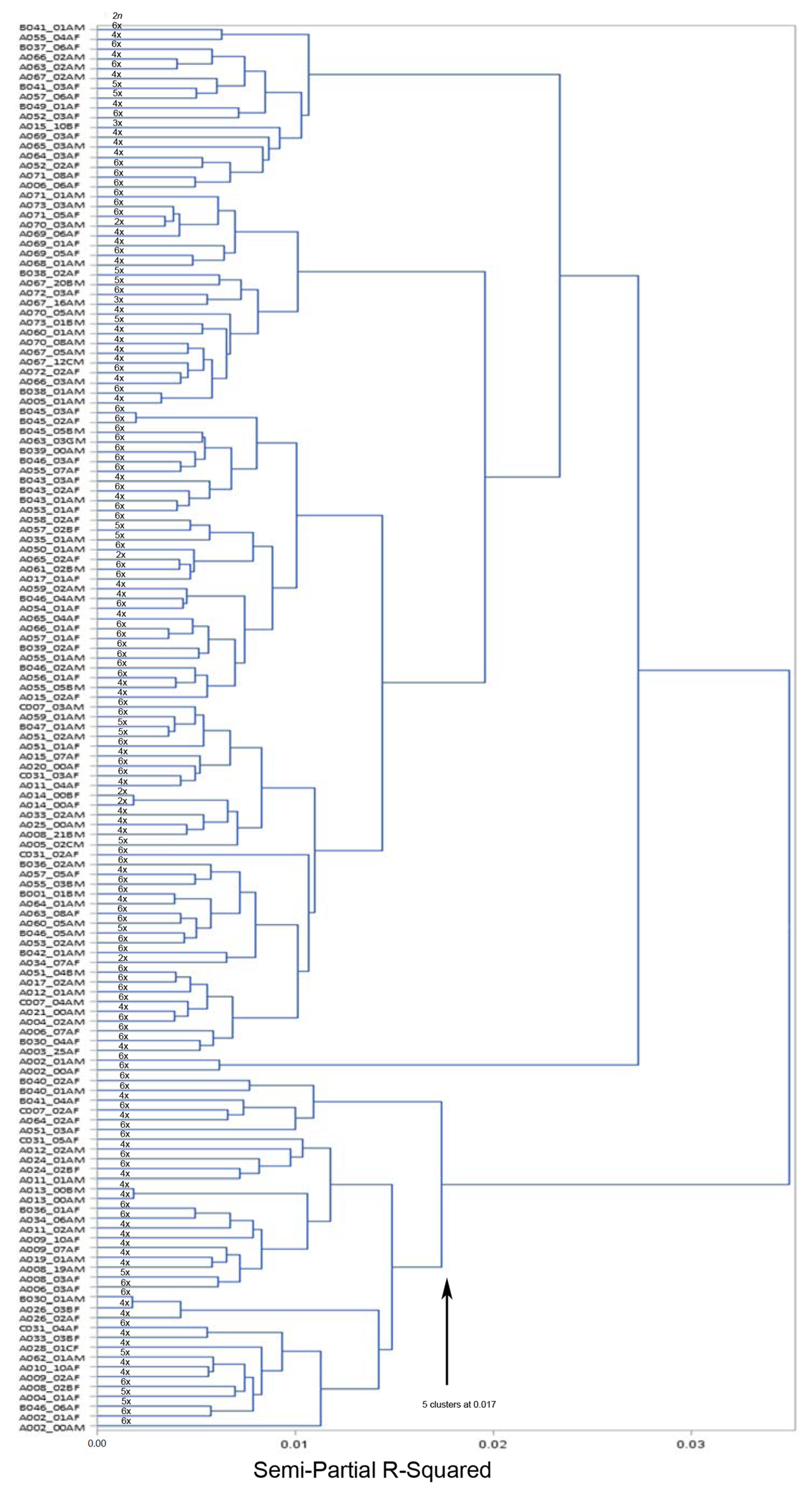
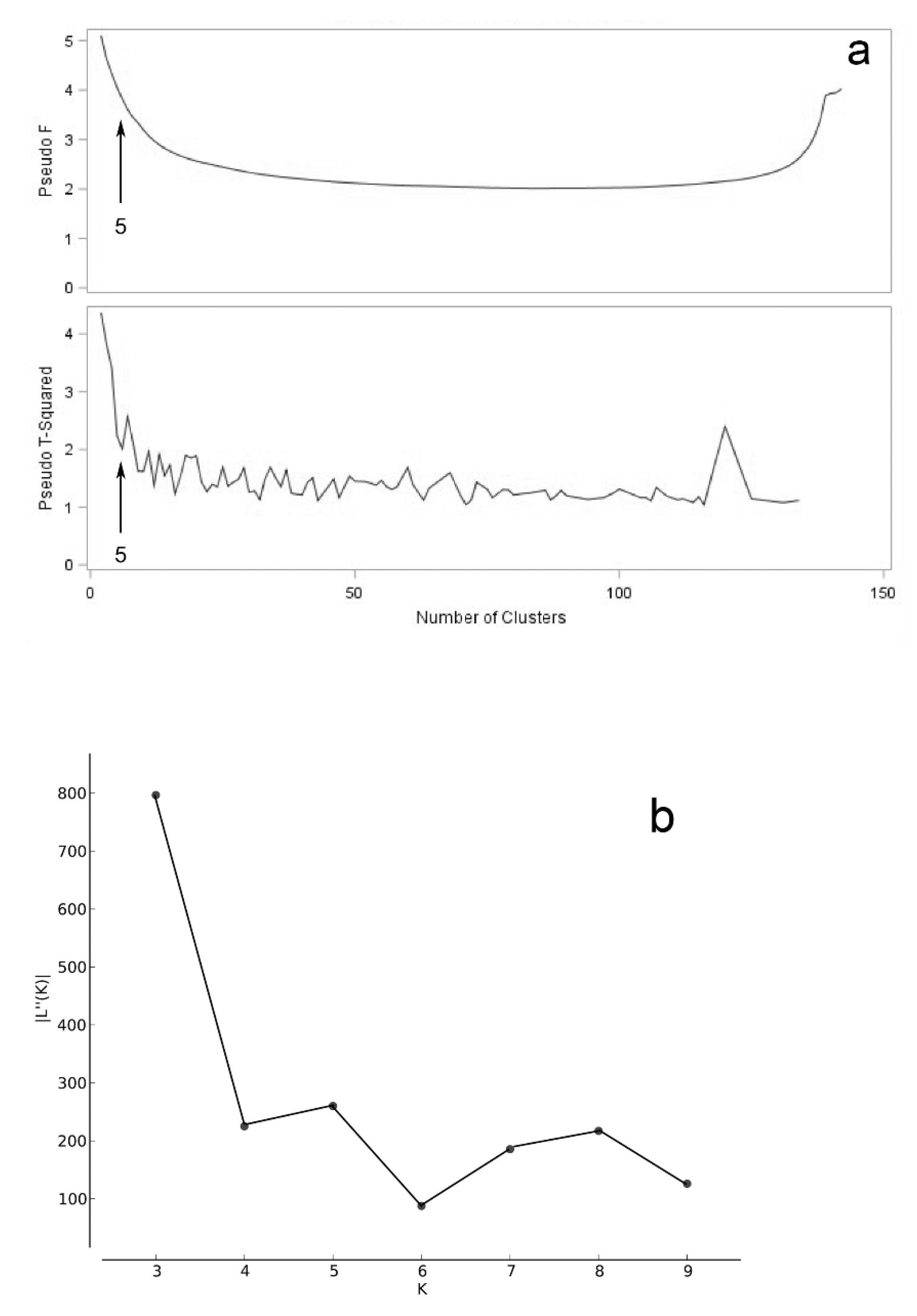
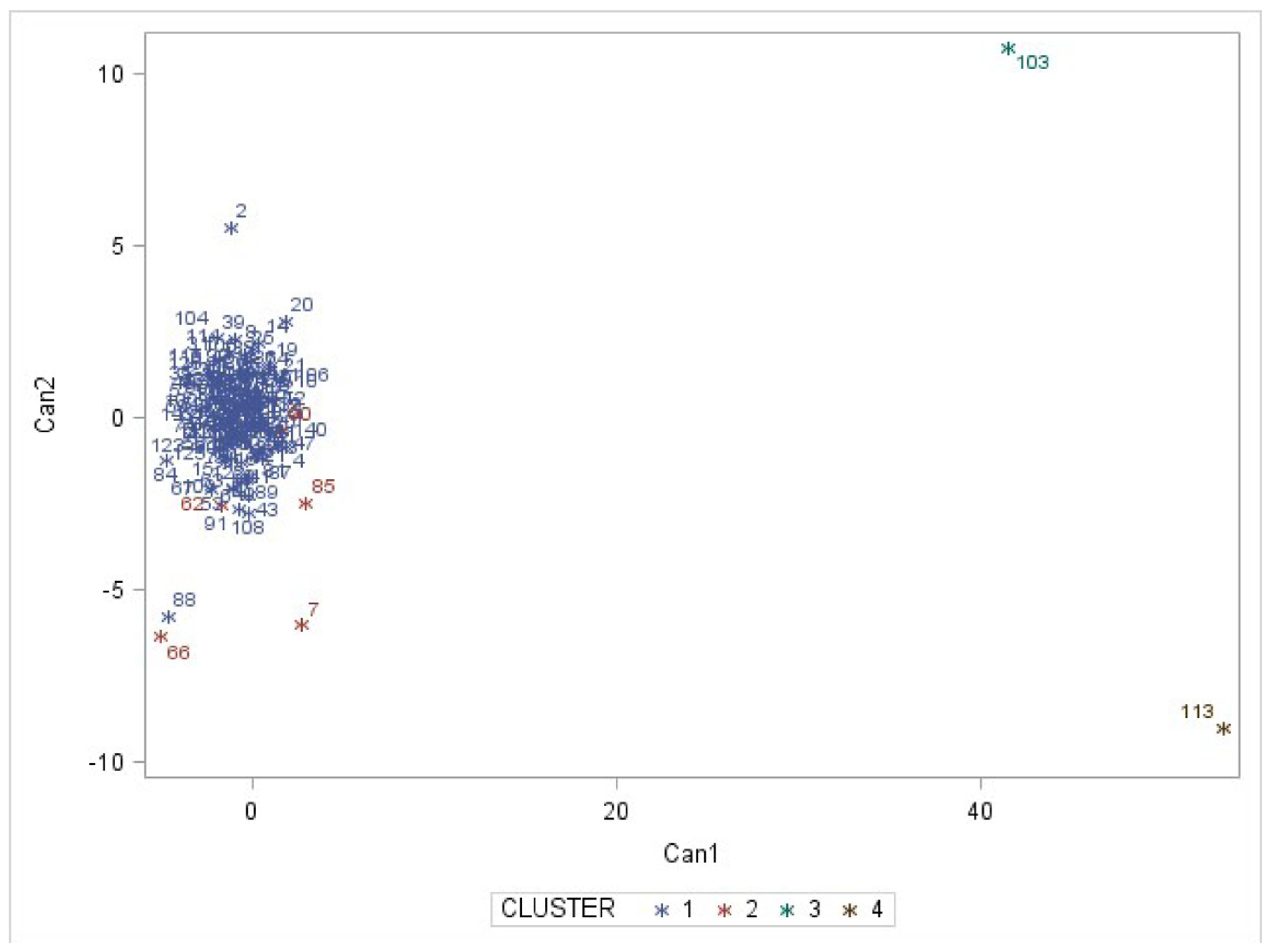
3.2. Accession Grouping
3.3. Phenotype Analysis
3.4. Core Germplasm Construction
4. Results and Discussion
4.1. Genetic Diversity
4.2. Accession Grouping
4.3. Phenotype Evaluation
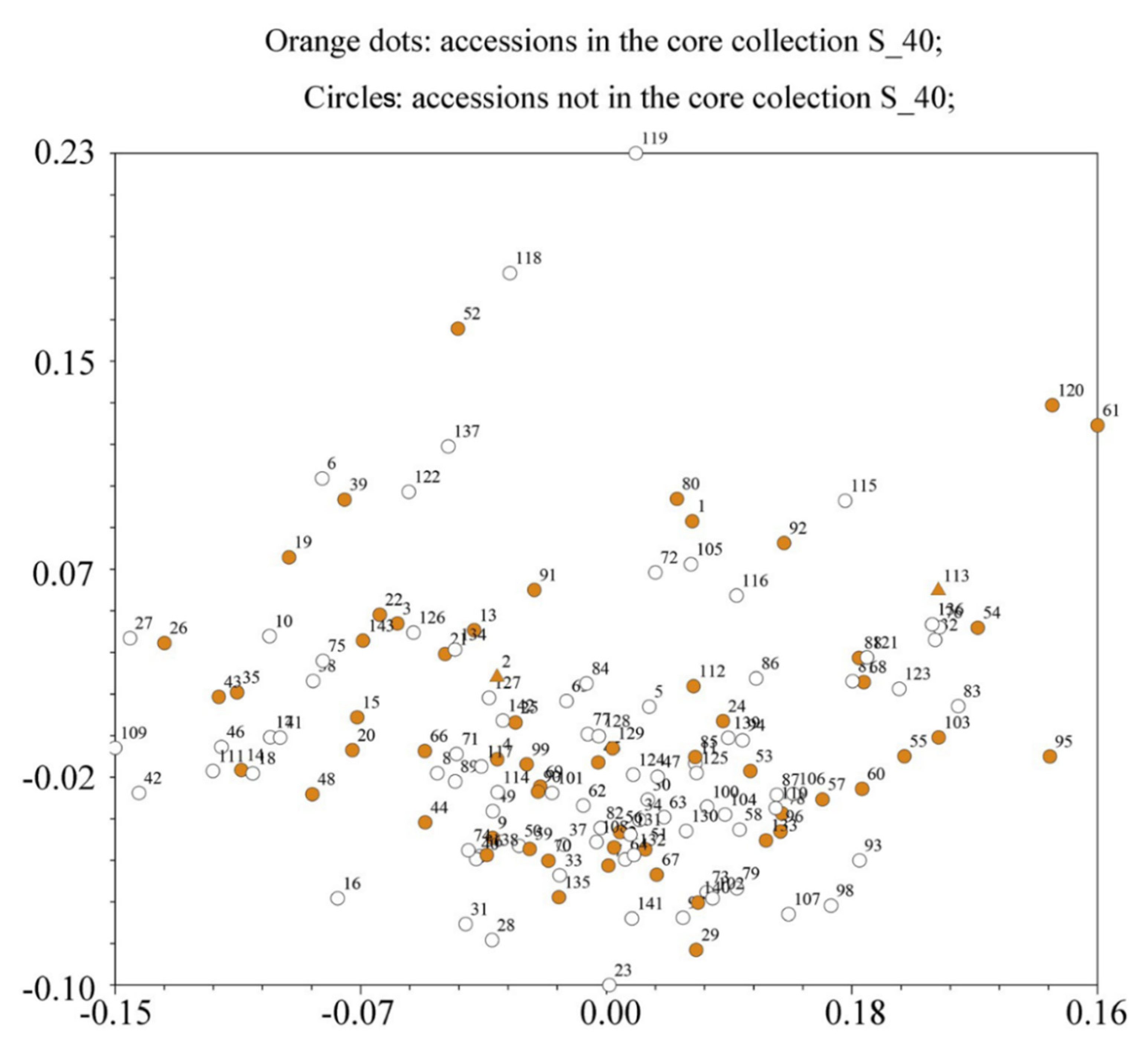
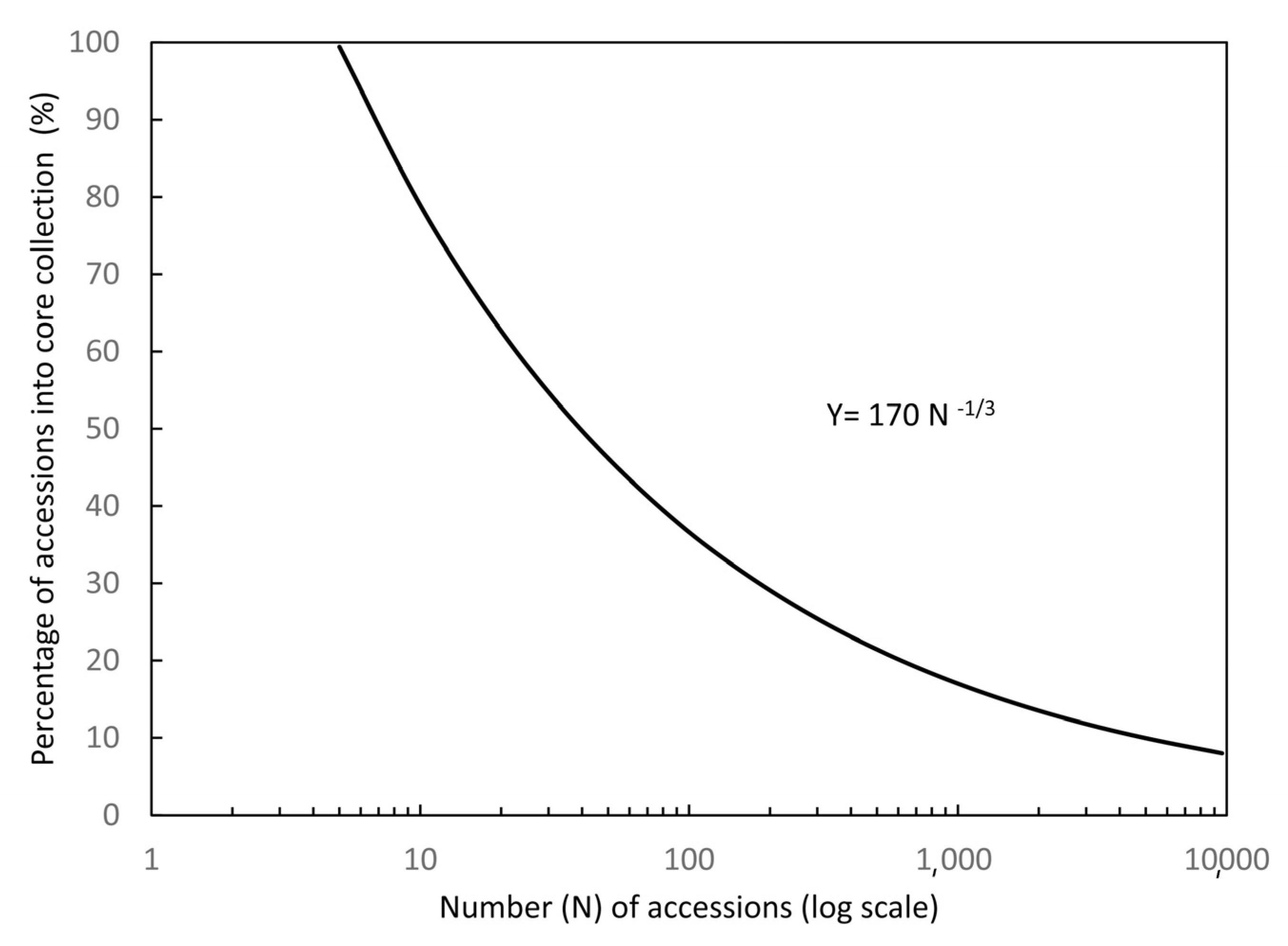
4.4. Core Germplasm Construction
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Johnson, P.G.; Riordan, T.P.; Arumuganathan, K. Ploidy level determinations in buffalograss clones and populations. Crop Sci. 1998, 38, 478–482. [Google Scholar] [CrossRef]
- Johnson, P.G.; Kenworthy, K.E.; Auld, D.L.; Riordan, T.P. Distribution of Buffalograss Polyploid Variation in the Southern Great Plains. Crop Sci. 2001, 41, 909–913. [Google Scholar] [CrossRef]
- Frankel, O.H.; Brown, A.H. Current plant genetic resources—A critical appraisal. In Genetics: New Frontiers, Proceedings of the XV International Congress of Genetics, New Delhi, India, 12–21 December 1984; Oxford & IBH Publishing Co.: New Delhi, India, 1984; pp. 3–13. [Google Scholar]
- Davies, L.R.; Allender, C.J. Who is sowing our seeds? A systematic review of the use of plant genetic resources in research. Genet. Resour. Crop Evol. 2017, 64, 1999–2008. [Google Scholar] [CrossRef]
- Pascual, L.; Fernández, M.; Aparicio, N.; López-Fernández, M.; Fité, R.; Giraldo, P.; Ruiz, M. Development of a Multipurpose Core Collection of Bread Wheat Based on High-Throughput Genotyping Data. Agronomy 2020, 10, 534. [Google Scholar] [CrossRef]
- Pathirana, R.; Carimi, F. Management and Utilization of Plant Genetic Resources for a Sustainable Agriculture. Plants 2022, 11, 2038. [Google Scholar] [CrossRef]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Egan, L.M.; Conaty, W.C.; Stiller, W.N. Core Collections: Is There Any Value for Cotton Breeding? Front. Plant Sci. 2022, 13, 895155. [Google Scholar] [CrossRef]
