A Comparison of the Physiological Traits and Gene Expression of Brassinosteroids Signaling under Drought Conditions in Two Chickpea Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Physiological Traits
2.2. The Identification and Analysis of CaBES1 Genes
2.3. Expression Analysis
2.4. RNA Extraction
2.5. Primer Design
2.6. Real Time PCR Analysis
2.7. Statistical Analysis
3. Results
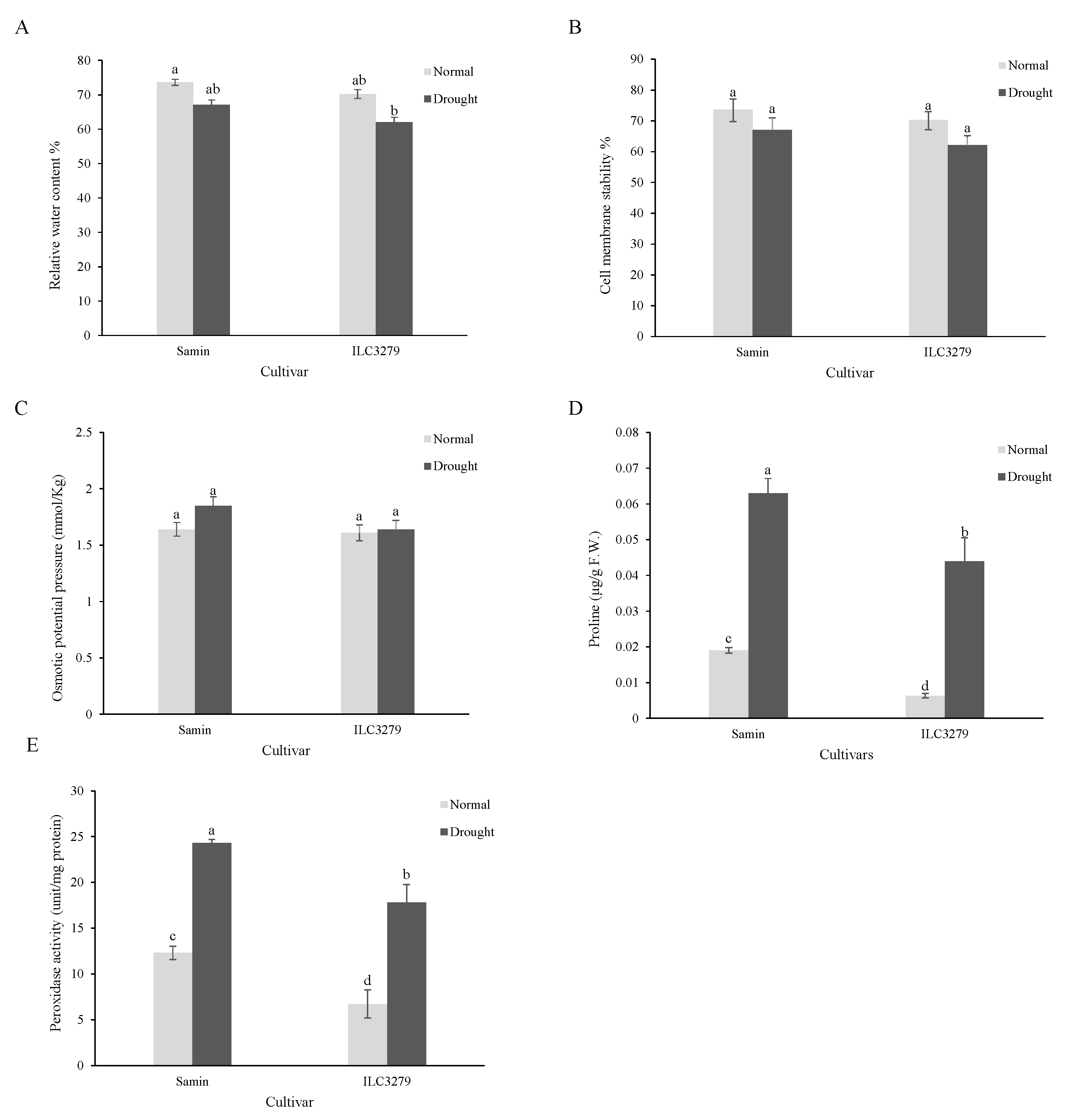
3.1. The Physiological Characters of Two Chickpea Cultivars under Drought Conditions
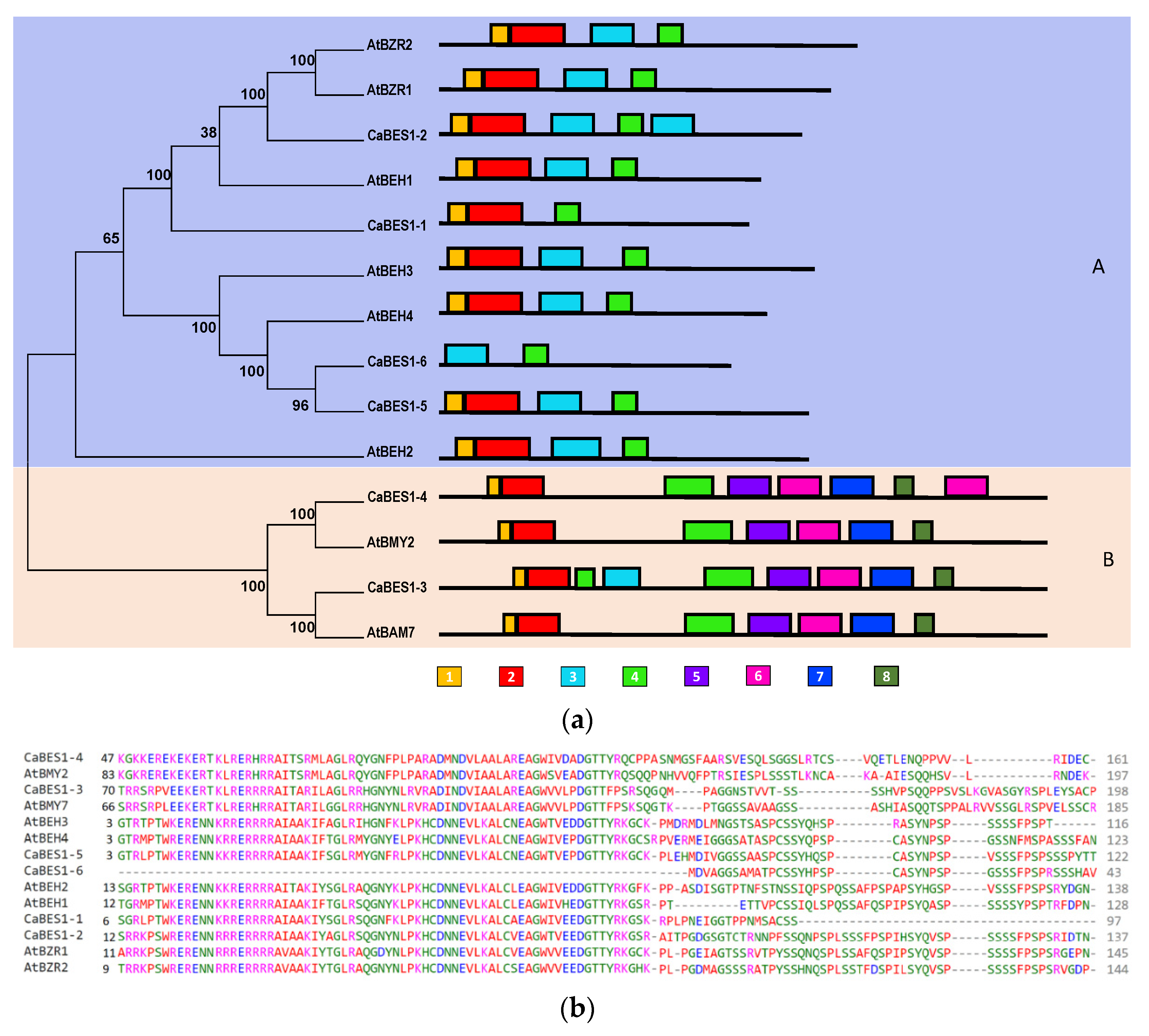
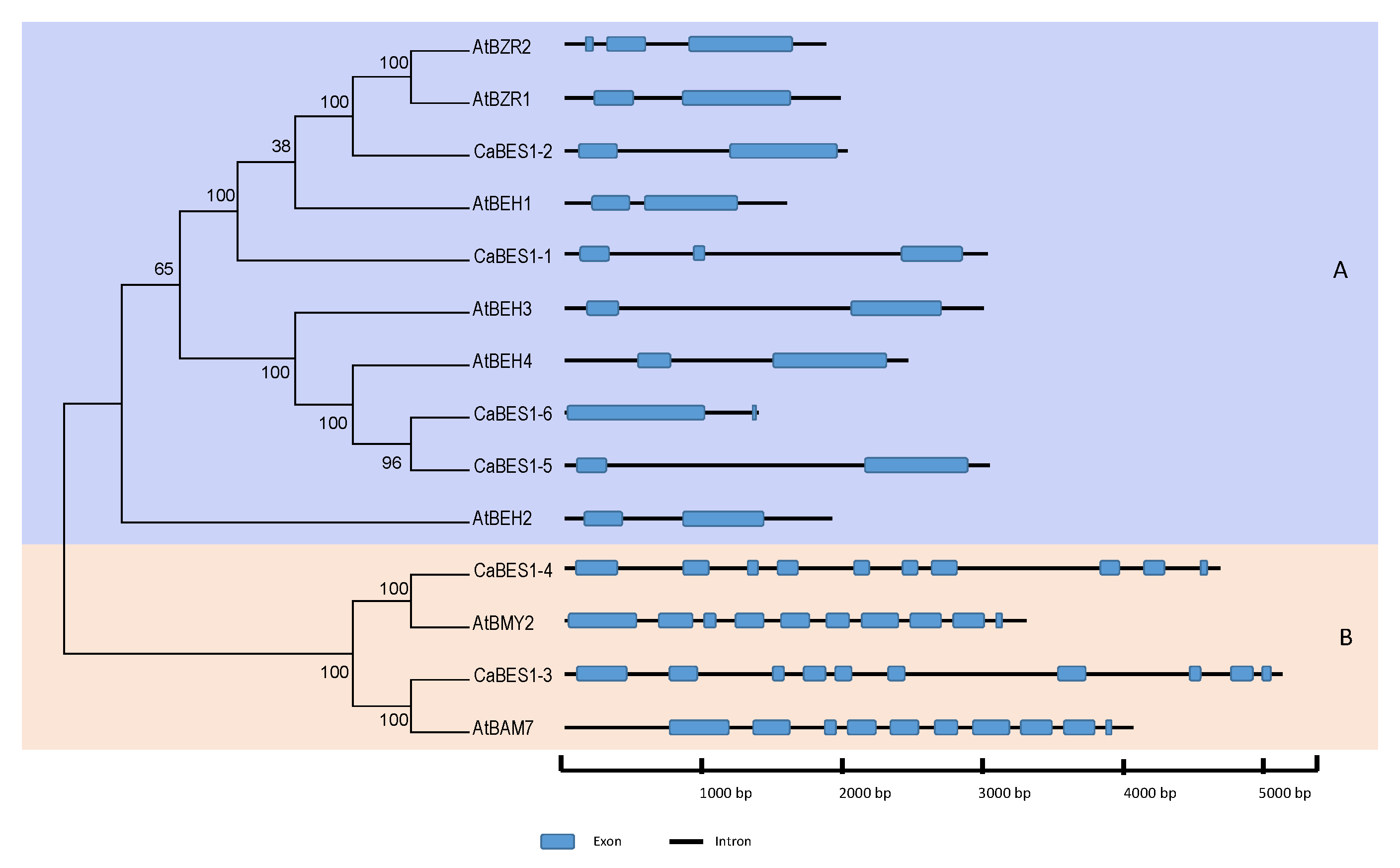
3.2. The Identification of CaBES1 Genes in Chickpea Plants
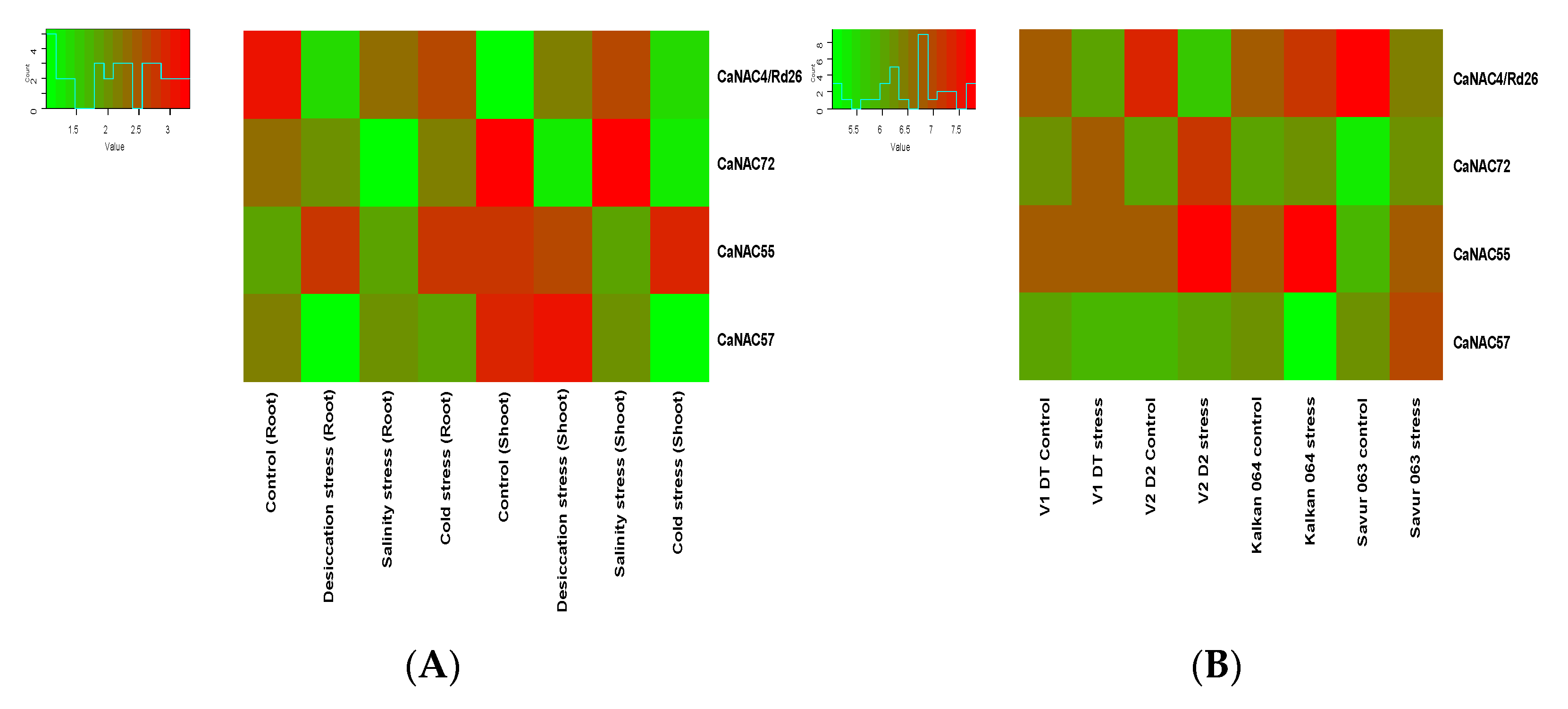
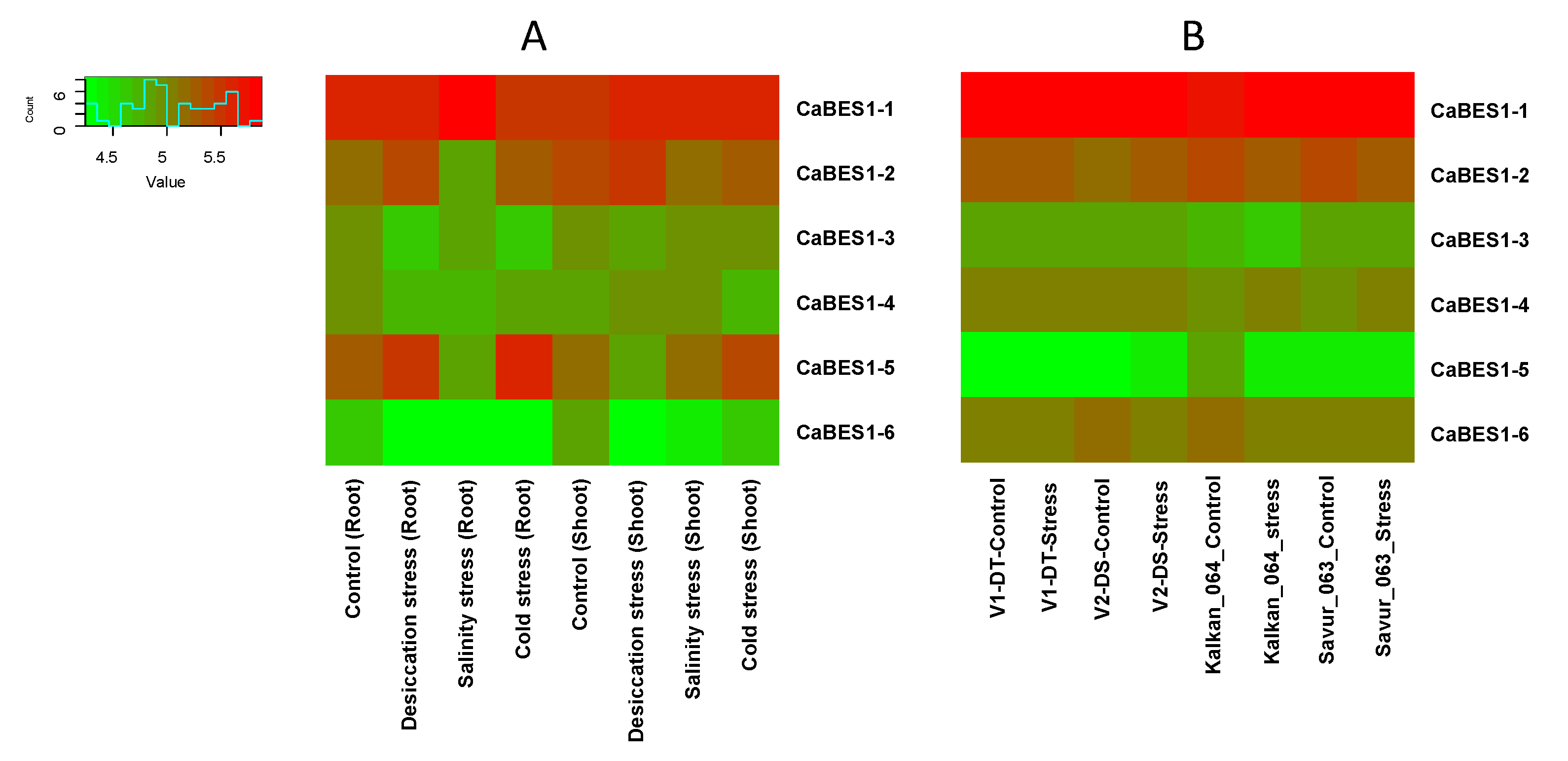
3.3. RNA-Seq Expression of BES1 Genes
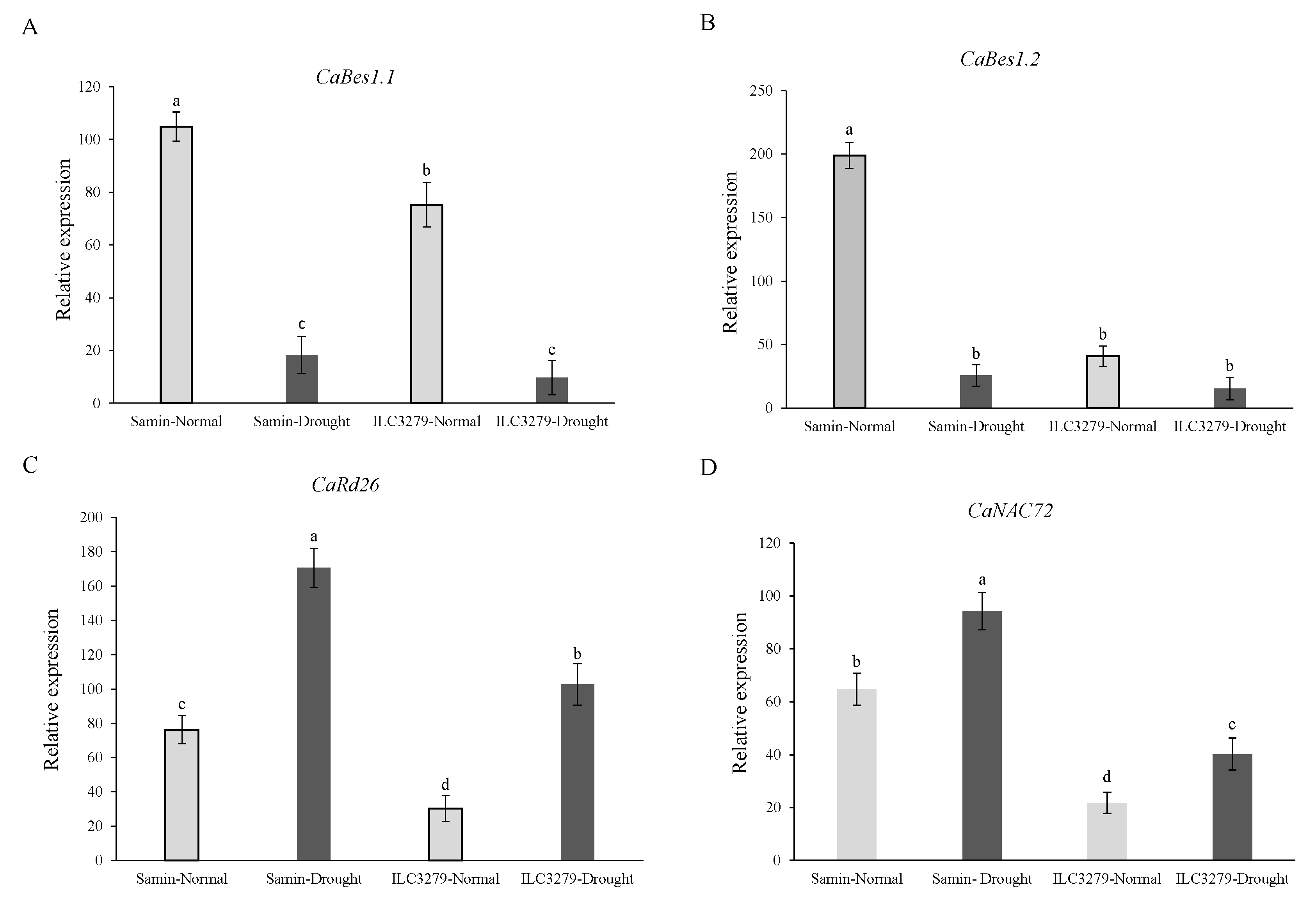
3.4. The Expression of CaBES1.1, CaBES1.2, CaRD26 and CaNAC72 under Drought Stress
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BR | Brassinosteroids |
| BES1 | BRI1-ethylmethylsulfone-suppressor1 |
| NAC | (NAM, ATAF and CUC) |
| RD26 | Response to desiccation 26 |
| FC | Field capacity |
| RWC | Relative water contents |
| qRT-PCR | Quantitative real-time PCR |
| EC | Electrical conductivity |
| MEGA | Molecular Evolution Genetic Analysis |
References
- FAOSTAT. FAOSTAT Statistical Database. Food and Agriculture Organization of the United Nations, Rome—References—Scientific Research Publishing. 2020. Available online: https://www.scirp.org/(S(351jmbntvnsjt1aadkposzje))/reference/referencespapers.aspxreferenceid=3058619 (accessed on 26 November 2023).
- Bisht, N.; Tiwari, S.; Singh, P.C.; Niranjan, A.; Singh Chauhan, P. A multifaceted rhizobacterium Paenibacillus lentimorbus alleviates nutrient deficiency-induced stress in Cicer arietinum L. Microbiol. Res. 2019, 223–225, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.N.; Tiwari, S.; Sapre, S.; Tripathi, N.; Payasi, D.K.; Singh, M.; Thakur, S.; Sharma, M.; Tiwari, S.; Tripathi, M.K. Prioritization of physio-biochemical selection indices and yield-attributing traits toward the acquisition of drought tolerance in chickpea (Cicer arietinum L.). Plants 2023, 12, 3175. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, J.; Krishnamurthy, L.; Purushothaman, R.; Upadhyaya, H.D.; Gaur, P.M.; Gowda, C.L.L.; Ito, O.; Varshney, R.K. Scope for improvement of yield under drought through the root traits in chickpea (Cicer arietinum L.). Field Crops Res. 2015, 170, 47–54. [Google Scholar] [CrossRef]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response mechanism of plants to drought Stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Manivannan, P.; Jaleel, C.A.; Sankar, B.; Kishorekumar, A.; Somasundaram, R.; Lakshmanan, G.M.A.; Panneerselvam, R. Growth, biochemical modifications and proline metabolism in Helianthus Annuus L. as Induced by drought Stress. Colloids Surf. B Biointerfaces 2007, 59, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol. 1993, 125, 27–58. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Khan, M.S.; Ahmad, D.; Khan, M.A. Utilization of genes encoding osmoprotectants in transgenic plants for enhanced abiotic stress tolerance. Electron. J. Biotechnol. 2015, 18, 257–266. [Google Scholar] [CrossRef]
- Bergonci, T.; Ribeiro, B.; Ceciliato, P.H.O.; Guerrero-Abad, J.C.; Silva-Filho, M.C.; Moura, D.S. Arabidopsis thaliana RALF1 Opposes brassinosteroid effects on root cell elongation and lateral root formation. J. Exp. Bot. 2014, 65, 2219–2230. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 298–318. [Google Scholar] [CrossRef]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.Y.; Li, L.; et al. RD26 mediates crosstalk between drought and brassinosteroid signaling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Beste, L.; Nahar, N.; Dalman, K.; Fujioka, S.; Jonsson, L.; Dutta, P.C.; Sitbon, F. Synthesis of hydroxylated sterols in transgenic Arabidopsis plants alters growth and steroid metabolism. Plant Physiol. 2011, 157, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Yin, Y.; Fei, S. Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Sci. 2015, 234, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Gao, Y.; Guo, J.; Yu, T.F.; Zheng, W.J.; Liu, Y.W.; Chen, J.; Xu, Z.S.; Ma, Y.Z. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A Brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive Cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Li, Q.F.; He, J.X. BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 2016, 9, 113–125. [Google Scholar] [CrossRef]
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef]
- Zhu, W.; Jiao, D.; Zhang, J.; Xue, C.; Chen, M.; Yang, Q. Genome-wide identification and analysis of BES1/BZR1 transcription factor family in potato (Solanum tuberosum. L). Plant Growth Regul. 2020, 92, 375–387. [Google Scholar] [CrossRef]
- Wu, P.; Song, X.M.; Wang, Z.; Duan, W.K.; Hu, R.; Wang, W.L.; Li, Y.; Hou, X. Genome-Wide Analysis of the BES1 Transcription factor family in Chinese cabbage (Brassica rapa ssp. Pekinensis). Plant Growth Regul. 2016, 80, 291–301. [Google Scholar] [CrossRef]
- Li, Q.; Guo, L.; Wang, H.; Zhang, Y.; Fan, C.; Shen, Y. In silico genome-wide identification and comprehensive characterization of the BES1 gene family in soybean. Heliyon 2019, 5, e01868. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ji, T.; Liang, M.; Li, S.; Tian, Y.; Gao, L. Genome-wide identification, structural, and gene expression analysis of BRI1-EMS-suppressor 1 transcription factor family in Cucumis sativus. Front. Genet. 2020, 11, 583996. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sun, Y.Q.; Li, G.L.; Zhang, S.Y. Genome-wide identification, characterization, and expression patterns of the BZR transcription factor family in sugar beet (Beta vulgaris L.). BMC Plant Biol. 2019, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ruperao, P.; Batley, J.; Edwards, D.; Khan, T.; Colmer, T.D.; Pang, J.; Siddique, K.H.M.; Sutton, T. Investigating drought tolerance in chickpea using genome-wide association mapping and genomic selection based on whole-genome resequencing data. Front. Plant Sci. 2018, 9, 190. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Elston, J. Genotypic differences in leaf water relations between Brassica juncea and B. napus. Ann. Bot. 1992, 70, 3–9. [Google Scholar] [CrossRef]
