Genotype-by-Environment Interaction in Tepary Bean (Phaseolus acutifolius A. Gray) for Seed Yield
Abstract
1. Introduction
2. Materials and Methods
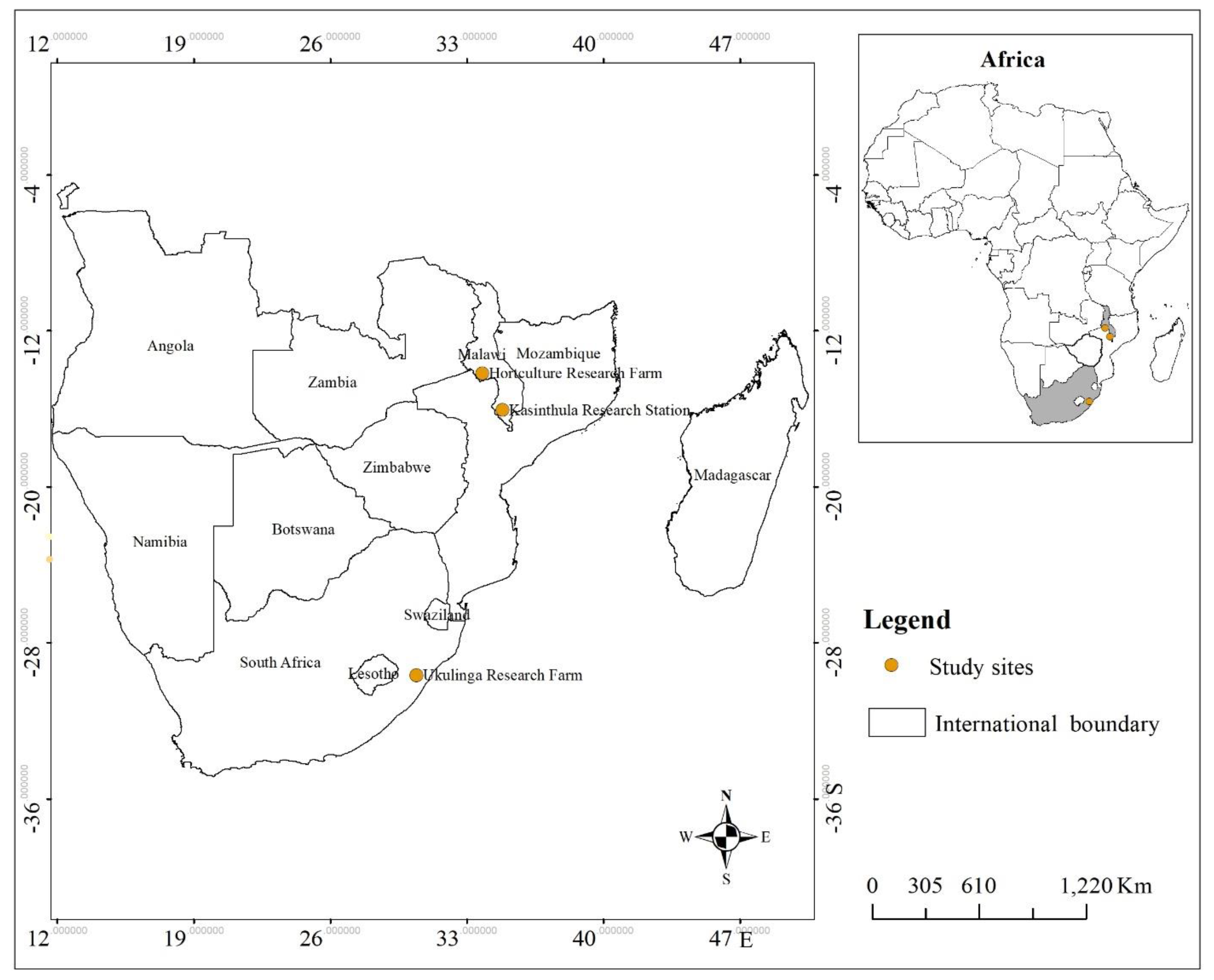
2.1. Study Sites
2.2. Plant Materials
2.3. Trial Design and Management
2.4. Data Collection
2.5. Data Analysis
2.5.1. Analysis of Variance
2.5.2. AMMI Analysis
2.5.3. AMMI Stability Value
2.5.4. Yield Stability Index
2.5.5. Cultivar Superiority Measure
2.5.6. BLUPs and BLUEs Estimation
2.5.7. BLUP-Based Stability Parameter and Multi-Trait Stability Index
2.5.8. Broad-Sense Heritability
3. Results
3.1. The Combined Analysis of Variance
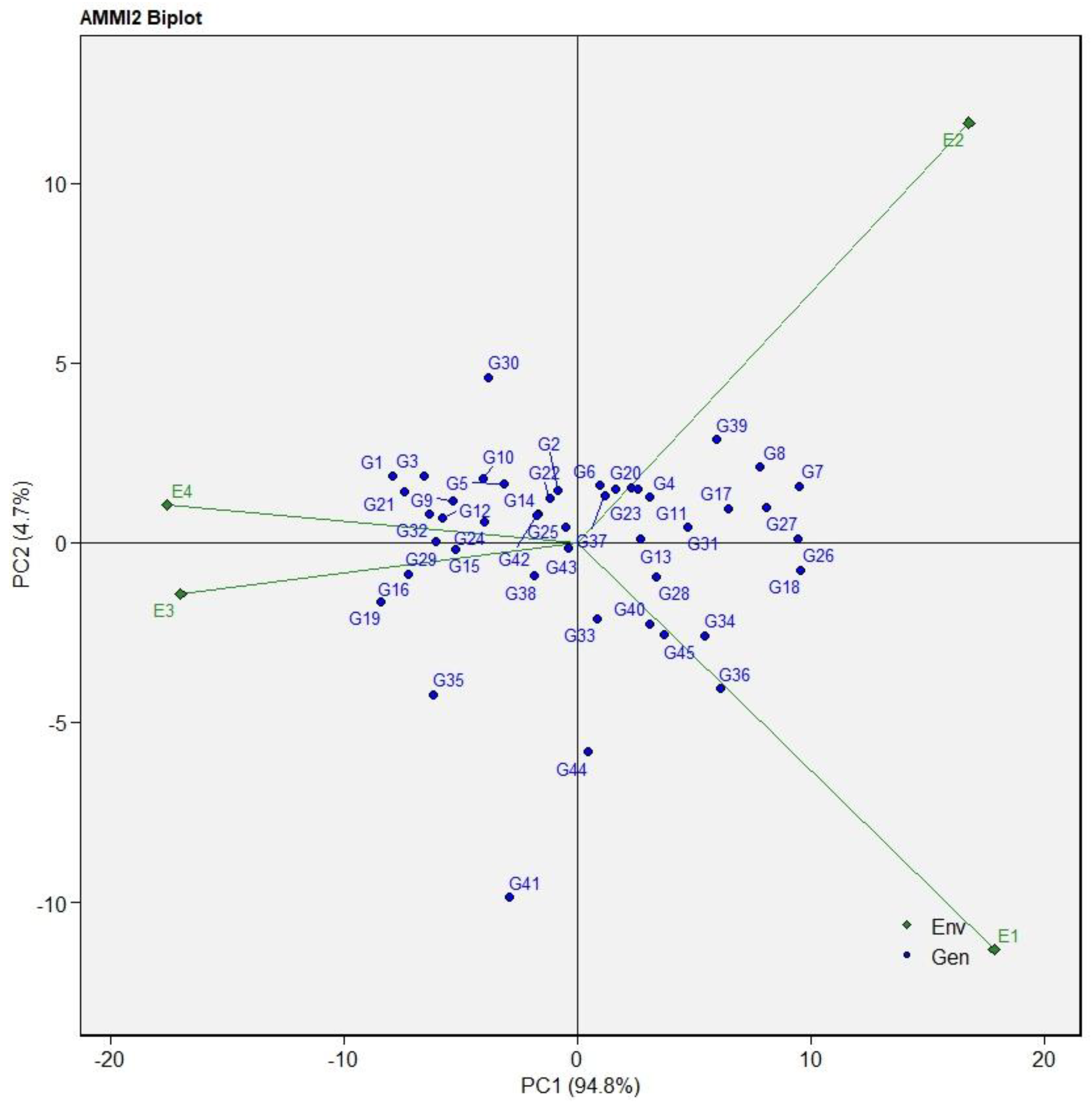
3.2. AMMI Analysis for Seed Yield
AMMI 2 Biplot
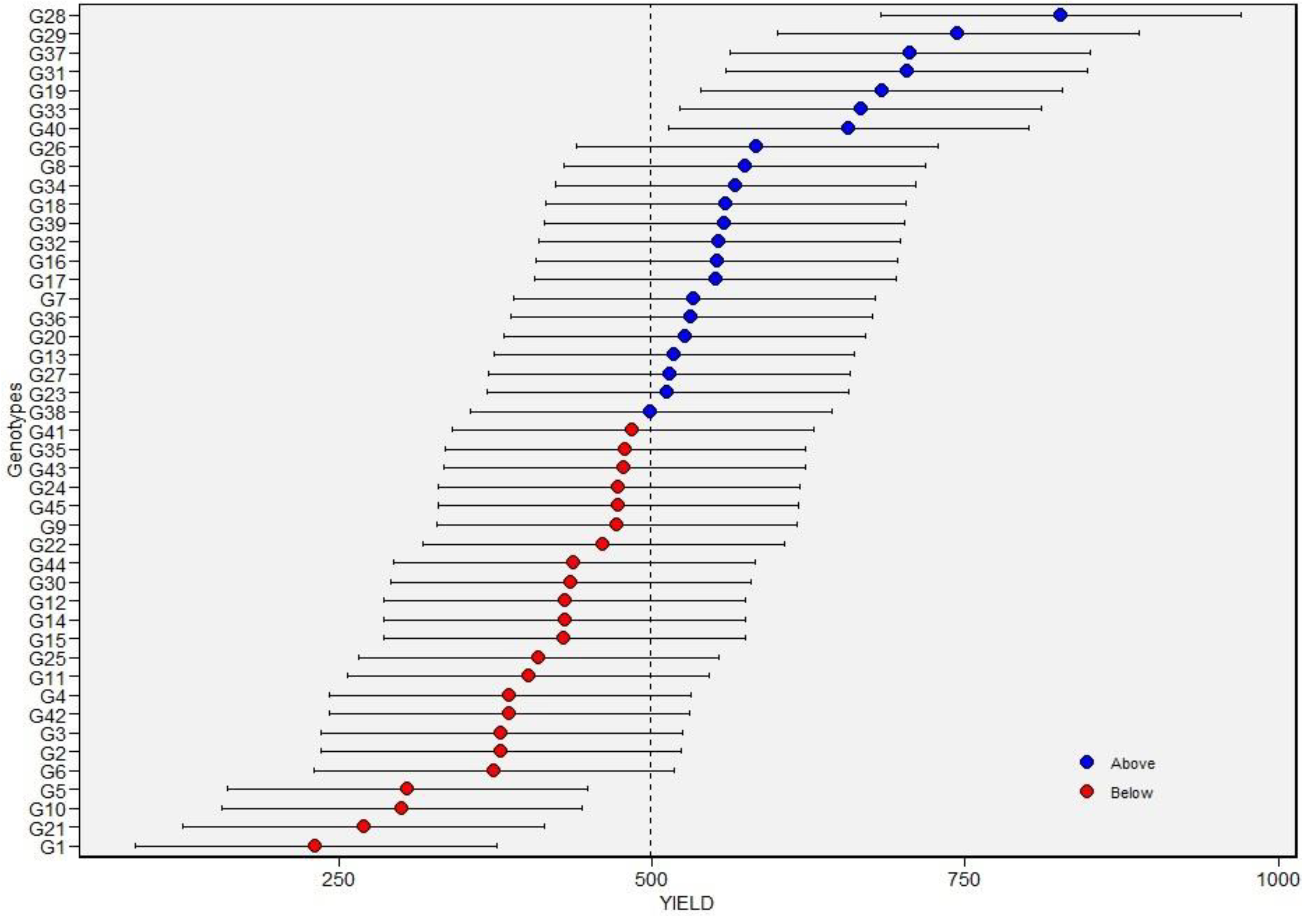
3.3. Estimation of BLUPs and BLUEs for Seed Yield among Tepary Bean Genotypes
3.4. Mean Performance of Tepary Bean Genotypes in Days to 90% Maturity
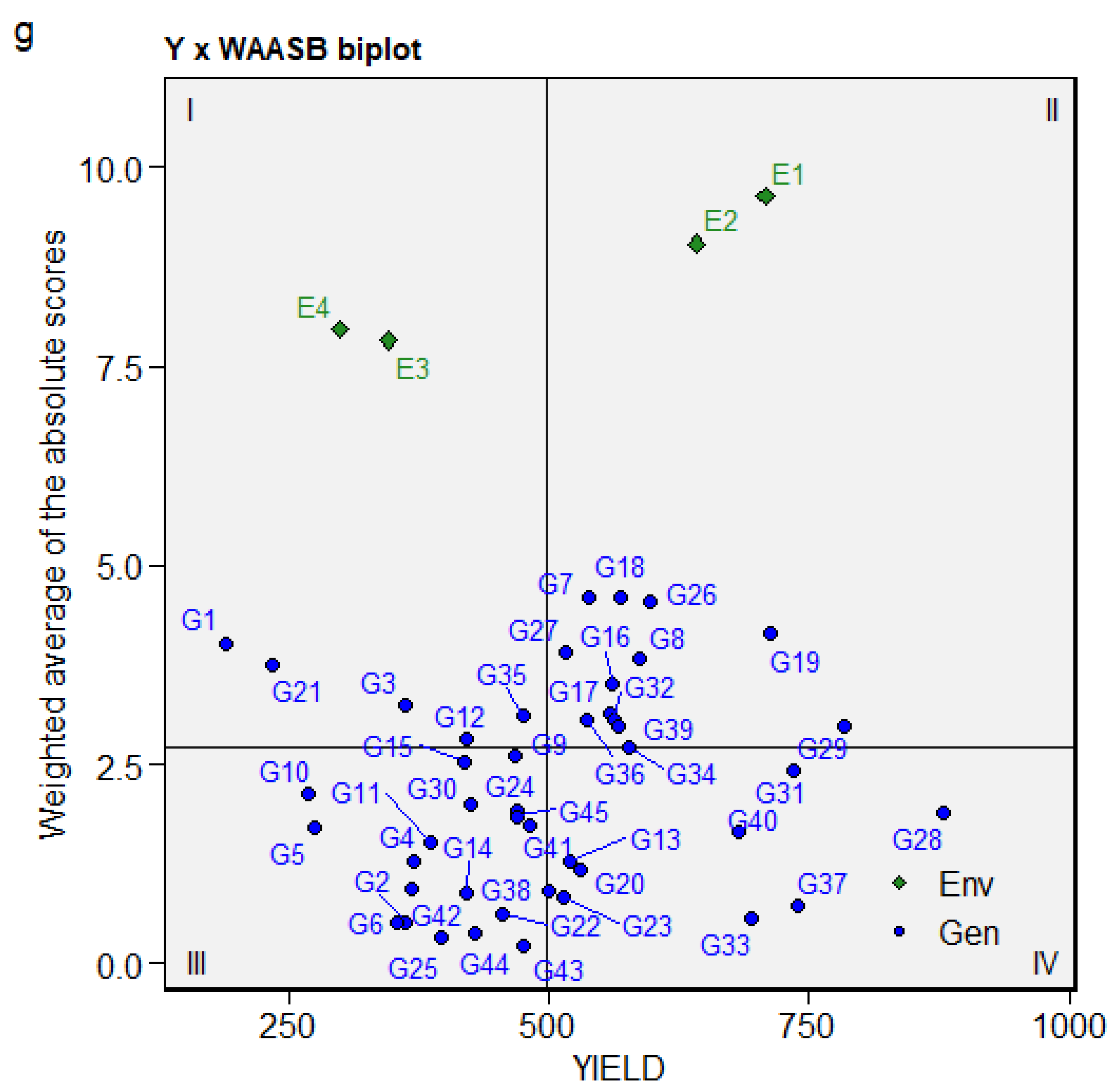
3.5. Identification of Stable Genotypes Using ASV, YSI, and WAASB Biplot
3.6. MTSI Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://sdgs.un.org/goals (accessed on 15 August 2022).
- Maggio, G.; Sitko, N.; Ignaciuk, A. Cropping system diversification in Eastern and Southern Africa: Identifying policy options to enhance productivity and build resilience. In FAO Agricultural Development Economics Working Paper; FAO: Rome, Italy, 2018. [Google Scholar]
- Moghaddam, S.M.; Oladzad, A.; Koh, C.; Ramsay, L.; Hart, J.; Mamidi, S.; Hoopes, G.; Sreedasyam, A.; Wiersma, A.; Zhao, D.; et al. The tepary bean genome provides insight into evolution and domestication under heat stress. Nat. Commun. 2021, 12, 2638. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, F.; Mohamed, M.; Schmitz-Eiberger, N.; Keutgen, N.; Noga, G. Comparative drought postponing and tolerance potentials of two tepary bean lines in relation to seed yield. Afr. Crop. Sci. J. 2005, 13, 49–60. [Google Scholar]
- Beebe, S.E.; Rao, I.M.; Blair, M.W.; Acosta-Gallegos, J.A. Phenotyping common beans for adaptation to drought. Front. Physiol. 2013, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Amarteifio, J.O.; Moholo, D. The chemical composition of four legumes consumed in Botswana. J. Food Comp. Anal. 1998, 11, 329–332. [Google Scholar] [CrossRef]
- Bhardwaj, H.L.; Hamama, A.A. Oil and fatty acid composition of tepary bean seed. HortScience 2005, 40, 1436–1438. [Google Scholar] [CrossRef]
- Porch, T.G.; Cichy, K.; Wang, W.; Brick, M.; Beaver, J.S.; Santana-Morant, D.; Grusak, M.A. Nutritional composition and cooking characteristics of tepary bean (Phaseolus acutifolius Gray) in comparison with common bean (Phaseolus vulgaris L.). Genet. Resour. Crop Evol. 2017, 64, 935–953. [Google Scholar] [CrossRef]
- López-Ibarra, C.; Ruiz-López, F.d.J.; Bautista-Villarreal, M.; Báez-González, J.G.; Romero, B.A.R.; González-Martínez, B.E.; López-Cabanillas, L.M.; Vázquez-Rodríguez, J.A. Protein Concentrates on Tepary Bean (Phaseolus acutifolius Gray) as a Functional Ingredient: In silico Docking of Tepary Bean Lectin to Peroxisome Proliferator-Activated Receptor Gamma. Front. Nutr. 2021, 8, 661463. [Google Scholar] [CrossRef]
- Kusolwa, P.; Myers, J. Seed storage proteins ARL2 and its variants from the apalocus of wild tepary bean G40199 confers resistance to Acanthoscelides obtectus when expressed in common beans. Afr. Crop. Sci. J. 2011, 19, 255–265. [Google Scholar]
- Singh, S.P.; Muñoz, C.G. Resistance to common bacterial blight among Phaseolus species and common bean improvement. Crop Sci. 1999, 39, 80–89. [Google Scholar] [CrossRef]