- Raturi, D.; Chaudhary, M.; Bhat, V.; Goel, S.; Raina, S.N.; Rajpal, V.R.; Singh, A. Overview of developed core and mini core collections and their effective utilization in cultivated rice and its related species (Oryza sp.)—A review. Plant Breed. 2022, 141, 501–512. [Google Scholar] [CrossRef]
- Brown, A.H.D. The case for core collections. In The Use of Plant Genetic Resources; Brown, A.H.D., Frankel, O.H., Marshall, D.R., Williams, J.T., Eds.; Cambridge University Press: Cambridge, UK, 1989; pp. 135–156. [Google Scholar]
- Cuevas, H.E.; Prom, L.K. Assessment of molecular diversity and population structure of the Ethiopian sorghum [Sorghum bicolor (L.) Moench] germplasm collection maintained by the USDA-ARS National Plant Germplasm System using SSR markers. Genet. Resour. Crop Evol. 2013, 60, 1817–1830. [Google Scholar] [CrossRef]
- Reeves, P.A.; Tetreault, H.M.; Richards, C.M. Bioinformatic Extraction of Functional Genetic Diversity from Heterogeneous Germplasm Collections for Crop Improvement. Agronomy 2020, 10, 593. [Google Scholar] [CrossRef]
- Gulsen, O.; Sherman, R.C.; Vogel, K.P.; Lee, D.J.; Baenziger, P.S.; Heng-Miss, T.M.; Budak, H. Nuclear genome diversity and relationships among naturally occurring buffalograss genotypes determined by sequence-related amplified polymorphism markers. HortScience 2005, 40, 537–541. [Google Scholar] [CrossRef]
- Huff, D.R.; Peakall, R.; Smouse, P.E. Rapd variation within and among natural populations of outcrossing buffa-lograss [Buchloë dactyloides (nutt.) Engelm.]. Theor. Appl. Genet. 1993, 86, 927–934. [Google Scholar] [CrossRef]