- Sabaghpour, S.H.; Ferayedi, Y.; Kamel, M.; Mahmoodi, A.A.; Mahdeyeh, M.; Mahmoodi, F.; Saeed, A.; Kanoni, H.; Pouralibaba, H.R.; Khaledahmadi, M.; et al. Samin, a new drought tolerance, large seed size and high potential yield chickpea cultivar for spring planting on cold dryland condition of Iran. Res. Achiev. Field Hortic. Crops 2017, 6, 111–121. [Google Scholar]
- Ball, R.A.; Oosterhuis, D.M. Measurement of root and leaf osmotic potential using the vapor-pressure osmometer. Environ. Exp. Bot. 2005, 53, 77–84. [Google Scholar] [CrossRef]
- Tripathy, J.N.; Zhang, J.; Robin, S.; Nguyen, T.T.; Nguyen, H.T. QTLs for cell-membrane stability mapped in rice (Oryza sativa L.) under drought stress. Theor. Appl. Genet. 2000, 100, 1197–1202. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophyll fluorescence signatures of leaves during the autumnal chlorophyll breakdown. J. Plant Physiol. 1987, 131, 101–110. [Google Scholar] [CrossRef]
- Herzog, V.; Fahimi, H.D. A new sensitive colorimetric assay for peroxidase using 3,3′-Diaminobenzidine as hydrogen donor. Anal. Biochem. 1973, 55, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. S2), W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jia, L.; Tian, G.; Dong, Y.; Zhang, X.; Zhou, Z.; Luo, X.; Li, Y.; Yao, W. shinyCircos-V2.0: Leveraging the creation of Circos plot with enhanced usability and advanced features. iMeta 2023, 2, e109. [Google Scholar] [CrossRef]
- Chen, F.; Mackey, A.J.; Stoeckert, C.J.; Roos, D.S. OrthoMCL-DB: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006, 34, D363–D368. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sabaghpour, S.H.; Mahmodi, A.A.; Saeed, A.; Kamel, M.; Malhotra, R. Study on chickpea drought tolerance lines under dryland condition of Iran. Indian J. Crop Sci. 2006, 1, 70–73. [Google Scholar]