- Miklas, P.N.; Kelly, J.D.; Beebe, S.E.; Blair, M.W. Common bean breeding for resistance against biotic and abiotic stresses: From classical to MAS breeding. Euphytica 2006, 147, 105–131. [Google Scholar] [CrossRef]
- Salgado, M.O.; Schwartz, H.F.; Brick, M.A.; Pastor-Corrales, M.A. Resistance to Fusarium oxysporum f.sp. phaseoli in tepary bean (Phaseolus acutifolius). Plant Dis. 1994, 78, 357–360. [Google Scholar]
- Miklas, P.N.; Schwartz, H.F.; Salgado, M.O.; Nina, R.; Beaver, J.S. Reaction of select tepary bean to ashy stem blight and fusarium wilt. HortScience 1998, 33, 136–139. [Google Scholar]
- Porch, T.G.; Beaver, J.S.; Brick, M.A. Registration of tepary germplasm with multiple-stress tolerance, TARS-Tep 22 and TARS-Tep 32. J. Plant Reg. 2013, 7, 358–364. [Google Scholar] [CrossRef]
- Mhlaba, Z.B.; Amelework, B.; Shimelis, H.A.; Modi, A.T.; Mashilo, J. Variance components and heritability of yield and yield-related traits in tepary bean (Phaseolus acutifolius). South Afr. J. Plant Soil 2018, 36, 117–128. [Google Scholar] [CrossRef]
- Muñoz, L.C.; Debouck, D.; Rivera, M.; Muñoz, J.; Alpala, D.; Sarsu, F.; Rao, I. Mutation breeding for heat and drought tolerance in tepary bean (Phaseolus acutifolius A. Gray). Aust. J. Crop Sci. 2021, 15, 60–68. [Google Scholar] [CrossRef]
- Suárez, J.C.; Contreras, A.T.; Anzola, J.A.; Vanegas, J.I.; Rao, I.M. Physiological Characteristics of Cultivated Tepary Bean (Phaseolus acutifolius A. Gray) and Its Wild Relatives Grown at High Temperature and Acid Soil Stress Conditions in the Amazon Region of Colombia. Plants 2022, 11, 116. [Google Scholar] [CrossRef]
- Pratt, R.; Grant, L.; Velasco-Cruz, C.; Lauriault, L. Field performance of selected and landrace tepary bean varieties in diverse southwestern USA irrigated production environments. Legume Sci. 2022, e157. [Google Scholar] [CrossRef]
- Mhlaba, Z.B.; Amelework, B.; Shimelis, H.A.; Modi, A.T.; Mashilo, J. Genetic interrelationship among tepary bean (Phaseolus acutifolius A. Gray) genotypes revealed through SSR markers. Aust. J. Crop Sci. 2018, 12, 1587–1595. [Google Scholar] [CrossRef]
- Blair, M.W.; Pantoja, W.; Muñoz, L.C. First use of microsatellite markers in a large collection of cultivated and wild accessions of tepary bean (Phaseolus acutifolius A. Gray). Theor. Appl. Genet. 2012, 125, 1137–1147. [Google Scholar] [CrossRef]
- Federici, C.T.; Ehdaie, B.; Waines, J.G. Domesticated and wild tepary bean: Field performance with and without drought stress. Agron. J. 1990, 82, 896–900. [Google Scholar] [CrossRef]
- Souter, J.R.; Gurusamy, V.; Porch, T.G.; Bett, K.E. Successful introgression of abiotic stress tolerance from wild tepary bean to common bean. Crop Sci. 2017, 57, 1160–1171. [Google Scholar] [CrossRef]
- Rao, I.; Beebe, S.; Polania, J.; Ricaurte, J.; Cajiao, C.; Garcia, R.; Riverra, M. Can tepary bean be a model for improvement of drought resistance in common bean? Afr. Crop Sci. J. 2013, 21, 265–281. [Google Scholar]
- Mhlaba, Z.B.; Mashilo, J.; Shimelis, H.; Assefa, A.B.; Modi, A.T. Progress in genetic analysis and breeding of tepary bean (Phaseolus acutifolius A. Gray): A review. Sci. Hortic. 2018, 237, 112–119. [Google Scholar] [CrossRef]
- Gurmu, F.; Shimelis, H.; Laing, M. Genotype-By-Environment Interaction and Stability of Sweetpotato Genotypes for Root Dry Matter, Β-carotene and Fresh Root Yield. Open Agric. 2017, 2, 473–485. [Google Scholar] [CrossRef]