- Wu, L.; Lin, H. Identifying Buffalograss [Buchloe dactyloides (Nutt.) Engelm.] Cultivar Breeding Lines Using Random Amplified Polymorphic DNA (RAPD) Markers. J. Am. Soc. Hortic. Sci. 1994, 119, 126–130. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E.; Huff, D.R. Evoluationary implications of allozyme and RAPD variation in diploid populations of dioecious buffalograss Buchloe dactyloides. Mol. Ecol. 1995, 4, 135–147. [Google Scholar] [CrossRef]
- Budak, H.; Shearman, R.C.; Gulsen, O.; Dweikat, I. Understanding ploidy complex and geographic origin of the Buchloe dactyloides genome using cytoplasmic and nuclear marker system. Theor. Appl. Genet. 2005, 111, 1545–1552. [Google Scholar] [CrossRef]
- Budak, H.; Shearman, R.C.; Parmaksiz, I.; Gaussoin, R.E.; Riordan, T.P.; Dweikat, I. Molecular characterization of buffalograss germplasm using sequence-related amplified polymorphism markers. Theor. Appl. Genet. 2004, 108, 328–334. [Google Scholar] [CrossRef]
- Hothem, S.D.; Marley, K.A.; Larson, R.A. Photochemistry in hoagland’s nutrient solution. J. Plant Nutr. 2003, 26, 845–854. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Li, G.; Quiros, G.L.F. Sequence-related amplified polymorphism (srap), a new marker system based on a simple pcr reaction: Its application to mapping and gene tagging in brassica. Theor. Appl. Genet. 2001, 103, 455–461. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Hennink, S.; Zeven, A.C. The interpretation of Nei and Shannon-Weaver within population variation indices. Euphytica 1991, 51, 235–240. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Hu, J.; Zhu, J.; Xu, H.M. Methods of constructing core collections by stepwise clustering with three sampling strategies based on the genotypic values of crops. Theor. Appl. Genet. 2000, 101, 264–268. [Google Scholar] [CrossRef]
- Wang, J.C.; Hu, J.; Xu, H.M.; Zhang, S. A strategy on constructing core collections by least distance stepwise sampling. Theor. Appl. Genet. 2007, 115, 1–8. [Google Scholar] [CrossRef]