- Garg, R.; Shankar, R.; Thakkar, B.; Kudapa, H.; Krishnamurthy, L.; Mantri, N.; Varshney, R.K.; Bhatia, S.; Jain, M. Transcriptome analyses reveal genotype- and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Sci. Rep. 2016, 6, srep19228. [Google Scholar] [CrossRef] [PubMed]
- Mafakheri, A.; Siosemardeh, A.; Bahramnejad, B.; Struik, P.; Sohrabi, Y. Effect of drought stress on yield, proline and chlorophyll Contents in Three Chickpea Cultivars. Aust. J. Crop Sci. 2010, 4, 580–585. [Google Scholar]
- Kaur, D.; Grewal, S.K.; Kaur, J.; Singh, S.; Singh, I. Water Deficit Stress Tolerance in Chickpea Is Mediated by the Contribution of integrative defence systems in different tissues of the plant. Funct. Plant Biol. 2016, 43, 903–918. [Google Scholar] [CrossRef] [PubMed]
- Çevik, S.; Akpinar, G.; Yildizli, A.; Kasap, M.; Karaosmanoğlu, K.; Ünyayar, S. Comparative physiological and leaf proteome analysis between drought-tolerant chickpea Cicer reticulatum and drought-sensitive chickpea C. arietinum. J. Biosci. 2019, 44, 20. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Asthir, B.J.B.P. Proline: A key player in plant abiotic stress tolerance. Biol. Plant 2015, 59, 609–619. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Drought stress enhances nutritional and bioactive compounds, phenolic acids and antioxidant capacity of amaranthus leafy vegetable. BMC Plant Biol. 2018, 18, 258. [Google Scholar] [CrossRef]
- Mafakheri, A.; Siosemardeh, A.; Bahramnejad, B.; Struik, P.; Sohrabi, Y. Effect of drought stress and subsequent recovery on protein, carbohydrate contents, catalase and peroxidase activities in three chickpea (‘Cicer arietinum’) cultivars. Aust. J. Crop Sci. 2011, 5, 1255–1260. [Google Scholar]
- Patel, P.K.; Hemantaranjan, A. Salicylic acid induced alteration in dry matter partitioning, antioxidant defence system and yield in chickpea (Cicer arietinum L.) under drought stress. Asian J. Crop Sci. 2012, 4, 86–102. [Google Scholar] [CrossRef]