- Gerrano, A.; van Rensburg, W.; Mathew, I.; Shayanowako, A.; Bairu, M.; Venter, S.; Swart, W.; Mofokeng, A.; Mellem, J.; Labuschagne, M. Genotype and genotype × environment interaction effects on the grain yield performance of cowpea genotypes in dryland farming system in South Africa. Euphytica 2020, 216, 80. [Google Scholar] [CrossRef]
- Ligarreto–Moreno, G.; Pimentel–Ladino, C. Grain yield and genotype x environment interaction in bean cultivars with different growth habits. Plant Prod. Sci. 2022, 25, 232–241. [Google Scholar] [CrossRef]
- Mwiinga, B.; Sibiya, J.; Kondwakwenda, A.; Musvosvi, C.; Chigeza, G. Genotype x environment interaction analysis of soybean (Glycine max (L.) Merrill) grain yield across production environments in Southern Africa. Field Crops Res. 2020, 256, 107922. [Google Scholar] [CrossRef]
- Mushoriwa, H.; Mathew, I.; Gwata, E.T.; Tongoona, P.; Derera, J. Grain Yield Potential and Stability of Soybean Genotypes of Different Ages across Diverse Environments in Southern Africa. Agronomy 2022, 12, 1147. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P. GGE Biplot vs. AMMI analysis of genotype by environment data. Crop Sci. 2007, 47, 643–655. [Google Scholar] [CrossRef]
- Gauch, H.G. Statistical analysis of yield trials by AMMI and GGE. Crop Sci. 2006, 46, 1488–1500. [Google Scholar] [CrossRef]
- Gauch, H.G.; Piepho, H.P.; Annicchiarico, P. Statistical analysis of yield trials by AMMI and GGE: Further considerations. Crop Sci. 2008, 48, 866–889. [Google Scholar] [CrossRef]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef]
- Babic, V.B.; Babic, M.M.; Ivanovic, M.R.; Filipovic, M.R. Pattern in interaction in the maize yield trial. J. Agric. Sci. 2011, 56, 101–110. [Google Scholar] [CrossRef]
- Bose, L.K.; Jambhulkar, N.N.; Singh, O.N. Additive main effects and multiplicative interaction (AMMI) analysis of grain yield stability in early duration rice. J. Anim. Plant Sci. 2014, 24, 1885–1897. [Google Scholar]
- Zali, H.; Farshadfar, E.; Sabaghpour, S.H.; Karimizadeh, R. Evaluation of genotype × environment interaction in chickpea using measures of stability from AMMI model. Ann. Biol. Res. 2012, 3, 3126–3136. [Google Scholar]
- Ajay, B.C.; Bera, S.K.; Singh, A.L.; Kumar, N.; Gangadhar, K.; Kona, P. Evaluation of Genotype × Environment Interaction and Yield Stability Analysis in Peanut Under Phosphorus Stress Condition Using Stability Parameters of AMMI Model. Agric. Res. 2020, 9, 477–486. [Google Scholar] [CrossRef]
- Burbano-Erazo, E.; León-Pacheco, R.I.; Cordero-Cordero, C.C.; López-Hernández, F.; Cortés, A.J.; Tofiño-Rivera, A.P. Multi-environment yield components in advanced common bean (Phaseolus vulgaris L.) tepary bean (P. acutifolius A. Gray) interspecific lines for heat and drought tolerance. Agronomy 2021, 11, 1978. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Khalili, M.; Poczai, P.; Olivoto, T. Stability Indices to Deciphering the Genotype-by-Environment Interaction (GEI) Effect: An Applicable Review for Use in Plant Breeding Programs. Plants 2022, 11, 414. [Google Scholar] [CrossRef]
- Gerrano, A.S.; Thungo, Z.G.; Shimelis, H.; Mashilo, J.; Mathew, I. Genotype-by-Environment Interaction for the Contents of Micro-Nutrients and Protein in the Green Pods of Cowpea (Vigna unguiculata L. Walp.). Agriculture 2022, 12, 531. [Google Scholar] [CrossRef]
- Molosiwa, O.O.; Baleseng, L.; Chirwa, L. Effect of Plant Spacing on Agronomic Performance and Fodder Quality of Four Tepary Bean (Phaseolus acutifolius A. Gray) Cultivars. Hindawi Int. J. Agron. 2022, 2022, 6590608. [Google Scholar] [CrossRef]