- Tamasauskas, D.; Sakalauskas, V.; Kriksciuniene, D. Evaluation Framework of Hierarchical Clustering Methods for Binary Data. In Proceedings of the 12th International Conference on Hybrid Intelligent Systems (HIS), Pune, India, 4–7 December 2012; pp. 421–426. [Google Scholar] [CrossRef]
- Abeyo, B.; Shearman, R.C. Buffalograss (Buchloe dactyloides) turfgrass performance and seed yield characteristics. Int. Turfgrass Soc. Res. J. 2009, 11, 519–531. [Google Scholar]
- Johnson, P.G.; Riordan, T.P.; Johnson-Cicalese, J. Low-mowing tolerance in buffalograss. Crop Sci. 2000, 40, 1339–1343. [Google Scholar] [CrossRef]
- Barker, R.E. Statistics of cultivar discrimination: Are differences real. Intern. Turfgrass Soc. Res. J. 1997, 8, 215–227. [Google Scholar]
- Diwan, N.; McIntosh, M.S.; Bauchan, G.R. Methods of developing a core collection of annual Medicago species. Theor. Appl. Genet. 1995, 90, 755–761. [Google Scholar] [CrossRef]
- Oliveira, M.F.; Nelson, R.L.; Geraldi, I.O.; Cruz, C.D.; de Toledo, J.F.F. Establishing a soybean germplasm core collection. Field Crops Res. 2010, 119, 277–289. [Google Scholar] [CrossRef]
- Charmet, G.; Balfourier, F. The use of geostatistics for sampling a core collection of perennial ryegrass populations. Genet. Resour. Crop Evol. 1995, 42, 303–309. [Google Scholar] [CrossRef]
- Rao, V.R.; Hodgkin, T. Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tissue Organ Cult. (PCTOC) 2002, 68, 1–19. [Google Scholar] [CrossRef]

| Origin | Climate † | Accessions | Accession Code (Name) |
|---|---|---|---|
| Yellowstone National Park | Alpine (Dfc/Dfb), a.p ‡. 380 to 2000 mm | 7 | 137 (C007_02AF), 138 (C007_03AM), 139 (C007_04AM), 140 (C031_02AF), 141 (C031_03AF), 142 (C031_04AF), 143 (C031_05AF) |
| East Wyoming | Semi-arid and continental (BSk), a.p. 300 mm | 11 | 13 (A008_02BF), 14 (A008_03AF), 15 (A008_19AM), 16 (A008_21BM), 17 (A009_02AF), 18 (A009_07AF), 19 (A009_10AF), 20 (A010_10AF), 21 (A011_01AM), 22 (A011_02AM), 23 (A011_04AF) |