- Talebi, R.; Ensafi, M.H.; Baghebani, N.; Karami, E.; Mohammadi, K. Physiological Responses of Chickpea (Cicer arietinum) Genotypes to Drought Stress. Available online: https://www.researchgate.net/publication/236008585_Physiological_responses_of_chickpea_Cicer_arietinum_genotypes_to_drought_stress (accessed on 27 December 2022).
- Macar, T.; Ekmekçi, Y. Alterations in photochemical and physiological activities of chickpea (Cicer arietinum L.) cultivars under drought stress. J. Agron. Crop Sci. 2009, 195, 335–346. [Google Scholar] [CrossRef]
- Herbinger, K.; Tausz, M.; Wonisch, A.; Soja, G.; Sorger, A.; Grill, D. Complex interactive effects of drought and ozone stress on the antioxidant defence systems of two wheat cultivars. Plant Physiol. Biochem. 2002, 40, 691–696. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, X.; Zhu, W.; Lin, H.; Chen, X.; Li, Y.; Ye, W.; Yin, Z. Molecular traits and functional exploration of BES1 gene Family in plants. Int. J. Mol. Sci. 2022, 23, 4242. [Google Scholar] [CrossRef]
- Yang, J.; Wu, Y.; Li, L.; Li, C. Comprehensive analysis of the BES1 gene family and its expression under abiotic stress and hormone treatment in Populus trichocarpa. Plant Physiol. Biochem. 2022, 173, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Feng, W.; Sun, F.; Zhang, Y.Y.; Qu, J.T.; Liu, B.; Lu, F.; Yang, L.; Fu, F.; Li, W. Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Growth Regul. 2018, 86, 235–249. [Google Scholar] [CrossRef]
- Saha, G.; Park, J.I.; Jung, H.J.; Ahmed, N.U.; Kayum, M.A.; Kang, J.G.; Nou, I.S. Molecular characterization of BZR transcription factor family and abiotic stress induced expression profiling in Brassica rapa. Plant Physiol. Biochem. 2015, 92, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Cao, D. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yina, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Liu, Y.; Cao, Y.; Zhao, Y.; Zhang, H.; Sun, F.; Yang, Q.; Li, W.; Lu, Y.; Zhang, X.; et al. Maize ZmBES1/BZR1-3 and-9 transcription factors negatively regulate drought tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2022, 23, 6025. [Google Scholar] [CrossRef]