- Molosiwa, O.O.; Kgokong, S.B.; Makwala, B.; Gwafila, C.M.; Ramokapane, M.G. Genetic diversity in tepary bean (Phaseolus acutifolius) landraces grown in Botswana. J. Plant Breed. Crop Sci. 2014, 6, 194–199. [Google Scholar] [CrossRef]
- Konvalina, P. Alternative Crops and Cropping Systems; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef]
- Polania, J.; Rao, I.M.; Cajiao, C.; Grajales, M.; Rivera, M.; Velasquez, F.; Raatz, B.; Beebe, S.E. Shoot and root traits contribute to drought resistance in recombinant inbred lines of MD 23–24 × SEA 5 of common bean. Front. Plant Sci. 2017, 8, 296. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Veneklaas, E.; Polania, J.; Rao, I.M.; Beebe, S.E.; Merchant, A. Field drought conditions impact yield but not nutritional quality of the seed in common bean (Phaseolus vulgaris L.). PLoS ONE. 2019, 14, e0217099. [Google Scholar] [CrossRef] [PubMed]
- Ambachew, D.; Mekbib, F.; Asfaw, A.; Beebe, S.E.; Blair, M.W. Trait associations in common bean genotypes grown under drought stress and field infestation by BSM bean fly. Crop J. 2015, 3, 305–316. [Google Scholar] [CrossRef]
- Parker, A.; Namuth-Covert, D. Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability; International Union for the Protection of New Varieties of Plants (UPOV): Geneva, Switzerland, 2017. [Google Scholar]
- Payne, R.; Murray, D.; Harding, S. An Introduction to the GenStat Command Language; VSN International: Hempstead, UK, 2017. [Google Scholar]
- Bartlett, M.S. Properties of sufficiency and statistical tests. Proc. R. Soc. Lond. 1937, 160, 268–282. [Google Scholar]
- Zobel, R.W.; Wright, M.G.; Gauch, H.G. Statistical analysis of yield trial. Agron. J. 1988, 80, 388–393. [Google Scholar] [CrossRef]
- Purchase, J.L.; Hatting, H.; Van Deventer, C.S. Genotype × environment interaction of winter wheat (Triticum aestivum L.) in South Africa: II. Stability analysis of yield performance. South Afr. J. Plant Soil 2000, 17, 101–107. [Google Scholar] [CrossRef]
- Kang, M.S. Simultaneous selection for yield and stability in crop performance trials: Consequences for growers. Agron. J. 1993, 85, 754–757. [Google Scholar] [CrossRef]
- Lin, C.S.; Binns, M.R. A superiority measure of cultivar performance for cultivar x location data. Can. J. Plant Sci. 1988, 68, 193–198. [Google Scholar] [CrossRef]
- Alvarado, G.; Roodríguez, F.M.; Pacheco, A.; Burgueño, J.; Crossa, J.; Vargas, M.; Pérez-Rodríguez, P.; Lopez-Cruz, M.A. META-R: A Software to analyse data from multi-environment plant breeding trials. Crop J. 2020, 8, 745–756. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D. Metan: An R package for multi-environment trial analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef]
- Tukamuhabwa, P.; Asiimwe, M.; Nabasirye, M.; Kabayi, P.; Maphosa, M. Genotype by environment interaction of advanced generation soybean lines for grain yield in Uganda. Afr. Crop Sci. J. 2011, 20, 107–115. [Google Scholar]
- Assefa, T.; Mahama, A.A.; Brown, A.V.; Cannon, E.K.S.; Rubyogo, J.C.; Rao, I.M.; Blair, M.W.; Cannon, S.B. A review of breeding objectives, genomic resources, and marker-assisted methods in common bean (Phaseolus vulgaris L.). Mol. Breed. 2019, 39, 20. [Google Scholar] [CrossRef]
- Temesgen, T.; Keneni, G.; Sefera, T.; Jarso, M. Yield stability and relationships among stability parameters in faba bean (Vicia faba L.) genotypes. Crop J. 2015, 3, 258–268. [Google Scholar] [CrossRef]
- Yohane, E.N.; Shimelis, H.; Laing, M.; Mathew, I.; Shayanowako, A. Genotype-by-environment interaction and stability analyses of grain yield in pigeonpea [Cajanus cajan (L.) Millspaugh]. Acta Agric. Scand. Sect. B Soil Plant Sci. 2021, 71, 145–155. [Google Scholar] [CrossRef]