| Southeast North Dakota | Humid continental (Dfa/Dfb), a.p. 574 mm | 1 | 108 (B001_01BM) |
| Central North Dakota | Humid continental (Dfb), a.p. 427 mm | 5 | 1 (A002_00AM), 2 (A002_00AF), 3 (A002_01AM), 4 (A002_01AF), 43 (A028_01CF) |
| West North Dakota | Semi-arid (BSK), a.p. 381 mm | 8 | 5 (A003_25AF), 6 (A004_01AF), 7 (A004_02AM), 8 (A005_01AM), 9 (A005_02CM), 10 (A006_03AF), 11 (A006_06AF), 12 (A006_07AF) |
| Central South Dakota | Humid continental (Dfa), a.p. 508 mm | 11 | 38 (A024_01AM), 39 (A024_02BF), 40 (A025_00AM), 41 (A026_02AF), 42 (A026_03BF), 44 (A033_02AM), 45 (A033_03BF), 46 (A034_06AM), 47 (A034_07AF), 48 (A035_01AM), 109 (B030_01AM) |
| Northwest South Dakota | Steppe (BSk), a.p. 414 mm | 5 | 33 (A017_01AF), 34 (A017_02AM), 35 (A019_01AM), 36 (A020_00AF), 37 (A021_00AM) |
| West Central Nebraska | Semi-arid (BSk), a.p. 350 mm | 9 | 24 (A012_01AM), 25 (A012_02AM), 26 (A013_00AM), 27 (A013_00BM), 28 (A014_00AF), 29 (A014_00BF), 30 (A015_02AF), 31 (A015_07AF), 32 (A015_10BF) |
| Southeast Nebraska | Humid continental (Dfa), a.p. 800 mm | 11 | 110 (B030_04AF), 113 (B037_06AF), 114 (B038_01AM), 115 (B038_02AF), 116 (B039_00AM), 117 (B039_02AF), 118 (B040_01AM), 119 (B040_02AF), 120 (B041_01AM), 121 (B041_03AF), 122 (B041_04AF) |
| Iowa | Humid continental (Dfa), a.p. 970 mm | 2 | 111 (B036_01AF), 112 (B036_02AM) |
| North Central Kansas | Humid continental (Dfa), a.p. 930 mm | 24 | 49 (A050_01AM), 50 (A051_01AF), 51 (A051_02AM), 52 (A051_03AF), 53 (A051_04BM), 54 (A052_02AF), 55 (A052_03AF), 56 (A053_01AF), 57 (A053_02AM), 58 (A054_01AF), 123 (B042_01AM), 124 (B043_01AM), 125 (B043_02AF), 126 (B043_03AF), 127 (B045_02AF), 128 (B045_03AF), 129 (B045_05BM), 130 (B046_02AM), 131 (B046_03AF), 132 (B046_04AM), 133 (B046_05AM), 134 (B046_06AF), 135 (B047_01AM), 136 (B049_01AF), |