| Gene Name | Primer Name | Primer Sequence | Tm (°C) | Amplicon Length (bp) |
|---|---|---|---|---|
| IF4 | IF4A-F | GAAGACCAACGCATTCTATCAAG | 61.6 | 128 |
| IF4A-R | TGTTTGTGTTAGATGAGGCTGA | |||
| GAPDH | GAPDH-F | TGTCTCAGTTGTTGACCTTACAG | 60.1 | 158 |
| GAPDH-R | CGACTTCATTGGTGATACTAGGT | |||
| NAC72 | NAC72-F | GGTTCTGTGTCGCATATACAAGA | 60.1 | 201 |
| NAC72-R | GAATGCTTCAACAGGAGGAGAA | |||
| RD26 | RD26-F | ACACCGGCACCAAGTTAGAT | 60.1 | 168 |
| RD26-R | GTGATTGTAGAACGTCGTCGAG | |||
| BES1.1 | BES1.1-F | AAGCCTTCTCTTCCTCGTGA | 60.1 | 198 |
| BES1.1-R | CTCTCTCCTTCCACGTTGGT | |||
| BES1.2 | BES1.2-F | CGAGTCCAATCCATTCATACC | 60.1 | 181 |
| BES1.2-R | GGTGATGAGATAGGCGGTGT |
| Name | Gene ID | Length (aa) | Mw (Da) | pI | Chr | Location | Localization |
|---|---|---|---|---|---|---|---|
| CaBES1.1 | Ca_02075.1 | 271 | 29,580.08 | 9.01 | 8 | 4,822,047–4,825,123 | Nucleus |
| CaBES1.2 | Ca_04963.1 | 316 | 34,709.73 | 9.38 | 5 | 32,553,971–32,555,767 | Nucleus |
| CaBES1.3 | Ca_06210.1 | 671 | 74,967.30 | 5.45 | 3 | 22,988,642–22,995,557 | Nucleus |
| CaBES1.4 | Ca_10650.1 | 650 | 73,714.08 | 5.70 | 8 | 7,413,853–7,419,598 | Nucleus |
| CaBES1.5 | Ca_12925.1 | 321 | 34,887.85 | 8.19 | 1 | 47,282,645–47,285,551 | Nucleus |
| CaBES1.6 | Ca_12924.1 | 252 | 26,346.96 | 5.83 | 1 | 47,290,877–47,291,866 | Nucleus |
| Motif Name | Discovered Motifs | E-Value | Width | Biological Process | Molecular Function | Cellular Component |
|---|---|---|---|---|---|---|
| MEME-1 | RKPTWKERENNKRRE | 2.1 × 10−82 | 15 | None predicted | None predicted | None predicted |
| MEME-2 | RRRRAIAAKIYAGLRAQGNYNLPKHCDNNEVLKALCREAGWIVEEDGTTY | 1.5 × 10−434 | 50 | Transcription, DNA-templated, Brassinosteroid mediated signaling pathway | DNA-binding transcription factor activity | None predicted |
| MEME-3 | GSSANASPCSSYFPSPIPSYNPSPSSSSFPSPSRSDYNNN | 6.8 × 10−90 | 40 | Transcription, DNA-templated, Brassinosteroid mediated signaling pathway | DNA-binding transcription factor activity | None predicted |