- Bernardo, R. Reinventing quantitative genetics for plant breeding: Something old, something new, something borrowed, something BLUE. Heredity 2020, 125, 375–385. [Google Scholar] [CrossRef]
- Rukundo, P.; Shimelis, H.; Laing, M.; Mashilo, J. Genotype-by-environment interaction for dual-purpose traits in sweetpotato. J. Crop Improv. 2020, 34, 800–823. [Google Scholar] [CrossRef]
- Gujaria-Verma, N.; Ramsay, L.; Sharpe, A.G.; Sanderson, L.A.; Debouck, D.G.; Taran, K.E. Gene-based SNP discovery in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris) for diversity analysis and comparative mapping. BMC Genom. 2016, 17, 2. [Google Scholar] [CrossRef]
- Bocianowski, J.; Niemann, J.; Nowosad, K. Genotype-by environment interaction for seed quality traits in interspecific cross-derived Brassica lines using additive main effects and multiplicative interaction model. Euphytica 2019, 215, 7. [Google Scholar] [CrossRef]
- Balestre, M.; Von Pinho, R.G.; Souza, J.C.; Oliveira, R.L. Genotypic stability and adaptability in tropical maize based on AMMI and GGE biplot analysis. Genet. Mol. Res. 2009, 8, 1311–1322. [Google Scholar] [CrossRef]
- Tigabu, D.A.; Tadesse, Z.; Zegeye, H.; Assefa, A. Seasonal variability and genetic response of elite bread wheat lines in drought prone environments of Ethiopia. J. Plant Breed. Genet. 2017, 5, 15–21. [Google Scholar]
- White, J.W.; Singh, S.P. Sources and inheritance of earliness in tropically adapted indeterminate common bean. Euphytica 1991, 55, 15–19. [Google Scholar] [CrossRef]
- Beebe, S.; Rao, I.M.; Devi, M.; Polania, J. Common beans, biodiversity, and multiple stress: Challenges of drought resistance in tropical soils. Crop Pasture Sci. 2014, 65, 667–675. [Google Scholar] [CrossRef]

| Average Temperature (°C) | |||||||
|---|---|---|---|---|---|---|---|
| Site and Country | Year | Rainfall (mm) | Tmax (°C) | Tmin (°C) | Latitude | Longitude | Altitude († m.a.s.l) |
| Bunda, Lilongwe, Malawi | 2020/2021 | - | 32.7 | 11.6 | 14° 12′ S | 33° 46′ E | 1200 |
| Bunda, Lilongwe, Malawi | 2021/2022 | 24.33 | 10.8 | ||||
| Kasinthula, Chikwawa, Malawi | 2020/2021 | - | 40 | 11.2 | 16° S | 34° 5′ E | 60 |
| Kasinthula, Chikwawa, Malawi | 2021/2022 | 6.8 | 32.7 | 12.6 | |||
| BUNDA a | Kasinthula b | |
|---|---|---|
| Soil type | Loamy clay | Sandy loam |
| EC | 65.6 µS/cm | - |
| PH | 5.7 | 7.4 |
| N | 0.17% | 0.03% |
| K | 8.369 ppm | 5.4 |
| P | 0.02% | - |
| Organic matter | 4.48% | 0.03% |
| Organic carbon | 2.60% | 0.38% |
| Genotype Code | Genotype Designation/Name | Seed Coat Color | Genotype Code | Genotype Designation/Name | Seed Coat Color |
|---|---|---|---|---|---|
| G1 | G40001 | Cream | G24 | G40129 | Cream |
| G2 | G40005 | Cream | G25 | G40132 | Cream |
| G3 | G40013 | Black speckled | G26 | G40133 | Cream |
| G4 | G40014 | Cream | G27 | G40134 | Cream |
| G5 | G40017 | Cream | G28 | G40135 | Cream |
| G6 | G40019 | Black | G29 | G40136 | Cream |
| G7 | G40020 | Cream | G30 | G40137 | Cream |
| G8 | G40022 | Light brown | G31 | G40138 | Cream |
| G9 | G40023 | Cream | G32 | G40139 | Cream |
| G10 | G40032 | Grey | G33 | G40140 | Cream |
| G11 | G40035 | Black | G34 | G40143 | Cream |
| G12 | G40036 | Dark yellow | G35 | G40144A | Light brown |
| G13 | G40042 | Cream | G36 | G40147 | Cream |
| G14 | G40059 | Black | G37 | G40148 | Cream |
| G15 | G40062 | Cream | G38 | G40150 | Cream |
| G16 | G40063 | Cream | G39 | G40157 | Light brown |
| G17 | G40065 | Cream | G40 | G40158 | Cream |
| G18 | G40066A | Light brown | G41 | G40173A | Light brown |