| Southwest Kansas | Semi-arid steppe (BSk), a.p. 410 mm | 14 | 59 (A055_01AM), 60 (A055_03BM), 61 (A055_04AF), 62 (A055_05BM), 63 (A055_07AF), 64 (A056_01AF), 65 (A057_01AF), 66 (A057_02BF), 67 (A057_05AF), 68 (A057_06AF), 104 (A072_02AF), 105 (A072_03AF), 106 (A073_01BM), 107 (A073_03AM) |
| Oklahoma | Humid subtropical (Cfa), a.p. 928 mm | 7 | 69 (A058_02AF), 70 (A059_01AM), 71 (A059_02AM), 72 (A060_01AM), 73 (A060_05AM), 74 (A061_02BM), 75 (A062_01AM) |
| Northwest Oklahoma | Semi-arid (BSk), a.p. 438 mm | 3 | 101 (A071_01AM), 102 (A071_05AF), 103 (A071_08AF) |
| Northeast New Mexico | Semi-arid (BSk), a.p. 363 mm | 3 | 98 (A070_03AM), 99 (A070_05AM), 100 (A070_08AM) |
| Northwest Texas | Semi-arid (BSk), a.p. 520 mm | 22 | 76 (A063_02AM), 77 (A063_03GM), 78 (A063_08AF), 79 (A064_01AM), 80 (A064_02AF), 81 (A064_03AF). 82 (A065_02AF), 83 (A065_03AM), 84 (A065_04AF), 85 (A066_01AF), 86 (A066_02AM), 87 (A066_03AM), 88 (A067_02AM), 89 (A067_05AM), 90 (A067_12CM), 91 (A067_16AM), 92 (A067_20BM), 93 (A068_01AM), 94 (A069_01AF), 95 (A069_03AF), 96 (A069_05AF), 97 (A069_06AF) |
| Primer Combinations | Forward Sequence (5′-3′) | Reverse Sequence (3′–5′) | Polymorphic Bands |
|---|---|---|---|
| Me1-Em15 | TGA GTC CAA ACC GGA TA | GAC TGC GTA CGA ATT TAG | 161 |
| Me1-Em16 | TGA GTC CAA ACC GGA TA | GAC TGC GTA CGA ATT TGG | 123 |
| Me6-Em16 | TGA GTC CAA ACC GGT AA | GAC TGC GTA CGA ATT TCG | 64 |
| Me9-Em18 | TGA GTC CAA ACC GGT AG | GAC TGC GTA CGA ATT GGT | 120 |
| Me10-Em19 | TGA GTC CAA ACC GGT TG | GAC TGC GTA CGA ATT CCG | 98 |
| Me5-Em1 | TGA GTC CAA ACC GGA AG | GAC TGC GTA CGA ATT TAT | 105 |
| Me12-Em12 | TGA GTC CAA ACC GGT CA | GAC TGC GTA CGA ATT ATG | 77 |
| Me10-Em15 | TGA GTC CAA ACC GGT TG | GAC TGC GTA CGA ATT TAG | 85 |
| Me7-Em19 | TGA GTC CAA ACC GGT CC | GAC TGC GTA CGA ATT CCG | 84 |
| Me2-Em15 | TGA GTC CAA ACC GGA GG | GAC TGC GTA CGA ATT TAG | 116 |