| MEME-4 | NVDGVVVDCWWGIVEGWSPQEYNW SGYRELFNMIRDFKLKLQVVMAFHEC | 3.3 × 10−88 | 50 | Polysaccharide catabolic process | Beta-amylase activity | None predicted |
| MEME-5 | QWVLEIGKENPDIFFTDREGRRNPEC LSWGIDKERVLRGRTGIEVYFDFM | 1.2 × 10−83 | 50 | Polysaccharide catabolic process | Beta-amylase activity | None predicted |
| MEME-6 | EFDDFFEDGLISAVEIGLGPSGELKY PSFPEKHGWRYPGIGEFQCYDKY | 8.3 × 10−70 | 49 | Polysaccharide catabolic process | Beta-amylase activity | None predicted |
| MEME-7 | GHTFWARGPDNAGQYNSQPHETGFFCDGGDYDGYYGRFFLNWYSQVLIDH | 2.0 × 10−74 | 50 | Polysaccharide catabolic process | Beta-amylase activity | None predicted |
| MEME-8 | LRISNSAPVTPPLSSPTSRBP | 2.0 × 10−76 | 21 | None predicted | None predicted | None predicted |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felagari, K.; Bahramnejad, B.; Siosemardeh, A.; Mirzaei, K.; Lei, X.; Zhang, J. A Comparison of the Physiological Traits and Gene Expression of Brassinosteroids Signaling under Drought Conditions in Two Chickpea Cultivars. Agronomy 2023, 13, 2963. https://doi.org/10.3390/agronomy13122963
Felagari K, Bahramnejad B, Siosemardeh A, Mirzaei K, Lei X, Zhang J. A Comparison of the Physiological Traits and Gene Expression of Brassinosteroids Signaling under Drought Conditions in Two Chickpea Cultivars. Agronomy. 2023; 13(12):2963. https://doi.org/10.3390/agronomy13122963
Chicago/Turabian StyleFelagari, Khatereh, Bahman Bahramnejad, Adel Siosemardeh, Khaled Mirzaei, Xiujuan Lei, and Jian Zhang. 2023. "A Comparison of the Physiological Traits and Gene Expression of Brassinosteroids Signaling under Drought Conditions in Two Chickpea Cultivars" Agronomy 13, no. 12: 2963. https://doi.org/10.3390/agronomy13122963
APA StyleFelagari, K., Bahramnejad, B., Siosemardeh, A., Mirzaei, K., Lei, X., & Zhang, J. (2023). A Comparison of the Physiological Traits and Gene Expression of Brassinosteroids Signaling under Drought Conditions in Two Chickpea Cultivars. Agronomy, 13(12), 2963. https://doi.org/10.3390/agronomy13122963