| G19 | G40068 | Light brown | G42 | G40201 | Cream |
| G20 | G40069 | Cream | G43 | G40237 | Cream |
| G21 | G40111 | Black | G44 | Uchokwane | Cream |
| G22 | G40125 | Cream | G45 | Zimbabwe-landrace | Cream |
| G23 | G40127 | Cream |
| Source of Variation | DF | SS | MS | % SS Accounted | % GEI Explained |
|---|---|---|---|---|---|
| Environment (ENV) | 3 | 34,500,000 | 11,500,000.00 *** | 52.62 | |
| Replication (ENV) | 8 | 204,000 | 25,500.00 | ||
| Genotype (GEN) | 44 | 22,000,000 | 500,000.00 *** | 33.56 | |
| GEN × ENV | 132 | 9,060,000 | 68,636.36 ** | 13.82 | |
| IPCA1 | 46 | 4,290,000 | 93,260.87 *** | 94.80 | |
| IPCA2 | 44 | 215,000 | 4886.36 | 4.70 | |
| Residual | 892 | 44,300,000 | 49,663.68 | ||
| GEI noise (GEIN) | 6,555,605.38 | ||||
| GEI signal (GEIs) | 2,504,395.00 |
| E1 | E2 | E3 | E4 | |||||
|---|---|---|---|---|---|---|---|---|
| Genotype | BLUPs | BLUE | BLUPs | BLUE | BLUPs | BLUE | BLUPs | BLUE |
| G1 | 370.76 | 237.67 | 349.06 | 221.00 | 205.57 | 158.26 | 180.71 | 140.58 |
| G10 | 477.44 | 386.47 | 449.23 | 365.00 | 220.37 | 178.07 | 185.06 | 146.41 |
| G11 | 657.08 | 637.07 | 609.11 | 594.83 | 213.76 | 169.22 | 182.24 | 142.63 |
| G12 | 572.07 | 518.49 | 525.35 | 474.42 | 359.80 | 364.80 | 315.88 | 321.70 |
| G13 | 757.24 | 776.79 | 689.41 | 710.27 | 328.17 | 322.44 | 279.59 | 273.07 |
| G14 | 624.06 | 591.00 | 573.32 | 543.38 | 299.42 | 283.94 | 270.48 | 260.86 |
| G15 | 585.22 | 536.83 | 524.67 | 473.45 | 355.91 | 359.59 | 304.90 | 306.98 |
| G16 | 665.94 | 649.43 | 594.68 | 574.09 | 494.31 | 544.94 | 428.98 | 473.24 |
| G17 | 825.96 | 872.65 | 766.71 | 821.38 | 306.05 | 292.81 | 261.79 | 249.22 |
| G18 | 885.11 | 955.17 | 795.78 | 863.17 | 282.47 | 261.24 | 220.68 | 194.14 |
| G19 | 766.85 | 790.20 | 679.18 | 695.56 | 614.25 | 705.57 | 568.01 | 659.53 |
| G2 | 587.64 | 540.20 | 547.23 | 505.87 | 242.36 | 207.52 | 217.08 | 189.31 |
| G20 | 746.82 | 762.26 | 704.83 | 732.43 | 350.61 | 352.50 | 281.85 | 276.10 |
| G21 | 412.52 | 295.92 | 382.01 | 268.37 | 234.34 | 196.78 | 206.34 | 174.93 |
| G22 | 651.42 | 629.18 | 607.89 | 593.08 | 327.51 | 321.56 | 282.31 | 276.72 |
| G23 | 727.31 | 735.04 | 684.46 | 703.15 | 340.66 | 339.17 | 285.09 | 280.44 |
| G24 | 631.66 | 601.61 | 579.57 | 552.37 | 371.28 | 380.17 | 331.56 | 342.70 |
| G25 | 624.50 | 591.62 | 567.84 | 535.50 | 273.07 | 248.65 | 229.93 | 206.53 |
| G26 | 897.33 | 972.22 | 821.57 | 900.24 | 304.04 | 290.12 | 243.89 | 225.23 |
| G27 | 816.38 | 859.29 | 755.87 | 805.80 | 249.14 | 216.60 | 213.18 | 184.09 |
| G28 | 1030.46 | 1157.92 | 937.19 | 1066.44 | 586.66 | 668.61 | 538.43 | 619.89 |
| G29 | 833.54 | 883.22 | 769.90 | 825.96 | 639.84 | 739.83 | 584.99 | 682.28 |
| G3 | 510.91 | 433.17 | 484.30 | 415.41 | 317.50 | 308.15 | 289.81 | 286.76 |
| G30 | 568.71 | 513.80 | 584.02 | 558.77 | 337.74 | 335.26 | 294.67 | 293.28 |
| G31 | 933.38 | 1022.50 | 865.84 | 963.89 | 470.34 | 512.84 | 406.34 | 442.91 |
| G32 | 665.66 | 649.04 | 619.62 | 609.94 | 478.21 | 523.38 | 424.54 | 467.29 |
| G33 | 874.10 | 939.81 | 770.42 | 826.72 | 490.87 | 540.33 | 423.82 | 466.33 |
| G34 | 855.68 | 914.11 | 738.45 | 780.76 | 329.88 | 324.73 | 292.34 | 290.16 |
| G35 | 645.30 | 620.64 | 520.29 | 467.15 | 421.06 | 446.84 | 349.86 | 367.23 |
| G36 | 845.54 | 899.97 | 706.44 | 734.74 | 301.53 | 286.76 | 243.15 | 224.25 |
| G37 | 884.37 | 954.13 | 833.20 | 916.96 | 507.58 | 562.70 | 464.97 | 521.46 |
| G38 | 690.71 | 683.98 | 614.65 | 602.79 | 384.56 | 397.96 | 308.83 | 312.25 |
| G39 | 808.38 | 848.13 | 783.73 | 845.85 | 332.50 | 328.23 | 259.25 | 245.82 |