| Total | 1033 | ||
| Average | 103.3 |
| Leaf Length (mm) | Internode Length (mm) | Canopy Height (cm) | Tiller Number | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | 4th | 5th | 1st | 2nd | 3rd | 4th | 5th | |||
| Max. | 25.50 | 24.30 | 28.70 | 25.40 | 28.60 | 8.50 | 8.10 | 7.00 | 7.60 | 7.20 | 29.2 | 10.0 |
| Min. | 6.60 | 6.20 | 5.90 | 7.30 | 7.10 | 0.96 | 0.86 | 0.79 | 0.80 | 0.88 | 8.4 | 2.0 |
| Avg. | 14.07 | 14.96 | 15.31 | 14.85 | 14.93 | 4.29 | 4.01 | 3.96 | 4.09 | 4.05 | 17.2 | 5.3 |
| SD | 4.05 | 4.35 | 4.73 | 4.07 | 4.06 | 1.60 | 1.38 | 1.30 | 1.36 | 1.34 | 4.6 | 1.04 |
| Core Size † | Admixture-Prioritized Clustering | Least Distance Stepwise Clustering | ||||||
|---|---|---|---|---|---|---|---|---|
| PPL‡ | Admixtures | Admixture Retained | H’§ | PPL | Admixtures | Admixture Retained | H’ | |
| Original | 99.81% | 807 | 100.0% | 44.44 | 99.81% | 807 | 100.0% | 44.44 |
| S_45 | 82.87% | 635 | 78.7% | 32.37 | 87.71% | 688 | 85.3% | 35.30 |
| S_40 | 80.83% | 614 | 76.1% | 30.50 | 86.06% | 668 | 82.8% | 33.39 |
| S_35 | 77.73% | 582 | 72.1% | 28.61 | 81.12% | 620 | 76.8% | 30.78 |
| S_30 | 75.80% | 563 | 69.8% | 26.77 | 77.83% | 586 | 72.6% | 28.30 |
| S_25 | 72.99% | 535 | 66.3% | 24.34 | 73.96% | 548 | 67.9% | 25.48 |
| S_20 | 67.96% | 485 | 60.1% | 21.45 | 68.25% | 492 | 61.0% | 22.33 |
| S_15 | 58.37% | 406 | 50.3% | 17.64 | 61.57% | 428 | 53.0% | 18.61 |
| S_10 | 51.21% | 332 | 41.1% | 11.04 | 51.89% | 348 | 43.1% | 13.67 |
| D_45 | 82.19% | 629 | 77.9% | 32.49 | 86.64% | 680 | 84.3% | 33.12 |
| D_40 | 80.83% | 615 | 76.2% | 30.84 | 83.93% | 649 | 80.4% | 31.00 |
| D_35 | 77.73% | 583 | 72.2% | 28.62 | 81.12% | 622 | 77.1% | 28.94 |
| D_30 | 75.70% | 563 | 69.8% | 26.49 | 76.57% | 577 | 71.5% | 26.43 |
| D_25 | 71.35% | 520 | 64.4% | 24.29 | 71.44% | 527 | 65.3% | 23.47 |
| D_20 | 66.60% | 472 | 58.5% | 20.91 | 66.21% | 476 | 59.0% | 20.90 |
| D_15 | 60.70% | 416 | 51.5% | 17.68 | 59.73% | 417 | 51.7% | 17.03 |
| D_10 | 50.73% | 332 | 41.1% | 13.01 | 48.40% | 318 | 39.4% | 12.15 |
| J_45 | 84.70% | 654 | 81.0% | 32.81 | 86.64% | 677 | 83.9% | 32.89 |
| J_40 | 81.32% | 619 | 76.7% | 30.58 | 84.22% | 652 | 80.8% | 31.06 |
| J_35 | 79.57% | 601 | 74.5% | 28.92 | 80.83% | 619 | 76.7% | 28.74 |
| J_30 | 75.67% | 571 | 70.8% | 26.65 | 76.28% | 575 | 71.3% | 26.26 |
| J_25 | 72.22% | 528 | 65.4% | 24.00 | 71.25% | 525 | 65.1% | 23.31 |
| J_20 | 67.47% | 481 | 59.6% | 21.18 | 65.54% | 469 | 58.1% | 20.29 |
| J_15 | 61.18% | 426 | 52.8% | 17.75 | 59.83% | 418 | 51.8% | 16.94 |
| J_10 | 50.63% | 331 | 41.0% | 12.99 | 48.40% | 318 | 39.4% | 12.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Y.; Jiang, M.; Zou, B.; Li, D. Core Germplasm Construction Based on Genetic and Phenotypic Diversity of Buffalograss (Bouteloua dactyloides (Nutt.) Columbus) from the Great Plains of America. Agronomy 2023, 13, 1382. https://doi.org/10.3390/agronomy13051382
Qian Y, Jiang M, Zou B, Li D. Core Germplasm Construction Based on Genetic and Phenotypic Diversity of Buffalograss (Bouteloua dactyloides (Nutt.) Columbus) from the Great Plains of America. Agronomy. 2023; 13(5):1382. https://doi.org/10.3390/agronomy13051382
Chicago/Turabian StyleQian, Yongqiang, Manhua Jiang, Bokun Zou, and Deying Li. 2023. "Core Germplasm Construction Based on Genetic and Phenotypic Diversity of Buffalograss (Bouteloua dactyloides (Nutt.) Columbus) from the Great Plains of America" Agronomy 13, no. 5: 1382. https://doi.org/10.3390/agronomy13051382
APA StyleQian, Y., Jiang, M., Zou, B., & Li, D. (2023). Core Germplasm Construction Based on Genetic and Phenotypic Diversity of Buffalograss (Bouteloua dactyloides (Nutt.) Columbus) from the Great Plains of America. Agronomy, 13(5), 1382. https://doi.org/10.3390/agronomy13051382