| G4 | 636.16 | 607.89 | 593.45 | 572.31 | 211.10 | 165.67 | 172.56 | 129.67 |
| G40 | 896.54 | 971.11 | 788.03 | 852.04 | 452.13 | 488.45 | 386.50 | 416.32 |
| G41 | 739.55 | 752.12 | 515.41 | 460.14 | 372.69 | 382.06 | 325.47 | 334.55 |
| G42 | 585.30 | 536.94 | 537.57 | 491.99 | 274.45 | 250.51 | 220.81 | 194.31 |
| G43 | 687.24 | 679.14 | 618.82 | 608.79 | 329.72 | 324.52 | 289.73 | 286.66 |
| G44 | 710.73 | 711.92 | 550.39 | 510.42 | 287.72 | 268.27 | 240.82 | 221.12 |
| G45 | 755.61 | 774.52 | 643.17 | 643.80 | 272.77 | 248.25 | 231.99 | 209.30 |
| G5 | 493.86 | 409.38 | 461.61 | 382.80 | 210.86 | 165.34 | 179.72 | 139.26 |
| G6 | 604.96 | 564.37 | 565.06 | 531.51 | 213.84 | 169.32 | 189.18 | 151.92 |
| G7 | 845.58 | 900.03 | 792.85 | 858.96 | 248.90 | 216.28 | 210.46 | 180.44 |
| G8 | 853.87 | 911.59 | 809.63 | 883.09 | 299.66 | 284.26 | 273.19 | 264.49 |
| G9 | 608.83 | 569.76 | 566.89 | 534.14 | 382.40 | 395.06 | 353.50 | 372.10 |
| NS environments | DS environments | |||||||
| H2 NS | 0.535 | H2 DS | 0.48 | |||||
| VG | 14,636.89 | VG | 11,345.09 | |||||
| VGXE | 8267.17 | VGXE | 8487.22 | |||||
| VE | 65,395.24 | VE | 58,454.31 | |||||
| Error | 51,406.23 | Error | 47,514.74 | |||||
| Grand Mean | 526.40 | Grand Mean | 470.26 | |||||
| LSD (5%) | 212.28 | LSD (5%) | 199.61 | |||||
| CV (%) | 43.07 | CV% | 46.35 | |||||
| Genotype | BLUPs | BLUEs | Genotype | BLUPs | BLUEs | Genotype | BLUPs | BLUEs |
|---|---|---|---|---|---|---|---|---|
| G1 | 231.37 | 189.38 | G23 | 512.26 | 514.45 | G37 | 706.12 | 738.81 |
| G10 | 300.16 | 268.99 | G24 | 473.17 | 469.21 | G38 | 499.12 | 499.24 |
| G11 | 401.22 | 385.94 | G25 | 409.54 | 395.58 | G39 | 557.67 | 567.01 |
| G12 | 430.52 | 419.85 | G26 | 583.55 | 596.95 | G4 | 386.48 | 368.88 |
| G13 | 517.61 | 520.64 | G27 | 513.98 | 516.45 | G40 | 657.01 | 681.98 |
| G14 | 430.47 | 419.80 | G28 | 826.58 | 878.22 | G41 | 484.41 | 482.22 |
| G15 | 429.97 | 419.21 | G29 | 744.15 | 782.82 | G42 | 386.09 | 368.44 |
| G16 | 551.98 | 560.42 | G3 | 379.56 | 360.87 | G43 | 477.98 | 474.78 |
| G17 | 550.77 | 559.02 | G30 | 435.21 | 425.28 | G44 | 437.50 | 427.93 |
| G18 | 558.90 | 568.43 | G31 | 703.29 | 735.53 | G45 | 472.96 | 468.96 |
| G19 | 683.57 | 712.71 | G32 | 553.70 | 562.41 | G5 | 304.66 | 274.19 |
| G2 | 379.43 | 360.73 | G33 | 666.79 | 693.30 | G6 | 373.86 | 354.28 |
| G20 | 526.40 | 530.82 | G34 | 566.69 | 577.44 | G7 | 533.41 | 538.93 |
| G21 | 269.93 | 234.00 | G35 | 478.57 | 475.46 | G8 | 573.96 | 585.86 |
| G22 | 461.01 | 455.13 | G36 | 531.25 | 536.43 | G9 | 471.92 | 467.77 |
| H2 | 0.79 | |||||||
| VG | 18,174.81 | |||||||
| VE | 3193.38 | |||||||
| VGE | 42,325.87 | |||||||
| Error | 49,460.49 | |||||||
| Grand mean | 498.33 | |||||||
| LSD (5%) | 148.15 | |||||||
| CV(%) | 44.63 |
| Genotype | Mean Seed Yield (kg/ha) | IPCA1 | IPCA2 | ASV | Rasv | YSI | rYSI |
|---|---|---|---|---|---|---|---|
| G1 | 189.38 | −0.80 | 0.19 | 16 | 40 | 85 | 45 |
| G2 | 360.73 | −0.08 | 0.15 | 2 | 5 | 45 | 40 |
| G3 | 360.87 | −0.67 | 0.19 | 13 | 36 | 75 | 39 |
| G4 | 368.88 | 0.26 | 0.15 | 5 | 14 | 51 | 37 |
| G5 | 274.19 | −0.32 | 0.17 | 6 | 19 | 61 | 42 |
| G6 | 354.28 | 0.10 | 0.16 | 2 | 6 | 47 | 41 |
| G7 | 538.93 | 0.96 | 0.16 | 19 | 44 | 60 | 16 |
| G8 | 585.86 | 0.79 | 0.21 | 16 | 39 | 48 | 9 |
| G9 | 467.77 | −0.54 | 0.12 | 11 | 27 | 55 | 28 |
| G10 | 268.99 | −0.41 | 0.18 | 8 | 24 | 67 | 43 |
| G11 | 385.94 | 0.31 | 0.13 | 6 | 17 | 53 | 36 |
| G12 | 419.85 | −0.59 | 0.07 | 12 | 29 | 61 | 32 |
| G13 | 520.64 | 0.27 | 0.01 | 5 | 15 | 34 | 19 |
| G14 | 419.80 | −0.17 | 0.08 | 3 | 10 | 43 | 33 |
| G15 | 419.21 | −0.53 | −0.02 | 11 | 26 | 60 | 34 |
| G16 | 560.42 | −0.73 | −0.09 | 15 | 37 | 51 | 14 |
| G17 | 559.02 | 0.65 | 0.10 | 13 | 35 | 50 | 15 |
| G18 | 568.43 | 0.96 | −0.08 | 19 | 45 | 56 | 11 |
| G19 | 712.71 | −0.86 | −0.17 | 17 | 42 | 47 | 5 |
| G20 | 530.82 | 0.23 | 0.15 | 5 | 13 | 31 | 18 |
| G21 | 234.00 | −0.75 | 0.14 | 15 | 38 | 82 | 44 |
| G22 | 455.13 | −0.12 | 0.13 | 2 | 8 | 37 | 29 |
| G23 | 514.45 | 0.16 | 0.15 | 3 | 9 | 30 | 21 |
| G24 | 469.21 | −0.40 | 0.06 | 8 | 23 | 49 | 26 |
| G25 | 395.58 | −0.05 | 0.04 | 1 | 2 | 37 | 35 |
| G26 | 596.95 | 0.96 | 0.01 | 19 | 43 | 51 | 8 |
| G27 | 516.45 | 0.82 | 0.10 | 16 | 41 | 61 | 20 |
| G28 | 878.22 | 0.34 | −0.10 | 7 | 20 | 21 | 1 |
| G29 | 782.82 | −0.61 | 0.00 | 12 | 31 | 33 | 2 |
| G30 | 425.28 | −0.39 | 0.47 | 8 | 22 | 53 | 31 |
| G31 | 735.53 | 0.48 | 0.04 | 10 | 25 | 29 | 4 |
| G32 | 562.41 | −0.64 | 0.08 | 13 | 34 | 47 | 13 |
| G33 | 693.3 | 0.08 | −0.21 | 2 | 4 | 10 | 6 |
| G34 | 577.44 | 0.55 | −0.26 | 11 | 28 | 38 | 10 |
| G35 | 475.46 | −0.63 | −0.43 | 13 | 33 | 57 | 24 |
| G36 | 536.43 | 0.62 | −0.41 | 12 | 32 | 49 | 17 |
| G37 | 738.81 | 0.12 | 0.13 | 2 | 7 | 10 | 3 |
| G38 | 499.24 | −0.19 | −0.09 | 4 | 12 | 34 | 22 |
| G39 | 567.01 | 0.6 | 0.29 | 12 | 30 | 42 | 12 |
| G40 | 681.98 | 0.31 | −0.23 | 6 | 18 | 25 | 7 |
| G41 | 482.22 | −0.3 | −1 | 6 | 16 | 39 | 23 |
| G42 | 368.44 | −0.18 | 0.08 | 4 | 11 | 49 | 38 |
| G43 | 474.78 | −0.04 | −0.02 | 1 | 1 | 26 | 25 |
| G44 | 427.93 | 0.05 | −0.59 | 1 | 3 | 33 | 30 |
| G45 | 468.96 | 0.38 | −0.26 | 8 | 21 | 48 | 27 |
| Genotype | Pi | Genotype | Pi | Genotype | Pi |
|---|---|---|---|---|---|
| G1 | 274,164.66 | G16 | 72,832.3 | G31 | 17,210 |
| G2 | 152,774.47 | G17 | 66,099.6 | G32 | 70,053.5 |
| G3 | 161,487.1 | G18 | 68,719.6 | G33 | 23,934.9 |
| G4 | 147,718.46 | G19 | 34,308.4 | G34 | 58,390.9 |
| G5 | 206,575.77 | G20 | 72,891.1 | G35 | 104,117 |
| G6 | 155,652.08 | G21 | 241,536 | G36 | 73,954.1 |
| G7 | 79,437.85 | G22 | 105,384 | G37 | 15,139.1 |
| G8 | 59,548.84 | G23 | 79,102.1 | G38 | 86,674.9 |
| G9 | 105,545.32 | G24 | 102,303 | G39 | 63,068.1 |
| G10 | 211,236.38 | G25 | 133,774 | G40 | 26,849.7 |
| G11 | 138,815.6 | G26 | 59,155.1 | G41 | 97,649.9 |
| G12 | 128,755.04 | G27 | 84,884.7 | G42 | 149,147 |
| G13 | 76,723.9 | G28 | 1120.56 | G43 | 95,960 |
| G14 | 122,552.1 | G29 | 16,661.5 | G44 | 117,890 |
| G15 | 127,853.1 | G30 | 123,453 | G45 | 98,875.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mwale, S.E.; Shimelis, H.; Nkhata, W.; Sefasi, A.; Fandika, I.; Mashilo, J. Genotype-by-Environment Interaction in Tepary Bean (Phaseolus acutifolius A. Gray) for Seed Yield. Agronomy 2023, 13, 12. https://doi.org/10.3390/agronomy13010012
Mwale SE, Shimelis H, Nkhata W, Sefasi A, Fandika I, Mashilo J. Genotype-by-Environment Interaction in Tepary Bean (Phaseolus acutifolius A. Gray) for Seed Yield. Agronomy. 2023; 13(1):12. https://doi.org/10.3390/agronomy13010012
Chicago/Turabian StyleMwale, Saul Eric, Hussein Shimelis, Wilson Nkhata, Abel Sefasi, Isaac Fandika, and Jacob Mashilo. 2023. "Genotype-by-Environment Interaction in Tepary Bean (Phaseolus acutifolius A. Gray) for Seed Yield" Agronomy 13, no. 1: 12. https://doi.org/10.3390/agronomy13010012
APA StyleMwale, S. E., Shimelis, H., Nkhata, W., Sefasi, A., Fandika, I., & Mashilo, J. (2023). Genotype-by-Environment Interaction in Tepary Bean (Phaseolus acutifolius A. Gray) for Seed Yield. Agronomy, 13(1), 12. https://doi.org/10.3390/agronomy13010012







